Abstract
We identified oral bacterial species in blood cultures following single-tooth extraction and tooth brushing. Sequence analysis of 16S rRNA genes identified 98 different bacterial species recovered from 151 bacteremic subjects. Of interest, 48 of the isolates represented 19 novel species of Prevotella, Fusobacterium, Streptococcus, Actinomyces, Capnocytophaga, Selenomonas, and Veillonella.
Periodontal disease is among the most common of the infectious diseases. Several studies suggest an association between periodontitis and systemic diseases such as coronary artery disease, low birth weight, and distant-site infections such as infective endocarditis (2-4, 7, 9, 10). Bacteremia from the oral cavity is common, whether from routine daily activities such as tooth brushing or invasive dental procedures. This led to the practice of giving prophylactic antibiotics to patients with specific cardiac valve problems or prosthetic joints undergoing invasive dental procedures. Much of the emphasis on dental procedures in these professional association management recommendations is based on the many studies of the incidence, duration, nature, and magnitude of bacteremia following various dental procedures. There is a wide diversity of data from these studies due to different study methodologies, and the accuracy of all of these studies is further limited by the use of conventional methods of identification of cultivable species. An estimated 700 oral bacterial species have been identified by clonal analysis, only 40% of which have been cultivated (1, 13). The identification of the other 60% in pure cultivable form has been hampered by the difficulty of culturing some bacteria on currently available bacteriological media and the inappropriate choice of selective and nonselective media. The purpose of this study was to employ a variety of selective and nonselective media for the isolation of both aerobic and anaerobic bacteria from the blood of patients undergoing two different dental procedures and to use 16S rRNA sequencing for bacterial species identification.
Patient recruitment.
We enrolled 290 healthy adults requiring the extraction of a tooth and having at least 10 remaining teeth and randomized them to one of three groups, (i) 2 min of tooth brushing, (ii) single-tooth extraction with the American Heart Association-recommended dose of oral amoxicillin administered 1 h prior to the procedure, and (iii) tooth extraction with a placebo (6).
Sampling.
One hour following a thorough clinical examination and administration of amoxicillin or a placebo (groups ii and iii), a large-bore angiocath needle was placed in the antecubital fossa. Blood specimens were taken at the following time points: draw 1, baseline before the procedure; draw 2, 90 s into the procedure; draw 3, 3.5 min after draw 2; draws 4, 5, and 6, 20, 40, and 60 min, respectively, following the completion of the dental procedure. Ten milliliters of blood from each draw was inoculated directly into aerobic and anaerobic BACTEC bottles.
Bacterial culturing and isolation.
Blood specimens were cultured in BACTEC Plus Aerobic/F and LYTIC 10 Anaerobic/F (BD & Co., Sparks, MD) and incubated for 2 weeks before being discarded as negative. All positive cultures were subcultured on blood agar and chocolate plates (BD & Co., Sparks, MD). If gram-negative rods were observed, MacConkey II agar was added. For anaerobe-positive cultures, an anaerobic blood agar plate was also included. If gram-positive organisms were also seen, Columbia CNA agar plates were used. In addition, blood agar plates supplemented with phenylethyl alcohol and kanamycin-vancomycin were used for the isolation of fastidious and slow-growing obligate anaerobic bacteria and obligate anaerobic gram-negative bacilli, respectively.
Amplification of the 16S rRNA gene by PCR and purification of the PCR products.
Cells were lysed with proteinase K (11). The following bacterial universal primers were used: 9F forward primer, GAGTTTGATYMTGGCTCAG; 1541R reverse primer, AAGGAGGTGWTCCARCC (12). The PCR was performed under standard conditions with Gene Amp PCR System 9700 (PE Applied Biosystems, Foster City, CA). The reaction mixture (50-μl total volume) contained 1 μl of cell lysate as the template, 20 nmol of each primer, 40 nmol of deoxynucleoside triphosphates, and 1 U of Platinum Taq Polymerase (Invitrogen, Carlsbad, CA). PCR conditions were as follows: denaturation at 95°C for 45 s, annealing at 60°C for 45 s, and elongation for 1.5 min. A total of 30 cycles were performed, followed by a final elongation for 10 min at 72°C. PCR products were purified with a QIAquick PCR purification kit according to the manufacturer's instructions (Qiagen, Valencia, CA).
Sequencing of the 16S rRNA gene and sequencing analysis.
Purified PCR products were sequenced with an ABI Prism cycle sequencing kit with a BigDye Terminator cycle sequencing reaction kit according to the manufacturer's instructions (PE Applied Biosystems, Foster City, CA). The primers used were 9F (see above) and 533R (TTACCGCGGCTGCTG) (14). Sequencing reactions were run on an ABI model 3100 DNA sequencer. Approximately 500 bases were obtained first to determine identity or approximate phylogenetic position. Full sequences (about 1,500 bases with five or six additional sequencing primers were obtained for novel species (11). The sequences were compared to the known sequences of >10,000 microorganisms in our database and 100,000 sequences in the Ribosomal Database Project (http://wdcm.nig.ac.jp/RDP/html/index.html) and GenBank databases by BLAST (http://www.ncbi.nlm.nih.gov). Phylogenetic trees were constructed by the neighbor-joining method of Saitou and Nei (15). TREECON, a software package for the Microsoft Windows environment, was used for the construction and drawing of evolutionary trees (16).
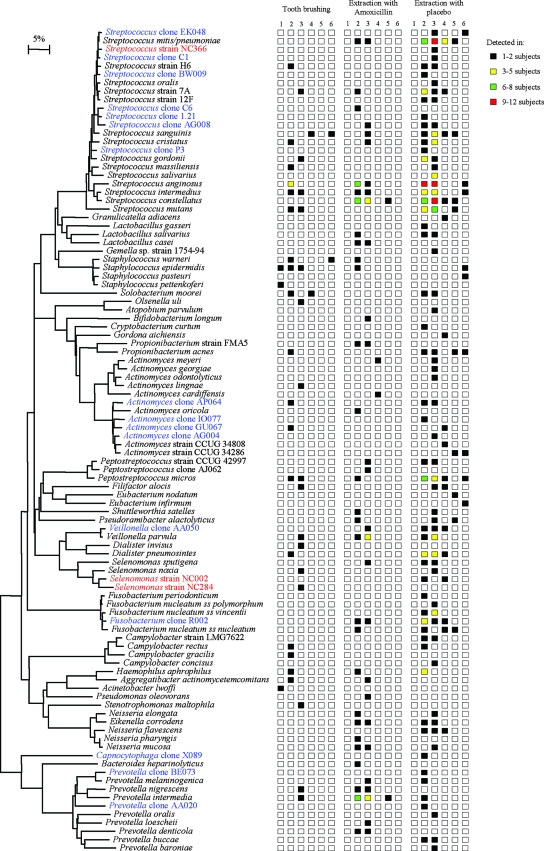
Our data demonstrate that the combination of BACTEC media for bacterial growth and 16S rRNA sequencing for bacterial identification allows the recovery of diverse and novel oral bacterial species from the blood following both tooth brushing and tooth extraction. With the BACTEC media for bacterial growth, we recovered 98 different species from the three groups (Fig. 1). As expected, the predominant species recovered were Streptococcus spp. and other low-G+C Firmicutes. In addition, we recovered representatives from four other bacterial phyla, namely, Fusobacteria, Actinobacteria, Bacteroidetes (e.g., Prevotella spp.), and Proteobacteria (e.g., including species of Eikenella, Campylobacter, and Haemophilus). The most predominant species recovered were Streptococcus spp., followed by Peptostreptococcus micros, Veillonella dispar or V. parvula and Dialister pneumosintes. Staphylococcus spp. were detected in only 9 (0.5%) of the ∼1,800 blood samples processed in this study, and they were most likely from skin contamination during blood drawing. Using the LYTIC 10 Anaerobic/F BACTEC media also allowed the detection of a high percentage of anaerobes, e.g., 9% genus Prevotella, 5% genus Actinomyces, and 5% genus Fusobacterium. We also identified 19 species or phylotypes which had not been cultivated prior to this study (highlighted in red and blue in Fig. 1), of which 3 were first recovered in this study (highlighted in red in Fig. 1).
FIG. 1.
Phylogenetic tree of bacterial species and phylotypes identified in the blood following tooth brushing and tooth extraction with or without antibiotic prophylaxis. Strains in red are putative new species from this study. Species in blue are cultivable representatives of phylotypes (species) or previously uncharacterized strains. The marker bar represents a 5% difference in nucleotide sequences. Columns 1 to 6 represent specific blood draws (see the text).
The advantage of BACTEC media for bacterial isolation from blood over the traditionally used lysis filtration method has also been documented in prior studies. For example, Lucas et al. reported obtaining a greater range of bacterial species by using the BACTEC system compared to the cell lysis filtration technique (8). In another study, Heimdahl et al. used lysis filtration for bacterial recovery from blood and found that bacteremia did not extend much beyond 10 min after a dental procedures (5). However, in our results with BACTEC media for 151 bacteremic subjects, 20% were still bacteremic at 20 min, 9% at 40 min, and 6% at 60 min. This suggests that the BACTEC method is superior to the lysis filtration method for detecting low levels of bacteremia.
Antibiotic prophylaxis reduced the incidence of bacteremia from dental extraction (Table 1). It also resulted in bacteremia with fewer bacterial species, which were cleared from the blood in a shorter time (i.e., mostly within 20 min) (Fig. 1). Although antibiotic prophylaxis reduced the bacteremia of several species of streptococci, as expected, it does not seem to affect species of proteobacteria (e.g., Eikenella corrodens) and Prevotella.
TABLE 1.
Bacterial species found (i.e., ≥2%) in blood samples following tooth brushing and dental extraction
| Bacterial species | No. of subjects (frequency [%])
|
|||
|---|---|---|---|---|
| Tooth brushing | Extraction with amoxicillin | Extraction with placebo | Total | |
| Streptococcus anginosus | 4 (8.5) | 14 (13.7) | 23 (8.8) | 41 (10.0) |
| Streptococcus mitis or S. pneumoniae | 4 (3.9) | 29 (11.1) | 33 (8.1) | |
| Streptococcus constellatus | 11 (10.8) | 18 (6.9) | 29 (7.1) | |
| Peptostreptococcus micros | 3 (6.4) | 2 (2.0) | 16 (6.2) | 21 (5.2) |
| Streptococcus intermedius | 3 (6.4) | 2 (2.0) | 13 (5.0) | 18 (4.4) |
| Streptococcus mutans | 3 (6.4) | 12 (4.6) | 15 (3.7) | |
| Prevotella intermedia | 1 (2.1) | 9 (8.8) | 3 (1.2) | 13 (3.2) |
| Veillonella dispar or V. parvula | 1 (2.1) | 5 (4.9) | 6 (2.3) | 12 (2.9) |
| Streptococcus sanguinis | 1 (2.1) | 1 (1.0) | 8 (3.1) | 10 (2.4) |
| Streptococcus sp. strain 7A | 2 (4.3) | 1 (1.0) | 7 (2.7) | 10 (2.4) |
| Streptococcus cristatus | 1 (2.1) | 2 (2.0) | 6 (2.3) | 9 (2.2) |
| Dialister pneumosintes | 1 (2.1) | 1 (1.0) | 6 (2.3) | 8 (2.0) |
Acknowledgments
This work was supported by NIDCR grant R01-DE13559-01 A2.
Footnotes
Published ahead of print on 23 April 2008.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 435721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck, J. D., P. Eke, G. Heiss, P. Madianos, D. Couper, D. Lin, K. Moss, J. Elter, and S. Offenbacher. 2005. Periodontal disease and coronary heart disease: a reappraisal of the exposure. Circulation 11219-24. [DOI] [PubMed] [Google Scholar]
- 3.Beck, J. D., P. Eke, D. Lin, P. Madianos, D. Couper, K. Moss, J. Elter, G. Heiss, and S. Offenbacher. 2005. Associations between IgG antibody to oral organisms and carotid intima-medial thickness in community-dwelling adults. Atherosclerosis 183342-348. [DOI] [PubMed] [Google Scholar]
- 4.Boggess, K. A., J. D. Beck, A. P. Murtha, K. Moss, and S. Offenbacher. 2006. Maternal periodontal disease in early pregnancy and risk for a small-for-gestational-age infant. Am. J. Obstet. Gynecol. 1941316-1322. [DOI] [PubMed] [Google Scholar]
- 5.Heimdahl, A., G. Hall, M. Hedberg, H. Sandberg, P.-O. Söder, K. Tunér, and C. E. Nord. 1990. Detection and quantitation by lysis-filtration of bacteremia after different oral surgical procedures. J. Clin. Microbiol. 282205-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lockhart, P. B., M. T. Brennan, H. Sasser, J. Noll, S. Coleman, J. Ashar, M. L. Kent, P. C. Fox, B. J. Paster, and F. K. Bahrani-Mougeot. 2007. Tooth extraction and tooth brushing both produce bacteremia of endocarditis-related pathogens. J. Am. Coll. Cardiol. 49301A. [Google Scholar]
- 7.Lockhart, P. B., and D. T. Durack. 1999. Oral microflora as a cause of endocarditis and other distant site infections. Infect. Dis. Clin. N. Am. 13833-850. [DOI] [PubMed] [Google Scholar]
- 8.Lucas, V. S., V. Lytra, T. Hassan, H. Tatham, M. Wilson, and G. J. Roberts. 2002. Comparison of lysis filtration and an automated blood culture system (BACTEC) for detection, quantification, and identification of odontogenic bacteremia in children. J. Clin. Microbiol. 403416-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Offenbacher, S., K. A. Boggess, A. P. Murtha, H. L. Jared, S. Lieff, R. G. McKaig, S. M. Mauriello, K. L. Moss, and J. D. Beck. 2006. Progressive periodontal disease and risk of very preterm delivery. Obstet. Gynecol. 10729-36. (Erratum, 107:1171.) [DOI] [PubMed] [Google Scholar]
- 10.Offenbacher, S., P. N. Madianos, C. M. Champagne, J. H. Southerland, D. W. Paquette, R. C. Williams, G. Slade, and J. D. Beck. 1999. Periodontitis-atherosclerosis syndrome: an expanded model of pathogenesis. J. Periodontal Res. 34346-352. [DOI] [PubMed] [Google Scholar]
- 11.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 1833770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paster, B. J., W. A. Falkler, Jr., C. O. Enwonwu, E. O. Idigbe, K. O. Savage, V. A. Levanos, M. A. Tamer, R. L. Ericson, C. N. Lau, and F. E. Dewhirst. 2002. Prevalent bacterial species and novel phylotypes in advanced noma lesions. J. Clin. Microbiol. 402187-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paster, B. J., I. Olsen, J. A. Aas, and F. E. Dewhirst. 2006. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2000 4280-87. [DOI] [PubMed] [Google Scholar]
- 14.Paster, B. J., M. K. Russell, T. Alpagot, A. M. Lee, S. K. Boches, J. L. Galvin, and F. E. Dewhirst. 2002. Bacterial diversity in necrotizing ulcerative periodontitis in HIV-positive subjects. Ann. Periodontol. 78-16. [DOI] [PubMed] [Google Scholar]
- 15.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 16.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10569-570. [DOI] [PubMed] [Google Scholar]