Abstract
Thermotolerant Campylobacter spp. (Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis) are leading causes of food-borne diarrhea in humans. In this study, the usefulness of fluorescence in situ hybridization (FISH) for the identification of Campylobacter isolates was investigated. A hierarchical FISH probe set that included six group-, genus-, and species-specific probes was developed and evaluated with 12 reference strains and 94 clinical isolates of Campylobacter, Arcobacter, and Helicobacter. FISH correctly identified all isolates to the genus level and detected all thermotolerant Campylobacter isolates. The assay showed high degrees of sensitivity for the identification of C. jejuni (90%), C. coli (97%), C. lari (81%), and C. upsaliensis (100%) to the species level.
Thermotolerant Campylobacter spp. (Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis) are leading causes of food-borne human gastroenteritis and the corresponding late-onset complications, such as reactive arthritis and Guillain-Barré syndrome (5). The phenotypic identification of Campylobacter spp. is complicated and of limited reliability (2, 5, 8, 11, 12). The identification of C. lari and C. upsaliensis to the species level and the discrimination of the close relative Arcobacter from Campylobacter are especially problematic, leading to uncertainty about the true clinical relevance of these organisms (2, 5, 8, 11, 12, 24). Various molecular methods have therefore been proposed as alternative diagnostic methods (5-7, 11-13, 20, 27). Among these, fluorescence in situ hybridization (FISH) has been described for the identification of Campylobacter (15, 25) and its relatives, Helicobacter and Arcobacter (3, 15, 16, 26), in environmental samples and chicken products. FISH is a microscopic method that uses fluorescently labeled oligonucleotide DNA probes that bind specifically to unique target sites on ribosomal RNA (10, 18, 23). The advantages of FISH are its simple methodology, high speed, low cost, and minimal equipment requirements (only a fluorescent microscope is needed) (10, 18, 23). The aim of this study was to establish and evaluate a FISH assay for the identification of thermotolerant Campylobacter in a clinical setting. A hierarchical set of six FISH probes (Table 1) was designed with ARB software (http://www.arb-home.de). One probe covers all Campylobacter and its relatives, Arcobacter and Helicobacter (the HelCArc probe). One group-specific probe targets the four thermotolerant Campylobacter spp. (the Catherm probe). Species-specific probes were designed for C. jejuni (the Cajej probe), C. upsaliensis (the Cup probe), and C. lari (the Clar1 and Clar2 probes). A combination of two probes was implemented for C. lari, since it was not possible to cover this heterogeneous species with a single probe. We did not succeed in designing a probe for C. coli with sufficient sensitivity.
TABLE 1.
Oligonucleotide probes
| Probe | Target (position) | Target organism(s) | Sequence (5′-3′) | Reference or source |
|---|---|---|---|---|
| HelCArc | 23S rRNA (1760) | Campylobacter spp., Helicobacter spp., and Arcobacter spp. | AAC AGT CGG GAG GGA CTC | This study |
| Catherm | 23S rRNA (1419) | Thermotolerant Campylobacter spp | GCC CTA AGC GTC CTT CCA | This study |
| Cajej | 23S rRNA (1693) | C. jejuni | AGC TAA CCA CAC CTT ATA CCG | This study |
| Clar 1a | 16S rRNA (622) | C. lari | TCC CAA GCA GTT CAA CGG T | This study |
| Clar 2a | 16S rRNA (1126) | C. lari | GAA GTG TTA GCA ACT AAA T | This study |
| Cup | 16S rRNA (1695) | C. upsaliensis | CTC TAC AGA ATT TGT TGG AT | This study |
| EUB | 16S rRNA (338) | Bacterial kingdom | GCT GCC TCC CGT AGG AGT | 1 |
The two C. lari-specific probes are used simultaneously.
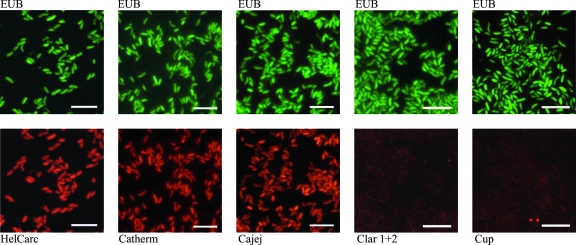
Probes were directly 5′ labeled with the fluorescent dye Cy3 (red) or 6-carboxyfluorescein (FAM; green) (Thermo, Ulm, Germany). Hybridization was performed as described previously with formamide at a concentration of 30% (10, 21, 23). A FAM-labeled eubacterial probe was always implemented as a control (1). Suspensions of bacteria were prepared in 0.9% saline from overnight cultures on agar plates. Ten microliters of the suspension was applied to glass slides. The slides were air dried and fixed for 20 min in 2% paraformaldehyde. Each slide was hybridized with one specific Cy3-labeled probe in combination with the FAM-labeled eubacterial probe (Fig. 1).
FIG. 1.
C. jejuni reference strain stained with FISH probes. The results obtained with C. jejuni ATCC 33560 are shown. Single slides were each stained simultaneously with the fluorescein isothiocyanate-labeled eubacterial probe (EUB) (green; upper row) and the Cy3-labeled Campylobacter-specific probes (red; lower row). Bars, 5 μm.
The assay was first evaluated with 12 bacterial reference strains (C. jejuni subsp. jejuni ATCC 33560, C. coli ATCC 33559, C. lari ATCC 35221, C. upsaliensis ATCC 43954, C. sputorum ATCC 35980, C. concisus ATCC 33237, C. fetus ATCC 27374, Arcobacter butzleri ATCC 49616, Arcobacter cryaerophilus ATCC 43158, Arcobacter nitrofigilis ATCC 33309, Helicobacter pylori ATCC 49396, and Helicobacter pylori DSMZ 4867). All probes correctly stained the corresponding target reference strains without any cross-reaction with nontarget reference strains.
The probes were further evaluated by using 94 isolates cultured from specimens from humans and animals (Table 2) from the Federal Institute for Risk Assessment, Berlin, Germany; the Animal Sciences Group, Wageningen-Lelystad, The Netherlands; and the Institute of Medical Microbiology, University of Ulm, Ulm, Germany. The isolates were phenotypically characterized by phase-contrast microscopy (characteristic morphology and motility) and Gram staining and by examination of catalase and oxidase production, growth at 25°C and 43°C, indoxyl acetate hydrolysis (22), hippurate hydrolysis (19), and susceptibility to nalidixic acid and cephalothin (5, 14). The identities of three C. coli isolates were confirmed by a previously published PCR approach (27).
TABLE 2.
Number and percentage of positive FISH results obtained with the isolates tested
| Species | No. (%) of the following species (probes):
|
|||||
|---|---|---|---|---|---|---|
| Total | Helicobacter, Campylobacter, Arcobacter (HelCArc) | Thermotolerant Campylobacter (Catherm) | C. jejuni (Cajej) | C. upsaliensis (Cup) | C. lari (Clar1 and Clar2) | |
| C. jejuni | 29 | 29 (100) | 29 (100) | 26 (90) | 0 (0) | 0 (0) |
| C. coli | 32 | 32 (100) | 32 (100) | 1 (3) | 0 (0) | 0 (0) |
| C. upsaliensis | 10 | 10 (100) | 10 (100) | 0 (0) | 10 (100) | 0 (0) |
| C. lari | 11 | 11 (100) | 11 (100) | 0 (0) | 0 (0) | 9 (81) |
| Helicobacter pylori | 12 | 12 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
The corresponding group-specific probe correctly detected all thermotolerant Campylobacter isolates without any cross-reaction (Table 2). For the species-specific identification of C. jejuni, the assay showed a sensitivity of 90% (26/29 isolates) (Table 2). The C. jejuni-specific probe showed one false-positive reaction (1/55 isolates) with a C. coli isolate and thus reached a specificity of 98%. The sensitivities of the C. lari-specific probe and the C. upsaliensis-specific probe were 81% (9/10 isolates) and 100% (11/11 isolates), respectively. The specificity of the C. lari- and C. upsaliensis-specific probes was 100% (Table 2).
The most striking capacity of the assay was the 100% reliable recognition of thermotolerant Campylobacter within less than 2 h with limited effort. From a clinical point of view, the identification of a Campylobacter as thermotolerant and, thus, pathogenic is critical in order to initiate adequate therapy and infection control measures. Our results extend a recent report of the successful application of a similar FISH probe for the detection of thermotolerant Campylobacter spp. in poultry (25).
We suggest the use of a two-step FISH procedure for the further differentiation of Campylobacter. In the first step, the C. jejuni-specific probe may be used in combination with the probe specific for thermotolerant Campylobacter (the Catherm probe) and with the probe specific for Campylobacter, Arcobacter, and Helicobacter (the HelCArc probe). This step identifies the most frequent isolate, C. jejuni, with minimal effort (Table 3). Strains that are negative with the C. jejuni-specific probe but positive with all other probes represent thermotolerant Campylobacter spp. other than C. jejuni. The corresponding strains may be further characterized with considerable reliability in a second step by using the species-specific probes (Table 2). Strains that are negative with the three available species-specific probes may be considered C. coli, with a sensitivity of 97% and a specificity of 92% according to the data obtained with our sample collection.
TABLE 3.
Algorithm for interpretation of FISH results
| Result obtained with the following probe (probe specificity):
|
Interpretation | |||
|---|---|---|---|---|
| EUB (all bacteria) | HelCArc (Helicobacter, Campylobacter, Arcobacter) | Catherm (thermotolerant Campylobacter) | Cajej (C. jejuni) | |
| + | + | + | + | C. jejuni |
| + | + | + | − | Thermotolerant Campylobacter other than C. jejuni |
| + | + | − | − | Arcobacter, Helicobacter, or nonthermotolerant Campylobacter |
| + | − | − | − | Some bacteria other than Arcobacter, Helicobacter, or Campylobacter |
| − | − | − | − | No result (the FISH procedure did not work) |
| + | − | + | − | No result (contradictory binding pattern) |
Strains that stain negative with the probe specific for thermotolerant Campylobacter spp. (the Catherm probe) but positive with the probe specific for Campylobacter and its relatives (the HelCArc probe) represent Arcobacter, Helicobacter, or nonthermotolerant Campylobacter spp. Recognition of these strains provides a considerable advantage, because Arcobacter in particular but also nonthermotolerant Campylobacter spp. may be confused with thermotolerant Campylobacter by biochemical methods (4, 9, 24). The corresponding strains may be further analyzed biochemically or by FISH with previously published probes specific for Campylobacter (15, 17), Arcobacter (15), and Helicobacter (3).
In summary, FISH is suitable for the rapid identification of cultured isolates of thermotolerant Campylobacter spp. in a routine laboratory.
Acknowledgments
This work was supported by a grant from the University of Ulm, Forschungsförderung, to Sven Poppert. This work was partially funded by Seapro Theranostics International B.V., Lelystad, The Netherlands.
We thank Jaap Wagenaar and Jeroen Dijkstra (Animal Sciences Group, Wageningen, Lelystad, The Netherlands) for support and for providing us with Campylobacter strains. We thank Damien Lynch, Steffen Stenger, and Len Cegielka for critical reading of the manuscript.
Footnotes
Published ahead of print on 2 April 2008.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 561919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barros-Velazquez, J., A. Jimenez, and T. G. Villa. 1999. Isolation and typing methods for the epidemiologic investigation of thermotolerant campylobacters. Int. Microbiol. 2217-226. [PubMed] [Google Scholar]
- 3.Chan, V., G. Crocetti, M. Grehan, L. Zhang, S. Danon, A. Lee, and H. Mitchell. 2005. Visualization of Helicobacter species within the murine cecal mucosa using specific fluorescence in situ hybridization. Helicobacter 10114-124. [DOI] [PubMed] [Google Scholar]
- 4.Diergaardt, S. M., S. N. Venter, A. Spreeth, J. Theron, and V. S. Brozel. 2004. The occurrence of campylobacters in water sources in South Africa. Water Res. 382589-2595. [DOI] [PubMed] [Google Scholar]
- 5.Fitzgerald, C., and I. Nachamkin. 2007. Campylobacter and Arcobacter, p. 933-946. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 6.Gorkiewicz, G., G. Feierl, C. Schober, F. Dieber, J. Kofer, R. Zechner, and E. L. Zechner. 2003. Species-specific identification of campylobacters by partial 16S rRNA gene sequencing. J. Clin. Microbiol. 412537-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill, J. E., A. Paccagnella, K. Law, P. L. Melito, D. L. Woodward, L. Price, A. H. Leung, L. K. Ng, S. M. Hemmingsen, and S. H. Goh. 2006. Identification of Campylobacter spp. and discrimination from Helicobacter and Arcobacter spp. by direct sequencing of PCR-amplified cpn60 sequences and comparison to cpnDB, a chaperonin reference sequence database. J. Med. Microbiol. 55393-399. [DOI] [PubMed] [Google Scholar]
- 8.Huysmans, M. B., J. D. Turnidge, and J. H. Williams. 1995. Evaluation of API Campy in comparison with conventional methods for identification of thermophilic campylobacters. J. Clin. Microbiol. 333345-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacob, J., I. Feuerpfeil, and E. Schulze. 1996. PCR-mediated DNA fingerprinting of atypical campylobacter strains isolated from surface and drinking water. Zentralbl. Bakteriol. Parasitenkd. Infektkrankh. Hyg. Abt. 1 Orig. 285106-112. [DOI] [PubMed] [Google Scholar]
- 10.Kempf, V. A., K. Trebesius, and I. B. Autenrieth. 2000. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 38830-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuijper, E. J., S. Stevens, T. Imamura, B. De Wever, and E. C. Claas. 2003. Genotypic identification of erythromycin-resistant Campylobacter isolates as Helicobacter species and analysis of resistance mechanism. J. Clin. Microbiol. 413732-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulkarni, S. P., S. Lever, J. M. Logan, A. J. Lawson, J. Stanley, and M. S. Shafi. 2002. Detection of Campylobacter species: a comparison of culture and polymerase chain reaction based methods. J. Clin. Pathol. 55749-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Logan, J. M., K. J. Edwards, N. A. Saunders, and J. Stanley. 2001. Rapid identification of Campylobacter spp. by melting peak analysis of biprobes in real-time PCR. J. Clin. Microbiol. 392227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luber, P., J. Wagner, H. Hahn, and E. Bartelt. 2003. Antimicrobial resistance in Campylobacter jejuni and Campylobacter coli strains isolated in 1991 and 2001-2002 from poultry and humans in Berlin, Germany. Antimicrob. Agents Chemother. 473825-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreno, Y., S. Botella, J. L. Alonso, M. A. Ferrus, M. Hernandez, and J. Hernandez. 2003. Specific detection of Arcobacter and Campylobacter strains in water and sewage by PCR and fluorescent in situ hybridization. Appl. Environ. Microbiol. 691181-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreno, Y., M. A. Ferrus, J. L. Alonso, A. Jimenez, and J. Hernandez. 2003. Use of fluorescent in situ hybridization to evidence the presence of Helicobacter pylori in water. Water Res. 372251-2256. [DOI] [PubMed] [Google Scholar]
- 17.Moreno, Y., M. Hernandez, M. A. Ferrus, J. L. Alonso, S. Botella, R. Montes, and J. Hernandez. 2001. Direct detection of thermotolerant campylobacters in chicken products by PCR and in situ hybridization. Res. Microbiol. 152577-582. [DOI] [PubMed] [Google Scholar]
- 18.Moter, A., and U. B. Gobel. 2000. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 4185-112. [DOI] [PubMed] [Google Scholar]
- 19.Nicholson, M. A., and C. M. Patton. 1995. Evaluation of disk method for hippurate hydrolysis by Campylobacter species. J. Clin. Microbiol. 331341-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.On, S. L., and P. J. Jordan. 2003. Evaluation of 11 PCR assays for species-level identification of Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 41330-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pernthaler, J., F. O. Gloeckner, W. Schoenhuber, and R. Amann. 2006. Fluorescence in situ hybridization with rRNA-targeted oligonucleotide probes. Fluorescence in situ hybridization. Methods Microbiol. Mar. Microbiol. 301-31. [Google Scholar]
- 22.Popovic-Uroic, T., C. M. Patton, M. A. Nicholson, and J. A. Kiehlbauch. 1990. Evaluation of the indoxyl acetate hydrolysis test for rapid differentiation of Campylobacter, Helicobacter, and Wolinella species. J. Clin. Microbiol. 282335-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poppert, S., A. Essig, B. Stoehr, A. Steingruber, B. Wirths, S. Juretschko, U. Reischl, and N. Wellinghausen. 2005. Rapid diagnosis of bacterial meningitis by real-time PCR and fluorescence in situ hybridization. J. Clin. Microbiol. 433390-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prouzet-Mauleon, V., L. Labadi, N. Bouges, A. Menard, and F. Megraud. 2006. Arcobacter butzleri: underestimated enteropathogen. Emerg. Infect. Dis. 12307-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid, M. W., A. Lehner, R. Stephan, K. H. Schleifer, and H. Meier. 2005. Development and application of oligonucleotide probes for in situ detection of thermotolerant Campylobacter in chicken faecal and liver samples. Int. J. Food Microbiol. 105245-255. [DOI] [PubMed] [Google Scholar]
- 26.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and K. H. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 632884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, G., C. G. Clark, T. M. Taylor, C. Pucknell, C. Barton, L. Price, D. L. Woodward, and F. G. Rodgers. 2002. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J. Clin. Microbiol. 404744-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]