Abstract
Few data on the molecular characteristics and epidemiology of Staphylococcus aureus from Indonesia are available. The purpose of the present study was to define S. aureus reservoirs in both the Indonesian community and hospital using a collection of 329 nasal carriage isolates obtained during a survey of 3,995 healthy individuals and patients from Java, Indonesia. Only one strain (0.3%) was identified as methicillin-resistant S. aureus by mecA gene PCR. The Panton-Valentine leukocidin (PVL) genes were detected in 35 methicillin-sensitive S. aureus strains (10.6%). Molecular typing by pulsed-field gel electrophoresis of the 329 isolates showed extensive genetic diversity among both PVL-positive and PVL-negative strains. In Surabaya, Indonesia, however, a cluster was identified that was strongly associated with the presence of the PVL locus (P < 0.0001). As determined by high-throughput amplified fragment length polymorphism, PVL-positive strains occurred throughout all major AFLP clusters (I to IV). Multilocus sequence typing of a subset of isolates showed that most PVL-positive strains belonged to sequence type (ST) 188, while most PVL-negative isolates belonged to ST45. The high prevalence of PVL-positive S. aureus strains in certain regions of Indonesia is of concern since these strains may cause severe infections in the community and in hospitals.
Humans are a natural reservoir of Staphylococcus aureus, with the moist squamous epithelium of the anterior nares acting as the primary ecological niche (12, 33). Nasal carriage of S. aureus has been identified as a major risk factor for the development of infections, including S. aureus bacteremia (13, 31).
In the past four decades, the incidences of both community-acquired and hospital-acquired S. aureus infections have increased, while antibiotic treatment options are increasingly hampered by the spread of methicillin-resistant S. aureus (MRSA) strains and, more recently, S. aureus strains resistant to other classes of antibiotics (26, 27). The prevalence of MRSA in some Asian countries, such as Taiwan and China, is among the highest in the world (2). Community-acquired MRSA (CA-MRSA) infections also appear to be an emerging phenomenon in some Asian countries (6, 10, 11). The majority of these CA-MRSA isolates carry the Panton-Valentine leukocidin (PVL), a virulence factor that is strongly associated with skin infections and severe necrotizing pneumonia (8, 9, 28).
Several studies have assessed local population structures of S. aureus in order to investigate clonality and associated virulence. In a previous analysis of over 1,000 S. aureus strains from The Netherlands, by using amplified fragment length polymorphism (AFLP) and multilocus sequence typing (MLST), evidence was provided showing that essentially any S. aureus genotype carried by humans can transform into a life-threatening human pathogen but that certain clones may be more virulent than others (20). A smaller study of 74 MRSA strains of unknown clinical or nonclinical origin from 12 Asian countries revealed two major genotypes with a distinct geographic distribution (14). Most of the Korean and Japanese isolates belonged to clonal complex (CC) 5, while most MRSA strains from other Asian countries, including seven strains from Indonesia, belonged to sequence type (ST) 239, a distinct lineage within CC8. This finding was recently corroborated by an analysis of 615 MRSA isolates from 11 Asian countries by Chongtrakool et al. (7). They found the same MLST profile (ST239) for three representative clinical isolates from Jakarta, Indonesia. However, further data on the genotypic characteristics of methicillin-susceptible S. aureus (MSSA) and MRSA from Indonesia are not available to our knowledge.
Recently, a population-based survey of 4,000 people in two cities on the island of Java (Surabaya and Semarang) in Indonesia was conducted by the Antimicrobial Resistance in Indonesia, Prevalence and Prevention (AMRIN) Study Group in order to quantify human carriage of resistant microorganisms (17). The purpose of the present study was to define S. aureus reservoirs in both the Indonesian community and hospital using S. aureus strains obtained in the AMRIN study.
MATERIALS AND METHODS
Bacterial isolates.
The AMRIN study is a population-based study among four groups of individuals in two cities, Semarang and Surabaya, Indonesia, aimed at investigating the level of carriage of resistant microorganisms. These groups involved patients on the day of admission to the hospital (group 1), patients on the day of discharge after five or more days of hospitalization (group 2), persons visiting a primary health center (group 3), and healthy relatives or household members of the group 1 patients (group 4). Nasal swabs were taken from patients or healthy individuals in the internal medicine, surgery, gynecology/obstetrics, and pediatrics departments (groups 1, 2, and 4). The specimens were collected from July to October 2001 in Surabaya and from January to May 2002 in Semarang. The medical ethics committees of both hospitals approved of the study protocol.
Cultures of the anterior nares were obtained with sterile cotton swabs from 3,995 persons after they gave informed consent. Within 24 h, these swabs were inoculated on phenol red mannitol agar (Becton Dickinson, Heidelberg, Germany). Colonies suspected of being S. aureus were stored in trypticase soy agar. This collection, comprising more than 1,200 putative S. aureus strains, was subsequently speciated with StaphaurexPlus rapid latex reagent (Abbott Murex, Chatillon, France) and the Vitek 2 system (bioMérieux, Inc., Hazelwood, MO). In case of doubt, an S. aureus-specific DNA hybridization test (AccuProbe; Gen-Probe, Inc., San Diego, CA) was performed. Susceptibility testing was performed in Indonesia, and the results have been published elsewhere (17). In the present analysis, we included the first 329 confirmed S. aureus isolates (Table 1).
TABLE 1.
Origin of the 329 S. aureus strains included in the study
| Population group and department | No. of isolates per city (%)
|
|
|---|---|---|
| Semarang | Surabaya | |
| Admission | 29 (23.2) | 45 (22.1) |
| Internal medicine | 4 | 4 |
| Surgery | 10 | 13 |
| Gynecology/obstetrics | 7 | 25 |
| Pediatrics | 8 | 3 |
| Discharge | 31 (25.6) | 60 (29.4) |
| Internal medicine | 3 | 13 |
| Surgery | 7 | 10 |
| Gynecology/obstetrics | 12 | 31 |
| Pediatrics | 9 | 6 |
| Primary health center | 33 (26.4) | 56 (27.4) |
| Relatives | 32 (24.8) | 43 (21.1) |
| Internal medicine | 4 | 6 |
| Surgery | 11 | 12 |
| Gynecology/obstetrics | 5 | 16 |
| Pediatrics | 12 | 9 |
| Total | 125 (100) | 204 (100) |
Pulsed-field gel electrophoresis.
Pulsed-field gel electrophoresis (PFGE) of SmaI digests of chromosomal DNA from all 329 strains was performed as described previously (15). Relatedness among the PFGE profiles was evaluated with Bionumerics software (version 3.0; Applied Maths, Ghent, Belgium). A dendrogram was produced using the Dice coefficient and an unweighted-pair group method using arithmetic averages (UPGMA). Band tolerance was set at 2.0%.
DNA isolation and detection of mecA and pvl genes.
Chromosomal DNA was extracted with MagNA Pure LC DNA isolation kit III (bacteria, fungi) using the MagNA Pure LC instrument (Roche Diagnostics, Almere, The Netherlands) (20). The DNA concentration was assessed spectrophotometrically, and samples were stored at −20°C. The presence of the mecA and pvl genes was determined by PCR (18, 24). mecA-positive strains and isolates containing the pvl genes (whether mecA-positive or not) were further subjected to a multiplex PCR to identify staphylococcal cassette chromosome mec (SCCmec) types I to V (5). Positive and negative control strains were included in each PCR run.
High-throughput AFLP.
A selection of 81 isolates was analyzed by high-throughput AFLP. We selected every fourth isolate when going from top to bottom through the PFGE dendrogram. DNA restriction, ligation of AFLP adapters, and amplification of the modified fragments were carried out as described previously (20). Briefly, bacterial DNA was digested with the enzymes MboI and Csp6I (New England Biolabs, Westburg, Leusden, The Netherlands). Ligation was performed by using specific linker oligonucleotide pairs (for MboI, 5′-CTCGTAGACTGCGTACC-3′ and 5′-ATCGGTACGCAGTCTAC-3′, and for Csp6I, 5′-ACGATGAGTCCTGAC-3′ and 5′-TAGTCAGGACTCAT-3′). Subsequently, a nonselective preamplification was performed using the MboI primer (5′-GTAGACTGCGTACCGATC-3′) and the Csp6I primer (5′-ACGATGAGTCCTGACTAC-3′). In the final amplification, a 33P-labeled MboI primer containing one selective nucleotide (either +C or +G) and a Csp6I primer containing two selective nucleotides (+TA) were used. The amplified material was analyzed by polyacrylamide gel electrophoresis and autoradiography.
AFLP database.
We compared the genetic structure of the 81 Indonesian S. aureus isolates with the (previously determined) natural population structure of the S. aureus carriage isolates from healthy individuals from the Rotterdam area (The Netherlands) (20). The AFLP database comprises high-throughput AFLP patterns from 829 nonclinical S. aureus isolates from the Dutch study (20).
AFLP data analysis.
For two-dimensional clustering of the AFLP genotype patterns in the AFLP database, an agglomerative (successive) hierarchical procedure was performed using the UPGMA distance algorithm (20). The Tanimoto method was used to calculate the similarity matrix (Spotfire DecisionSite 7.2; Spotfire, Göteborg, Sweden).
MLST.
MLST was carried out for 36 S. aureus strains from the AFLP set using DNA arrays (Affymetrix, Santa Clara, CA; bioMérieux, Marcy l'Etoile, France) (29). The selected isolates were equally distributed across the PFGE dendrogram by selecting alternately one out of two and one out of three strains that had been analyzed by AFLP, going from top to bottom through the PFGE dendrogram.
Statistical analysis.
Data were analyzed using statistical software packages SPSS version 11.0 (SPSS, Chicago, IL) and EpiInfo version 5.00 (Centers for Disease Control and Prevention, Atlanta, GA). Chi-square or Fisher's exact tests (two-tailed) were used when appropriate for comparisons of proportions. P values less than 0.05 were considered significant.
RESULTS
PFGE.
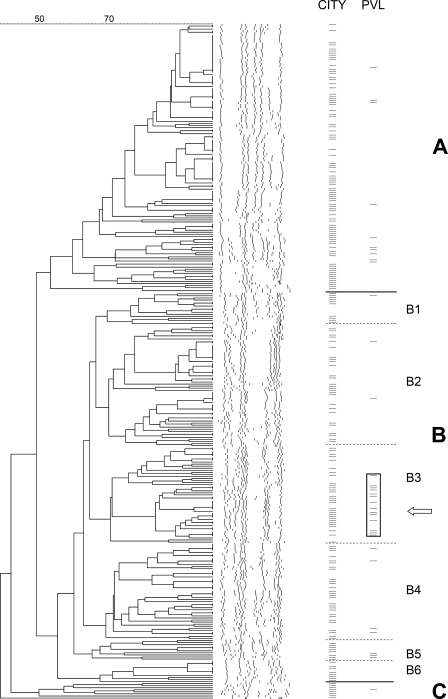
Molecular typing by PFGE revealed extensive strain heterogeneity (Fig. 1). At a similarity value of circa 50%; however, three PFGE clusters, designated A to C, could be distinguished (Fig. 1; Table 2). Strains from Semarang and Surabaya were equally distributed among clusters A and B. Cluster C consisted exclusively of strains from Surabaya (n = 8; P = 0.026) from patients visiting a primary health center (P < 0.001). However, the observed profiles within this cluster were variable. Therefore, it would be incorrect to define this cluster as a single clone. Both clusters A and B contained isolates from all four population groups and all four hospital departments, with similar distributions (data not shown). As shown in Fig. 1, cluster B could be further subdivided into six subclusters, B1 to B6, on the basis of segregated branches. Subcluster B3 was the most homogeneous, with a similarity level of >71%. Strains from Semarang were more frequently found in subcluster B2 (32/125 [25.6%]), compared with isolates from Surabaya (27/204 [13.2%]; P = 0.007). No other significant geographical differences in the distributions of the isolates were found.
FIG. 1.
Dendrogram based on PFGE SmaI restriction pattern analysis of 329 nares-colonizing S. aureus isolates. Similarity analysis was performed with the Dice coefficient and clustering by the UPGMA method. The scale on the top shows percentages of similarity. Further information about the strains is shown on the right side of the figure in two columns. In the first column (CITY), isolates from Surabaya are indicated by a dash, whereas isolates from Semarang are left blank. In the second column (PVL), only PVL-positive strains are marked by a dash. The single MRSA isolate is indicated by an arrow. Clusters are designated A to C, and subclusters within cluster B are designated B1 to B6. The rectangle highlights the 14 PVL-positive strains in subcluster B3.
TABLE 2.
Molecular characteristics of 329 Indonesian S. aureus isolates
| Characteristic | Total no. of isolates (%) | No. of isolates per city (%)
|
P value | |
|---|---|---|---|---|
| Semarang (n = 125) | Surabaya (n = 204) | |||
| PFGE cluster | ||||
| A | 131 (39.8) | 46 (36.8) | 85 (41.7) | 0.448 |
| B | 190 (57.8) | 79 (63.2) | 111 (54.4) | 0.147 |
| C | 8 (2.4) | 0 | 8 (3.9) | 0.026 |
| mecA presence | ||||
| Positive | 1 (0.3) | 0 | 1 (0.5) | 1.000 |
| Negative | 328 (99.7) | 125 (100) | 203 (99.5) | |
| PVL presence | ||||
| Positive | 35 (10.6) | 9 (7.2) | 26 (12.7) | 0.141 |
| Negative | 294 (89.4) | 116 (92.8) | 178 (87.3) | |
mecA- and PVL-positive strains.
Only 1/329 strains harbored the mecA gene (0.3%). This MRSA strain was isolated from the nose of a 41-year-old male patient upon discharge after 45 days of hospitalization in a surgery department in Surabaya. The discharge diagnosis was a malignancy. In the PFGE analysis, the strain clustered in B3. The SCCmec of this isolate was identified as type V.
The pvl genes were detected in 35 of 329 isolates (10.6%). The proportion of PVL-positive isolates from Surabaya was 12.6%, whereas 7.2% of the isolates from Semarang carried PVL (P = 0.141) (Table 2). The PVL-positive strains were evenly distributed among the four groups of patients and healthy persons (7, 9, 9, and 10 carriers in groups 1, 2, 3, and 4, respectively) and among individuals from the four departments (data not shown). In the PFGE analysis, the pvl genes were found in isolates from all three clusters. A cluster of PVL-positive strains could be identified within cluster B (Fig. 1). Strains from PFGE subcluster B3 were significantly enriched for the presence of PVL in comparison with the other PFGE clusters (14/47 [29.8%] cluster B3 isolates versus 21/282 [7.4%] non-B3 isolates; P < 0.0001). Twelve of these 14 PVL-positive PFGE subcluster B3 strains were isolated in Surabaya, and 4 of these were from patients who were discharged from the Department of Gynecology/Obstetrics. The single MRSA isolate did not harbor the pvl genes. In order to determine whether SCCmec elements were present in these 35 mecA-negative, PVL-positive isolates, a multiplex PCR for SCCmec types I to V was carried out. One strain (1/35 [2.9%]) was positive for SCCmec type I (IS1272). Other SCCmec types were not found. The SCCmec-positive isolate was cultured from a healthy relative accompanying a patient that was admitted to the gynecology/obstetrics department in Surabaya. In the PFGE analysis, the isolate clustered in B3.
AFLP.
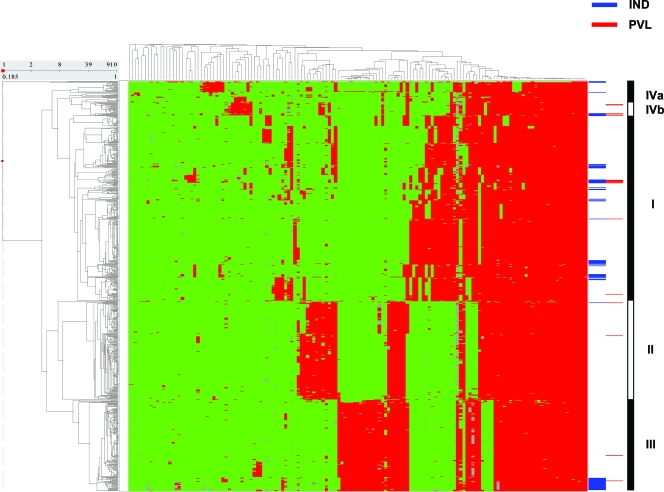
The AFLP patterns obtained for the 81 Indonesian strains, including 12 PVL-positive strains, were compared with those of S. aureus carriage isolates obtained from healthy individuals in The Netherlands (Fig. 2). Essentially, the Indonesian strains clustered within the previously defined AFLP clusters (I to IV) (Table 3) (20). However, in comparison with the representation of the Dutch isolates in AFLP cluster II (similar to CC30), the Indonesian strains were significantly underrepresented in this cluster (216/829 versus 1/81; P < 0.0001). The single strain in cluster II was isolated from a patient on the day of admission at the Department of Gynecology/Obstetrics in Semarang. Vice versa, the Indonesian strains were significantly overrepresented in cluster III (similar to CC45) (27/81 versus 176/829; P = 0.018).
FIG. 2.
Two-dimensional clustering of 81 carriage strains from Indonesia and 829 carriage strains from The Netherlands (20). The dendrogram on the y axis represents the phylogenetic clustering of the 910 strains. The dendrogram on the x axis shows the clustering of the AFLP markers. AFLP marker fragments are shown in red and green; red represents marker presence and green indicates absence. The blue horizontal bars at the right side of the figure indicate the position of the Indonesian isolates; the small red bars represent PVL-positive strains from both Indonesia and The Netherlands. The black-and-white bar at the far right of the figure represents the three main clusters (I, II, and III) and two minor clusters (IVa and IVb), as defined before (20).
TABLE 3.
Distribution of the Indonesian S. aureus collection of 81 strains and its subcollections in the five phylogenetic AFLP clusters and the distribution of the Dutch carriage strains in the five clusters for comparison
| Collection (total no. of strains) | No. of strains (%) in AFLP cluster:
|
||||
|---|---|---|---|---|---|
| I | II | III | IVa | IVb | |
| Dutch carriage strains (829)a | 367 (44) | 216 (26)b | 176 (21)c | 46 (6) | 24 (3) |
| Indonesia (81) | 44 (54) | 1 (1)b | 27 (33)c | 4 (5) | 5 (6) |
| Semarang (33) | 20 (61) | 1 (3) | 11 (33) | 1 (3) | 0 |
| Surabaya (48) | 24 (50) | 0 | 16 (33) | 3 (6) | 5 (10) |
| Admission (19) | 7 (37) | 1 (5) | 8 (42) | 1 (5) | 2 (11) |
| Discharge (25) | 17 (68) | 0 | 5 (20) | 2 (8) | 1 (4) |
| Primary health center (18) | 10 (56) | 0 | 7 (39) | 1 (6) | 0 |
| Relatives (19) | 10 (53) | 0 | 7 (37) | 0 | 2 (11) |
| PVL positive (12) | 7 (58) | 1 (8) | 1 (8) | 0 | 3 (25)d |
| PVL negative (69) | 37 (54) | 0 | 26 (38) | 4 (6) | 2 (3) |
| PFGE cluster A (32) | 2 (6) | 1 (3) | 23 (72)e | 2 (6) | 4 (13) |
| PFGE cluster B (47) | 40 (85)f | 0 | 4 (9) | 2 (4) | 1 (2) |
| PFGE cluster C (2) | 2 (100) | 0 | 0 | 0 | 0 |
From reference 20.
Underrepresentation of Indonesian strains in cluster II compared with the representation of Dutch strains in this cluster (P < 0.001).
Overrepresentation of Indonesian strains in cluster III compared with the representation of Dutch strains in this cluster (P = 0.018).
Cluster IVb was significantly enriched with PVL in comparison with the other AFLP clusters (P = 0.022).
Proportionately more PFGE cluster A strains compared with the other PFGE cluster strains (P < 0.001).
Proportionately more PFGE cluster B strains compared with the other PFGE cluster strains (P < 0.001).
The distributions of the isolates from Semarang and Surabaya across the major AFLP clusters were similar (Table 3). Strains from all four population groups and from the three nonpediatric departments were found in major clusters I, III, and IV, without any significant differences. AFLP cluster IV did not contain strains from the pediatrics department. However, this was not statistically significant (data not shown). All major clusters contained PVL-positive strains. Isolates from minor cluster IVb were significantly enriched for the presence of PVL in comparison with the isolates from the other AFLP clusters (3/5 versus 9/76; P = 0.022).
MLST.
We identified nine different STs among 30 strains and three new profiles for which no ST has been defined yet (Table 4). Three other isolates could not be typed by MLST, since no PCR product could be obtained repeatedly for the aroE gene. The strains that were classified in AFLP cluster I revealed seven different known STs and the three unknown STs. In contrast, AFLP clusters III and IVb were more homogeneous. These results were in agreement with those published before (20, 30). Cluster IVa harbored the three nontypeable strains. The six PVL-positive strains that were analyzed by MLST revealed three different STs: ST188/CC1 (n = 3), ST121/CC121 (n = 2), and ST45/CC45 (n = 1). The three isolates with ST188 were all from Surabaya and clustered in subcluster B3 in the PFGE analysis. Using the AFLP database (22), the STs of six other PVL-positive isolates could be deduced: ST188/CC1 (n = 3, including the SCCmec type I-positive strain), ST121/CC121 (n = 1), ST25/CC25 (n = 1), and ST30/CC30 (n = 1).
TABLE 4.
Sequence types of 36 S. aureus strains assigned by MLST
| AFLP cluster | MLST ST (no. of strains)
|
|
|---|---|---|
| Semarang | Surabaya | |
| I | ST5 (1), ST8 (1), ST9 (1), ST15 (2), unknown ST (2)a | ST1 (3), ST5 (1), ST15 (1), ST20 (1), ST188 (4), unknown ST (1)a |
| II | ||
| III | ST45 (6) | ST45 (6) |
| IVa | NDb (1) | ND (2) |
| IVb | ST121 (3) | |
Three different profiles were observed.
ND, not determinable, because no PCR product could be obtained for one of the housekeeping genes.
DISCUSSION
Nasal carriage of S. aureus plays a key role in the epidemiology and pathogenesis of S. aureus infections (12, 33). To define reservoirs of this pathogen in both the hospital and the community, it is necessary to study the molecular epidemiology of carriage isolates of patients and healthy individuals. This is the first report of such an analysis of a well-defined population-based collection of S. aureus isolates from Indonesia.
Interestingly, we found a low prevalence of MRSA (0.3%) but a high prevalence of PVL-positive MSSA (10.6%) in this collection of Indonesian carriage strains. Molecular typing by PFGE showed extensive genetic diversity among both PVL-positive and PVL-negative strains. In Surabaya, however, we identified a cluster that was strongly associated with the presence of the PVL locus. The high prevalence of PVL-positive commensal S. aureus is in contrast with the low prevalences in carriage isolates previously reported: 0.6% by Melles et al. (The Netherlands) (21), 0% by Prevost et al. (France) (25), and 1.4% by Von Eiff et al. (Germany) (32). In populations with high MRSA carriage rates, more PVL-positive isolates can be found. Kuehnert et al. reported the presence of PVL in 1.0% of 297 American MSSA nasal isolates but in 8.0% of 75 MRSA strains (16). In a recent study from Taiwan, 18 (6.0%) of 300 colonizing isolates from children carried the pvl genes, 15 of which were MRSA (19). Evidence was provided that linked these PVL-positive carriage strains from the community to CA-MRSA-infecting strains. The PVL locus seems to represent a genetic marker of CA-MRSA strains worldwide (28). In Indonesia, however, PVL is apparently not associated with MRSA. Although small numbers of PVL-positive MSSA strains have been reported from other countries, this is, to the best of our knowledge, a rare phenomenon (1, 16, 21). In only one study, from the Cape Verde islands, was a similar finding reported (3). The pvl genes were detected in 34.9% of 63 nosocomial MSSA strains isolated from nasal and wound swabs from patients and health care workers. Eighteen of the 22 PVL-positive strains were from nasal samples, but since it is unknown whether these carriers suffered from skin infections, a possible association with active disease cannot be ruled out. In our study, none of the patients from groups 1 and 2 that carried a PVL-positive isolate had a diagnosis of a skin infection at the moment of inclusion in the study.
It has been suggested by Vandenesch et al. that first, intercontinental exchange of MSSA or MRSA had occurred, which was then possibly followed by the introduction of a mecA gene harboring SCCmec in MSSA and the pvl genes in MSSA or MRSA (28). Since the two loci (PVL and SCCmec) are widely separated on the S. aureus chromosome, coacquisition on a single mobile genetic element is unlikely (4). In the present study, we found one mecA gene-positive strain that was PVL negative and one mecA gene-negative SCCmec type I-positive strain that was PVL positive. Therefore, we have shown that the SCCmec has only rarely been introduced in the Indonesian carriage S. aureus, but the pvl genes, on the other hand, have been integrated in distinct phylogenetic subpopulations, as demonstrated by PFGE and AFLP. Overall, in contrast with the Dutch strains, the Indonesian strains were virtually absent from AFLP cluster II (similar to CC30), but PVL-positive strains occurred throughout all the major AFLP clusters (I to IV). AFLP cluster IVb (similar to ST121) was significantly enriched with PVL compared with the other clusters. MLST of a subset of isolates showed that PVL-positive strains mainly belonged to ST188/CC1 and ST121/CC121, while most PVL-negative isolates belonged to ST45/CC45. PVL-positive MSSA of ST188 has not been described before, but ST121 has been detected among MSSA isolates carrying PVL in other countries, such as Portugal, Cape Verde, The Netherlands, and Germany (1, 3, 21, 23). CC239, to which previous MRSA strains from Jakarta reportedly belong, was not found, indicating that geographical differences may exist within Indonesia (7, 14). Three PVL-negative strains from AFLP cluster IVa were not typeable by MLST because no PCR product could be obtained for the aroE gene, which encodes shikimate dehydrogenase. We assume that this is caused by a primer binding site mutation, but this needs further research.
The PVL-positive MSSA strains seem to be successful commensals in Indonesia. Whether they are also successful pathogens needs to be corroborated by analyzing clinical isolates. Although the mecA gene may be transmitted to any MSSA strain, it is of concern that the MRSA isolate clustered in PFGE subcluster B3 together with the successful PVL-positive strains, because this may be an early warning for CA-MRSA emergence in Indonesia. Since this single MRSA strain from our study was isolated from a patient at the moment of discharge after a hospital stay of more than 5 days and the strain did not contain the pvl genes, we assume that the strain was nosocomially acquired. The SCCmec type of the isolate was identified as type V. The strain was resistant to erythromycin and chloramphenicol but sensitive to trimethoprim-sulfamethoxazole, tetracycline, and gentamicin (data not shown).
The low prevalence of MRSA in this collection of Indonesian carriage strains is comparable to the prevalence in the Dutch population (0.1% of 2,332 carriage strains isolated from 9,859 individuals) (34). The low prevalence in The Netherlands is ascribed to restrictive antibiotic use and a national search-and-destroy policy when dealing with MRSA. Since there is no search-and-destroy policy in Indonesia, the low MRSA prevalence in Java may be due to limited antibiotic consumption. On the other hand, a scenario of a low carriage rate of MRSA in the community and a high prevalence of invasive MRSA in the hospital, such as in Portugal, remains a possibility (1). This hypothesis, however, needs to be explored further.
In summary, our data provide a unique insight into the molecular characteristics and population structure of S. aureus carried by healthy individuals and patients from Java, Indonesia. There is good news with respect to the low prevalence of MRSA. The picture is less bright with respect to the presence of PVL. Nasal isolates that harbor the pvl genes may serve as an endogenous reservoir for infections or may be spread to other individuals. Further research is needed and continued surveillance is warranted, as the epidemiology of S. aureus is constantly changing.
Acknowledgments
We thank the directors of the Dr. Soetomo Hospital, Surabaya, Indonesia, and the directors of the Dr. Kariadi Hospital, Semarang, Indonesia, who facilitated our work in these hospitals. We also thank all staff members who have been involved in the isolation of bacteria. We also gratefully acknowledge the contributions of the medical students Diana Huis in ′t Veld, Suzanne Werter, Rianne de Jong, and Rozemarijn van der Meulen from the Radboud University Medical Center, Nijmegen, The Netherlands, who helped us collect the specimens in Indonesia. We thank Nicole Lemmens-den Toom for excellent technical assistance. The SCCmec type V S. aureus control strain (WIS) was kindly provided by Ad C. Fluit (University Medical Center Utrecht, Utrecht, The Netherlands).
This work was facilitated by grant number 99-MED-03 from the Royal Netherlands Academy of Sciences and Arts in the framework of its Scientific Program Indonesia-Netherlands (SPIN), Amsterdam, The Netherlands. The oligoarray-mediated MLST for S. aureus characterization was financially supported by bioMérieux (Alain Troesch and Corinne Jay).
Members of the AMRIN study group are as follows: Widjoseno Gardjito, Erni P. Kolopaking, Djoko Roeshadi, Eddy Rahardjo, Hari Parathon, Kuntaman Kuntaman, Ni Made Mertaniasih, Nun Zairina, Endang Isbandiati, Mariyatul Qibtiyah, Marijam Purwanta, and Usman Hadi from the Dr. Soetomo Hospital-School of Medicine, Airlangga University, Surabaya, Indonesia; Ariawan Soejoenoes, Budi Riyanto, Hendro Wahyono, Musrichan Adhisaputro, Bambang Triwara, Endang Sri Lestari, Bambang Wibowo, Muchlis A.U. Sofro, M.M.D.E.A.H. Hapsari, and Helmia Farida from the Dr. Kariadi Hospital-School of Medicine, Diponegoro University, Semarang, Indonesia; Peterhans van den Broek and D. Offra Duerink from the Leiden University Medical Center, Leiden, The Netherlands; Henri A. Verbrugh and Inge C. Gyssens from Erasmus MC, University Medical Center, Rotterdam, The Netherlands; and Monique Keuter from the Radboud University Medical Center, Nijmegen, The Netherlands.
AFLP is a registered trademark of Keygene N.V., and the AFLP technology is subject to patents and patent applications owned by Keygene N.V.
Footnotes
Published ahead of print on 23 April 2008.
REFERENCES
- 1.Aires de Sousa, M., T. Conceição, C. Simas, and H. de Lencastre. 2005. Comparison of genetic backgrounds of methicillin-resistant and -susceptible Staphylococcus aureus isolates from Portuguese hospitals and the community. J. Clin. Microbiol. 435150-5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., M. I. Crisóstomo, I. S. Sanches, J. S. Wu, J. Fuzhong, A. Tomasz, and H. de Lencastre. 2003. Frequent recovery of a single clonal type of multidrug-resistant Staphylococcus aureus from patients in two hospitals in Taiwan and China. J. Clin. Microbiol. 41159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aires-de-Sousa, M., T. Conceição, and H. de Lencastre. 2006. Unusually high prevalence of nosocomial Panton-Valentine leukocidin-positive Staphylococcus aureus isolates in Cape Verde Islands. J. Clin. Microbiol. 443790-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 3591819-1827. [DOI] [PubMed] [Google Scholar]
- 5.Boye, K., M. D. Bartels, I. S. Andersen, J. A. Moller, and H. Westh. 2007. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I-V. Clin. Microbiol. Infect. 13725-727. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C. J., Y. C. Huang, C. H. Chiu, L. H. Su, and T. Y. Lin. 2005. Clinical features and genotyping analysis of community-acquired methicillin-resistant Staphylococcus aureus infections in Taiwanese children. Pediatr. Infect. Dis. J. 2440-45. [DOI] [PubMed] [Google Scholar]
- 7.Chongtrakool, P., T. Ito, X. X. Ma, Y. Kondo, S. Trakulsomboon, C. Tiensasitorn, M. Jamklang, T. Chavalit, J. H. Song, and K. Hiramatsu. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 501001-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dufour, P., Y. Gillet, M. Bes, G. Lina, F. Vandenesch, D. Floret, J. Etienne, and H. Richet. 2002. Community-acquired methicillin-resistant Staphylococcus aureus infections in France: emergence of a single clone that produces Panton-Valentine leukocidin. Clin. Infect. Dis. 35819-824. [DOI] [PubMed] [Google Scholar]
- 9.Ellis, M. W., D. R. Hospenthal, D. P. Dooley, P. J. Gray, and C. K. Murray. 2004. Natural history of community-acquired methicillin-resistant Staphylococcus aureus colonization and infection in soldiers. Clin. Infect. Dis. 39971-979. [DOI] [PubMed] [Google Scholar]
- 10.Ho, P. L., C. Cheung, G. C. Mak, C. W. Tse, T. K. Ng, C. H. Cheung, T. L. Que, R. Lam, R. W. Lai, R. W. Yung, and K. Y. Yuen. 2007. Molecular epidemiology and household transmission of community-associated methicillin-resistant Staphylococcus aureus in Hong Kong. Diagn. Microbiol. Infect. Dis. 57145-151. [DOI] [PubMed] [Google Scholar]
- 11.Hsu, L. Y., Y. L. Koh, N. L. Chlebicka, T. Y. Tan, P. Krishnan, R. T. Lin, N. Tee, T. Barkham, and T. H. Koh. 2006. Establishment of ST30 as the predominant clonal type among community-associated methicillin-resistant Staphylococcus aureus isolates in Singapore. J. Clin. Microbiol. 441090-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kluytmans, J. A., A. van Belkum, and H. A. Verbrugh. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kluytmans, J. A., and H. F. Wertheim. 2005. Nasal carriage of Staphylococcus aureus and prevention of nosocomial infections. Infection 333-8. [DOI] [PubMed] [Google Scholar]
- 14.Ko, K. S., J. Y. Lee, J. Y. Suh, W. S. Oh, K. R. Peck, N. Y. Lee, and J. H. Song. 2005. Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J. Clin. Microbiol. 43421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koning, S., A. van Belkum, S. Snijders, W. van Leeuwen, H. Verbrugh, J. Nouwen, M. Op 't Veld, L. W. A. van Suijlekom-Smit, J. C. van der Wouden, and C. Verduin. 2003. Severity of nonbullous Staphylococcus aureus impetigo in children is associated with strains harboring genetic markers for exfoliative toxin B, Panton-Valentine leukocidin, and the multidrug resistance plasmid pSK41. J. Clin. Microbiol. 413017-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuehnert, M. J., D. Kruszon-Moran, H. A. Hill, G. McQuillan, S. K. McAllister, G. Fosheim, L. K. McDougal, J. Chaitram, B. Jensen, S. K. Fridkin, G. Killgore, and F. C. Tenover. 2006. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001-2002. J. Infect. Dis. 193172-179. [DOI] [PubMed] [Google Scholar]
- 17.Lestari, E. S., J. A. Severin, P. M. G. Filius, K. Kuntaman, D. O. Duerink, U. Hadi, H. Wahjono, and H. A. Verbrugh. 2008. Antimicrobial resistance among commensal isolates of Escherichia coli and Staphylococcus aureus isolates in the Indonesian population inside and outside hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 2745-51. [DOI] [PubMed] [Google Scholar]
- 18.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 291128-1132. [DOI] [PubMed] [Google Scholar]
- 19.Lo, W. T., W. J. Lin, M. H. Tseng, S. R. Wang, M. L. Chu, and C. C. Wang. 2006. Community-acquired methicillin-resistant Staphylococcus aureus in children, Taiwan. Emerg. Infect. Dis. 121267-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melles, D. C., R. F. Gorkink, H. A. Boelens, S. V. Snijders, J. K. Peeters, M. J. Moorhouse, P. J. van der Spek, W. B. van Leeuwen, G. Simons, H. A. Verbrugh, and A. van Belkum. 2004. Natural population dynamics and expansion of pathogenic clones of Staphylococcus aureus. J. Clin. Investig. 1141732-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melles, D. C., W. B. van Leeuwen, H. A. Boelens, J. K. Peeters, H. A. Verbrugh, and A. van Belkum. 2006. Panton-Valentine leukocidin genes in Staphylococcus aureus. Emerg. Infect. Dis. 121174-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melles, D. C., W. B. van Leeuwen, S. V. Snijders, D. Horst-Kreft, J. K. Peeters, H. A. Verbrugh, and A. van Belkum. 2007. Comparison of multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), and amplified fragment length polymorphism (AFLP) for genetic typing of Staphylococcus aureus. J. Microbiol. Methods 69371-375. [DOI] [PubMed] [Google Scholar]
- 23.Monecke, S., P. Slickers, M. J. Ellington, A. M. Kearns, and R. Ehricht. 2007. High diversity of Panton-Valentine leukocidin-positive, methicillin-susceptible isolates of Staphylococcus aureus and implications for the evolution of community-associated methicillin-resistant S. aureus. Clin. Microbiol. Infect. 131157-1164. [DOI] [PubMed] [Google Scholar]
- 24.Murakami, K., W. Minamide, K. Wada, E. Nakamura, H. Teraoka, and S. Watanabe. 1991. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 292240-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prevost, G., P. Couppie, P. Prevost, S. Gayet, P. Petiau, B. Cribier, H. Monteil, and Y. Piemont. 1995. Epidemiological data on Staphylococcus aureus strains producing synergohymenotropic toxins. J. Med. Microbiol. 42237-245. [DOI] [PubMed] [Google Scholar]
- 26.Ruef, C. 2004. Epidemiology and clinical impact of glycopeptide resistance in Staphylococcus aureus. Infection 32315-327. [DOI] [PubMed] [Google Scholar]
- 27.Steinberg, J. P., C. C. Clark, and B. O. Hackman. 1996. Nosocomial and community-acquired Staphylococcus aureus bacteremias from 1980 to 1993: impact of intravascular devices and methicillin resistance. Clin. Infect. Dis. 23255-259. [DOI] [PubMed] [Google Scholar]
- 28.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Leeuwen, W. B., C. Jay, S. Snijders, N. Durin, B. Lacroix, H. A. Verbrugh, M. C. Enright, A. Troesch, and A. van Belkum. 2003. Multilocus sequence typing of Staphylococcus aureus with DNA array technology. J. Clin. Microbiol. 413323-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Leeuwen, W. B., D. C. Melles, A. Alaidan, M. Al-Ahdal, H. A. Boelens, S. V. Snijders, H. Wertheim, E. van Duijkeren, J. K. Peeters, P. J. van der Spek, R. Gorkink, G. Simons, H. A. Verbrugh, and A. van Belkum. 2005. Host- and tissue-specific pathogenic traits of Staphylococcus aureus. J. Bacteriol. 1874584-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Von Eiff, C., K. Becker, K. Machka, H. Stammer, and G. Peters. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 34411-16. [DOI] [PubMed] [Google Scholar]
- 32.Von Eiff, C., A. W. Friedrich, G. Peters, and K. Becker. 2004. Prevalence of genes encoding for members of the staphylococcal leukotoxin family among clinical isolates of Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 49157-162. [DOI] [PubMed] [Google Scholar]
- 33.Wertheim, H. F., D. C. Melles, M. C. Vos, W. van Leeuwen, A. van Belkum, H. A. Verbrugh, and J. L. Nouwen. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5751-762. [DOI] [PubMed] [Google Scholar]
- 34.Wertheim, H. F., M. C. Vos, H. A. Boelens, A. Voss, C. M. Vandenbroucke-Grauls, M. H. Meester, J. A. Kluytmans, P. H. van Keulen, and H. A. Verbrugh. 2004. Low prevalence of methicillin-resistant Staphylococcus aureus (MRSA) at hospital admission in The Netherlands: the value of search and destroy and restrictive antibiotic use. J. Hosp. Infect. 56321-325. [DOI] [PubMed] [Google Scholar]