Abstract
We have developed a rapid real-time PCR method using fluorescence resonance energy transfer probes and the LightCycler (Roche Diagnostics), which will detect the presence of the tcdC gene of Clostridium difficile in stool samples. Our PCR method also will identify the presence of base pair deletions, one of which (18 bp) has been associated with the “epidemic” toxin-hyperproducing strains. We compared the results of this PCR with those of three C. difficile toxin-detecting enzyme immunoassays (EIAs), an EIA for the detection of glutamate dehydrogenase (GDH), and culture of C. difficile. A total of 200 stool specimens were studied by the methods under comparison. C. difficile was isolated from 49 specimens by culture, and 44 of these were confirmed as containing one of the genes associated with toxin production (“toxigenic culture”). Using toxigenic culture as the “gold standard”, the sensitivities, specificities, and positive and negative predictive values, respectively, of the assays were 48%, 98%, 88%, and 87% for the Premier toxin A and B test; 48%, 99%, 91%, and 87% for the ImmunoCard toxin A & B test; 48%, 84%, 46%, and 85% for the Xpect C. difficile toxin A/B test; 32%, 100%, 100%, and 84% for the Triage C. difficile panel (for toxin A); and 86%, 97%, 90%, and 96% for the LightCycler PCR. Thus, in comparison to the sensitivity of toxigenic culture, the sensitivities of the toxin immunoassays were unacceptably low, while the LightCycler real-time PCR assay for the detection of the tcdC gene of C. difficile is sensitive and specific.
Clostridium difficile is the most commonly identified cause of antibiotic-associated diarrhea, accounting for 15% to 25% of cases (3). There have been marked increases in the incidence and severity of C. difficile infection (CDI) in recent years (2). In 2001 there was a 42% increase in short-stay hospital discharge diagnoses of CDI in the United States (13), with a further increase of 25% in 2004 and another 10% in 2005 (4). The C. difficile-related mortality rates per million in the population of the United States rose from 5.7 in 1999 to 23.7 in 2004, and there were an estimated 26,642 deaths due to CDI from 1999 to 2004 (20). Severe CDI outbreaks occurred in Pittsburgh, Pennsylvania, in 2000 and Quebec, Canada, in 2003, resulting in an attributable mortality rate increase to 6.9% (11, 16). Many cases of this severe form of CDI have been shown to be caused by an “epidemic” strain of C. difficile which has been characterized as “BI” by restriction enzyme analysis (REA), “NAP1” (North American pulsed field type 1) by pulsed-field gel electrophoresis, “027” by PCR ribotyping (6), and “toxinotype III” by REA of toxin genes. This BI/NAP1/027 “epidemic” strain has been shown to be a hyperproducer of toxins A and B, which may be the major reason for its increased virulence (26).
The tcdC gene within the pathogenicity locus of C. difficile encodes the putative negative regulator of toxin A and B production (21). Base pair deletions (especially 18 bp) in the tcdC gene of the “epidemic” strain were initially thought to be responsible for the toxin hyperproduction (12, 21). It has recently been reported that a single-nucleotide mutation at position 117, causing a frameshift that introduces a stop codon resulting in the truncation of the tcdC gene product, is the more likely mechanism (8). However, we conclude that the detection of the base pair deletions can serve as a sensitive (98%) and specific (90%) associated marker of the altered tcdC structure, since 18-bp or 39-bp deletions were present in 98% of isolates with tcdC genotypes predicted to have highly mutated tcdC proteins. Thus, the base pair deletions are probably reliable surrogate markers of the presence of the “epidemic” strain. Resistance to fluoroquinolone antimicrobials and the presence of a binary toxin are also characteristic of this strain (9, 10, 12, 14, 22).
The cytotoxicity of stool samples in a variety of tissue cell lines (CTX) has been viewed by some as the “gold standard” for the laboratory diagnosis of CDI (15). The anaerobic culture of stools on selective and differential media is more sensitive than CTX but is time consuming and requires confirmation of the toxigenicity of isolates by another method, such as CTX (17) or the molecular detection of toxin-regulating genes (“toxigenic culture”). The direct detection of the antigens of toxin A or B or both by a variety of commercially produced enzyme immunoassays (EIAs) is rapid and simple and is the method most widely used now by clinical laboratories. Several studies reported ≥90% sensitivity of EIAs in comparison to the results for CTX (1, 15). However, since the latter is only 70% to 80% as sensitive as culture (the “real gold standard”), the true sensitivity of the EIAs is much lower than 90% (19) and has been reported to be as low as 70% (23, 24). An additional assay detects the “common antigen” of C. difficile (glutamate dehydrogenase [GDH]) and is viewed by some as a surrogate for culture methods (27). This assay does not differentiate toxigenic and nontoxigenic strains. Thus, at present, there is no single simple, rapid, adequately sensitive method for the diagnosis of CDI available to clinical laboratories.
Rapid real-time PCR methods used directly on stool specimens have been described for the diagnosis of CDI (5, 18, 25). These assays targeted the genes associated with toxin B or both toxin A and B (tcdA and tcdB) and showed a high concordance with the results from either CTX or toxigenic culture (18, 25). We have developed a rapid real-time PCR assay using the LightCycler (LC; Roche Diagnostics), which will detect the presence of the tcdC gene (and, indirectly, the presence of toxin A and/or B) and will also identify the presence of base pair deletions within the tcdC gene. One of these deletions (18 bp) has been associated with the “epidemic” strain. We compared the results of this PCR with those of three C. difficile toxin-detecting EIAs, an EIA for the detection of GDH, and culture of C. difficile.
MATERIALS AND METHODS
LC PCR assay.
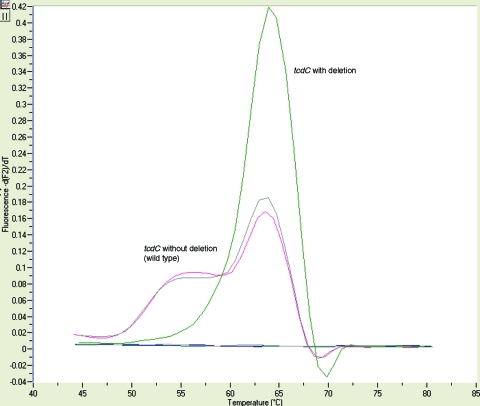
The LC PCR assay detects the presence of the tcdC gene, as well as the 18-bp, 39-bp, and other deletions found in the tcdC gene of C. difficile. Fifteen microliters of the “hot start” reaction mixture containing 1× LC FastStart DNA master hybridization probes (Taq DNA polymerase, reaction buffer, deoxynucleoside triphosphate mix with dUTP instead of dTTP, and 10 mM MgCl2), 3 mM MgCl2, and 1× LC CDD primer-probe set (CD1, CD2, CDD3, and CDD4, kit no. 296; TIB MolBiol LLC, Adelphia, NJ) was added to the LC cuvette (Table 1). Five microliters of the extracted DNA was added, and the reaction mixture was placed into the LC. The cycling parameters were denaturing of template at 95°C for 10 min; amplification of template, with 45 cycles of 10 s at 95°C, 10 s at 55°C (single acquisition), and 15 s at 72°C; and product detection by melt analysis for 0 s at 95°C, 20 s at 59°C, 20 s at 45°C (ramp, 0.2°C/s), and 0 s at 85°C (ramp, 0.2°C/s continuous acquisition). The differentiation of the toxin-containing strain (wild type) and the toxin strain with a deletion was accomplished by melt curve analysis (Fig. 1). Positive and negative controls were included with each run. The positive controls were C. difficile ATCC 9689 (wild type) and a CDC isolate with the 18-bp deletion. The negative control was Escherichia coli ATCC 25922.
TABLE 1.
Primer-probe sequences used in experiments for the detection of the tcdC, tcdA, and tcdB genes of C. difficile
| Gene | Name | Sequence (5′ to 3′) |
|---|---|---|
| Genes | ||
| tcdB | TB1 | GAGCTGCTTCAATTGGAGAGA |
| tcdB | TB2 | GTAACCTACTTTCATAACACCAG |
| tcdA | TA1 | ATGATAAGGCAACTTCAGTGG |
| tcdA | TA2 | TAAGTTCCTCCTGCTCCATCAA |
| Primer-probesa | ||
| tcdC | CD1 | ACC TCA TCA CCA TCT TCA ATA AC |
| tcdC | CD2 | TCA AAA TGA AAG ACG ACG AAA |
| tcdC | CDD3 | TTC AGC CTT TTT AGC TTC TTC AGC-FL |
| tcdC | CDD4 | LC Red640-TTA CGT TGA TTT TCA GCT TCA ATA GC-PH |
Kit no. 296; TIB MolBiol.
FIG. 1.
Melt curve analysis of the results of the LC PCR assay. The presence of the base pair deletions in the tcdC gene generates a curve at 64°C ± 2°C (green line), while the tcdC gene with no base pair deletions present (wild type) shows a curve with a double hump with a melt temperature of the larger peak of 63°C ± 2°C (red and gray lines representing patient and control results, respectively). The y axis represents the negative derivative of sample fluorescence/temperature.
Stool processing for PCR.
A swab was inserted into the stool sample at various locations and swirled into a tube containing 1 ml of sterile water (stool dilution of approximately 1:10), and the suspension allowed to settle. Two hundred microliters of the supernatant was placed into a sample cartridge for DNA extraction with a total nucleic acid isolation kit on a MagNA Pure system (Roche Diagnostics).
Verification of the LC PCR results.
A total of 50 strains of Clostridium were tested for the presence of the tcdC gene. Eleven were ATCC C. difficile strains, 7 were C. difficile clinical isolates, and 26 were clinical isolates of other Clostridium species (Table 2). Two C. difficile isolates provided by the CDC were also included; one isolate was toxinotype III with the 18-bp deletion, and the other isolate was a toxinotype V isolate with a 39-bp deletion. An additional four isolates, two representative BI strains and two nontoxigenic strains, were provided by Dale Gerding (Hines VA hospital, Hines, IL, and Loyola University Medical Center, Maywood, IL). Specimens submitted to the clinical microbiology laboratory at The Mayo Clinic that were positive for C. difficile toxin by an antigen detection assay (Premier toxins A and B; Meridian Bioscience Inc.) were also used to evaluate the performance of the LC PCR assay. We also performed conventional PCR on these antigen-positive stool samples, amplifying the whole (∼700 bp) tcdC gene by using primers which have been previously described (21). All tcdC gene gel-positive specimens were sequenced on an ABI 3730 DNA sequencer (Applied Biosystems, Foster City, CA) and analyzed by using Sequencher 4.2 software (Gene Codes Corp., Ann Arbor, MI). The sequence alignment was performed with Bioedit Sequence Alignment Editor software (Ibis Biosciences, Carlsbad, CA). Inhibition by the LC PCR assay was determined by spiking 92 C. difficile-negative DNA extracts with 100 targets/μl of a C. difficile wild-type plasmid. No inhibited samples were found in these extracted samples.
TABLE 2.
Clostridium species tested by the C. difficile tcdC PCR assay for which negative results were obtained
| Organism | Sourcea |
|---|---|
| Clostridium argentinense | Patient |
| Clostridium aminovalericum | Patient |
| Clostridium bifermentans | Proficiency (NYS) |
| Clostridium cadaveris | Patient |
| Clostridium clostriiforme | Proficiency |
| Clostridium cocleatum | Patient |
| Clostridium frigidicarnis | Patient |
| Clostridium glycolicum | Patient |
| Clostridium hathewayi | Patient |
| Clostridium histolyticum | ATCC 19401 |
| Clostridium hylemonae | Patient |
| Clostridium innocuum | Patient |
| Clostridium mangenotii | Patient |
| Clostridium novyii | Patient |
| Clostridium orbiscindens | Patient |
| Clostridium paraputrificum | Patient |
| Clostridium perfringens | ATCC 13124 |
| Clostridium ramosum | QC organism |
| Clostridium sordelli | ATCC 9714 |
| Clostridium sordelli | Proficiency (NYS) |
| Clostridium sordelli | Unknown |
| Clostridium sphenoides | Patient |
| Clostridium sporogenes | ATCC 11437 |
| Clostridium symbiosum | Patient |
| Clostridium tertium | Patient |
| Clostridium xylanolyticum | Patient |
Proficiency (NYS), proficiency testing organism provided by the New York State Department of Health.
Conventional PCR.
Amplification of the whole (∼700 bp) tcdC gene by conventional PCR was performed on 260 antigen-positive stool samples as has been previously described (21). The cycling parameters were denaturation for 5 min at 94°C and amplification consisting of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C for 30 cycles. The presence of an amplified product was confirmed by agarose gel detection. All tcdC gene gel-positive specimens (122/260) were sequenced and analyzed as described above.
Confirmation of melt temperature pattern.
Sequencing of the whole tcdC gene was performed on 61 antigen-positive and LC PCR-positive isolates to determine if the toxin melt temperature pattern generated by the C. difficile isolates was correct. The probe placement in the LC PCR assay allows for the generation of two distinct melt temperature patterns. Amplification, sequencing, and analysis of the entire tcdC gene for all gel-positive specimens were carried out as described above. The sample sequences included the C. difficile isolates from the 200 stool specimens included in this study along with an additional 17 samples. Twenty-two tcdC gene-positive specimens with a sequencing-documented deletion had a melt temperature of 64°C ± 2°C (mean ± standard deviation), and 39 tcdC gene-positive specimens without a deletion showed a double hump with a melt temperature of the larger peak at 63°C ± 2°C (Fig. 1).
Clinical specimens.
Two hundred stool specimens submitted to the clinical microbiology laboratory at The Mayo Clinic for the detection of C. difficile toxin were used in the comparison of test methods. The specimen criteria for use in this study were soft or liquid stools, one specimen per patient, and fresh or frozen stools less than 48 h old. Ten stools per day fitting the above criteria were tested by the methods being compared. The Premier toxin A and B test (Meridian Bioscience, Inc., Cincinnati, OH) was the routine test performed in the laboratory, and since these results were reported to the clinician, it was performed first. All methods were performed within 24 h of the routine test.
EIA, ImmunoCard, and rapid-screening assays.
The Premier toxin A and B EIA (Meridian Bioscience, Inc., Cincinnati, OH), the ImmunoCard toxin A & B test (Meridian Bioscience, Inc., Cincinnati, OH), the Xpect C. difficile toxin A/B test (Remel, Inc., Lenexa, KS), and the Triage C. difficile panel test (Biosite Diagnostics, San Diego, CA) were all performed on the 200 comparison test specimens according to the manufacturers' instructions.
Culture for C. difficile.
Anaerobic culture was performed on the 200 test specimen stools by plating the specimens onto prereduced fastidious anaerobic agar with sheep blood (FAASB) and taurocholate (1 mg/ml), cycloserine (0.5 mg/ml), cefoxitin (16 μg/ml), and fructose agar (TCCFA). The TCCFA plate was inoculated with fresh stool. The FAASB plate was inoculated with 10 μl of stool sample after alcohol shock (equal volumes of 95% ethanol to stool). After incubation in an anaerobic glove box (85% N, 10% H, 5% CO2) for 2 days at 35°C, presumptive C. difficile colonies were identified by the growth of irregular yellow colonies on TCCFA, characteristic odor and colonial morphology on nonselective medium with appropriate Gram stain characteristics, and 16S sequencing (MicroSeq; Applied Biosystems).
Confirmation of the presence of a toxin gene(s).
All C. difficile culture-positive specimens were tested for the presence of the toxin A (tcdA), toxin B (tcdB), and tcdC genes by both LC PCR and conventional PCR. The sequences of the primers used for amplification are found in Table 1, and the cycling parameters were the same as those listed above for the amplification of the entire tcdC gene. There was total agreement between the results of the two PCR methods, thus verifying the use of the LC PCR tcdC gene assay as a surrogate for the detection of the presence of toxin A/B genes in culture isolates.
RESULTS
C. difficile was isolated from 49 of the 200 specimens by culture, and 44 of these isolates were confirmed as containing the toxin genes (“toxigenic culture”) by using the LC PCR assay and another PCR assay detecting the toxin A and B genes. Using toxigenic culture as the “gold standard” for comparison, the sensitivities, specificities, and positive and negative predictive values, respectively, of the assays are listed in Table 3. The sensitivities of the toxin immunoassays were low (32% to 48%), although the specificities were high (84% to 100%). The LC PCR tcdC gene assay was both sensitive and specific in comparison to toxigenic culture, and the predictive values were both ≥90%. The Triage GDH assay detected 76% (37 of 49) of all culture-positive C. difficile isolates but only 32% (14 ToxA and GDH positive) of the 44 culture-positive isolates which had toxin genes detected. In our study, then, this GDH assay was not a sensitive alternative to culture for C. difficile, nor was it an accurate method for toxin detection.
TABLE 3.
Comparison of Clostridium difficile toxin immunoassay and real-time PCR results with those of toxigenic culture
| Assay | Result | Comparison to toxigenic culture resultsa
|
|||||
|---|---|---|---|---|---|---|---|
| No. of specimens
|
Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |||
| Positive | Negative | ||||||
| Premier toxins A and B | Positive | 21 | 3 | 48 | 98 | 88 | 87 |
| Negative | 23 | 153 | |||||
| Xpect C. difficile A/B | Positive | 21 | 25 | 48 | 84 | 46 | 85 |
| Negative | 23 | 131 | |||||
| Immunocard toxins A & B | Positive | 21 | 2 | 48 | 99 | 91 | 87 |
| Negative | 23 | 154 | |||||
| Triage C. difficile panel (toxin A) | Positive | 14 | 0 | 32 | 100 | 100 | 84 |
| Negative | 30 | 156 | |||||
| LC real-time PCR | Positive | 38 | 4 | 86 | 97 | 90 | 96 |
| Negative | 6 | 152 | |||||
Anaerobic culture was performed on the 200 test stools by plating the specimens onto prereduced fastidious anaerobic agar with sheep blood (FAASB) and taurocholate, cycloserine, cefoxitin, fructose agar (TCCFA). All C. difficile culture-positive specimens were tested for the presence of toxin production (as evidenced by detection of the tcdA, tcdB, or tcdC genes) by LC PCR and conventional PCR. PPV, positive predictive value; NPV, negative predictive value.
Base pair deletions of 18 bp or 39 bp in the tcdC gene (confirmed by sequence analysis) were detected in 12 of the 13 LC PCR deletion-positive specimens. One sample with the deletion produced a poor sequence, so the type of deletion could not be determined. Nine samples had the 18-bp deletion, while three had the 39-bp deletion. The sequences generated from the isolates with the 18-bp deletion were similar to that of an isolate identified as a BI strain by REA in the laboratory of Dale N. Gerding (Hines VA Hospital, Hines, IL, and Loyola University Medical Center, Maywood, IL; personal communication) and also correlated with the sequence of a C. difficile isolate provided by the CDC which was toxinotype III. The sequences generated from the isolates with the 39-bp deletion were similar to the sequence of another CDC isolate which was toxinotype V.
Forty-nine specimens were positive for C. difficile by culture, and 44 of those had a toxin gene(s) present as determined by our LC PCR assay for tcdC. To confirm that our PCR was accurately detecting the presence of the toxin gene(s) in these culture isolates, we tested all 49 of these specimens with a second PCR assay specific for toxin A (tcdA) and toxin B (tcdB), as described above. All 44 of the LC PCR tcdC gene-positive culture isolates were positive for the tcdA and tcdB genes (36 both toxin A and B, 5 toxin A only, and 3 toxin B only). Five culture isolates which were LC PCR negative for tcdC were also negative for the tcdA and tcdB genes. These results indicate that PCR detection of the tcdC putative negative regulator gene is equivalent to detection of the tcdA and tcdB genes for the demonstration of the presence of toxins A and/or B.
DISCUSSION
In a study of 200 stool samples, our PCR for the detection of the tcdC gene was 86% sensitive and 97% specific for the diagnosis of CDI, using toxigenic culture as the “gold standard.” Four specimens were PCR positive but were not detected by culture and had very low concentrations of gene target as evidenced by detection by melting curve analysis only, without crossing points above the baseline curve on the graph plot. Presumably, then, the very sensitive molecular assay was able to detect low concentrations of target nucleic acid when there were too few organisms present to be detected by culture. It is more difficult to understand why six culture-positive specimens were negative by PCR. Variations in specimen sampling or the efficiency of nucleic acid extraction may have played a role. Additionally, the PCR identified 13 specimens in which base pair deletions in the tcdC gene were detected; 9 of these were the 18-bp deletions which have been found in the BI or “epidemic” strain of C. difficile.
In comparison, the EIAs detecting toxins A and/or B had sensitivities of only 32% to 48%. The specificities of all the assays were ≥90% (except for the Remel Xpect at 84%). Interestingly, the Triage GDH (“common antigen”) assay was only 76% as sensitive as culture, calling into question the value of this assay as a surrogate marker for the presence of C. difficile. Of course, other versions of the “common antigen” (GDH) assay might have superior results. Similarly, other versions of EIAs which were not included in this study might have had higher sensitivities. Some studies using CTX as the “gold standard” have reported EIA sensitivities considerably higher than those we found (1, 15). However, toxigenic culture is the preferred “gold standard” because CTX is a subjective assay that is dependent upon the skill and experience of individual technologists and has been shown to be significantly less sensitive than toxigenic culture (19). Moreover, the results of a recent study showed that two toxin EIAs had sensitivities of 84% and 97% compared to the sensitivity of CTX but had a concordance of only 54% to 55% with toxigenic culture (25). In the same study, a tcdB PCR assay had a concordance of 71.4% with toxigenic culture, again demonstrating the superiority of the molecular method over immunoassays. Another recent study which reported results only from patients with significant diarrhea and a clinical diagnosis of CDI found that an EIA and CTX had similarly low sensitivities (73.3% and 76.7%) compared to the sensitivity of toxigenic culture; their toxin B PCR sensitivity was 93.3%. (18). These authors emphasized the importance of considering the number of diarrheal stools and the number of assay-positive stools in comparative studies. Although we did not utilize this type of information in our study, the similarity of our results to theirs suggests that we were dealing with similar specimens. The lower sensitivities of toxin immunoassays compared to the sensitivity of toxigenic culture in our and other studies strongly suggest that their use is not adequate for the accurate laboratory diagnosis of CDI. This is of concern because EIAs are the toxin assay method used by most clinical laboratories, accounting for 93% of the assays used in 2003 (13). Efforts to improve this situation have resulted in proposals for two-stage testing consisting of initial screening with a “common antigen” GDH test (considered equivalent to culture) and subsequent CTX of positive specimens for the confirmation of toxin production (23). This approach does not seem adequate since it is complex and time consuming and neither of these methods is sufficiently sensitive for its intended purpose.
The tcdC gene has been recognized as a putative negative regulator of tcdA and tcdB and thereby indicative of the presence of toxins A and B (21). We analyzed all of our culture-positive specimens which were also tcdC gene PCR positive by using a second PCR method which specifically detects the tcdA and tcdB genes. There was a 100% correlation between the results of the assays, indicating that the detection of the tcdC gene is a reliable indicator of the presence of the tcdA and tcdB genes. The presence of these genes was used as part of our definition of “toxigenic culture,” i.e., that the isolates recovered by culture were also toxin positive. An interesting finding was that the second PCR method identified five isolates with only the tcdA gene present, which presumably produced only toxin A. Such toxin A-positive/toxin B-negative isolates of C. difficile have rarely been recognized, and their significance here is undetermined. We can only state that characteristic gel bands were found in the conventional PCR method used.
A unique advantage of our PCR method is the ability to identify base pair deletions in the same assay that detects the tcdC gene. By virtue of melting curve analysis, the assay is able to distinguish isolates whose tcdC gene sequence contains the deletions. Some of these are 18-bp deletions which have been associated with the hyperproduction of toxins characteristic of the BI or “epidemic” strain of C. difficile. The accuracy of our tcdC PCR in detecting the 18-bp or 39-bp deletions was tested by sequencing the DNA amplified from specimens exhibiting the deletion-containing PCR melting curves. In all 13 instances, the deletions were identified within the nucleic acid sequences. It is interesting that three of these were 39-bp deletions, which have not previously been associated with human “epidemic” strains of C. difficile. This illustrates the fact that the detection of a “deletion”-type melting curve by our assay does not indicate what kind of deletion is present. Since it does not specifically identify the 18-bp deletion, the presence of a BI or “epidemic” strain cannot be assumed, although it may be suspected, especially if there is a consistent severe clinical picture.
Further experience should help to clarify and expand upon the observation that not all C. difficile isolates exhibiting the tcdC base pair deletions have a more severe clinical picture than is usually seen with “nonepidemic” strains causing CDI (21). In addition, no 18-bp deletion was detected in at least one “epidemic” strain (85% related to NAP1 by pulsed-field gel electrophoresis) from an outbreak of CDI in Ohio (7). The utility of detecting base pair deletions by PCR will ultimately be determined by studies correlating this finding with the clinical severity of CDI, and such studies are in progress. In addition, ongoing research should expand our understanding of the complex interrelationships between the gene structures, strain types, and toxinogenicity of C. difficile and the epidemiology and clinical severity of CDI.
Footnotes
Published ahead of print on 23 April 2008.
REFERENCES
- 1.Aldeen, W. E., M. Bingham, A. Aiderzada, J. Kucera, S. Jense, and K. C. Carroll. 2000. Comparison of the TOX A/B test to a cell culture cytotoxicity assay for the detection of Clostridium difficile in stools. Diagn. Microbiol. Infect. Dis. 36211-213. [DOI] [PubMed] [Google Scholar]
- 2.Archibald, L. K., S. N. Banerjee, and W. R. Jarvis. 2004. Secular trends in hospital-acquired Clostridium difficile disease in the United States. J. Infect. Dis. 1891585-1588. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett, J. G. 1994. Clostridium difficile: history of its role as an enteric pathogen and the current state of knowledge about the organism. Clin. Infect. Dis. 18(Suppl. 4)S265-S272. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett, J. G. 2002. Antibiotic associated diarrhea. N. Engl. J. Med. 346334-339. [DOI] [PubMed] [Google Scholar]
- 5.Belanger, S. D., M. Boissinot, N. Clairoux, F. J. Picard, and M. G. Bergeron. 2003. Rapid detection of Clostridium difficile in feces by real-time PCR. J. Clin. Microbiol. 41730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blossom, D. B., and L. C. McDonald. 2007. The challenges posed by reemerging Clostridium difficile infection. Clin. Infect. Dis. 45222-227. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2005. Severe Clostridium difficile-associated disease in populations previously at low risk: four states, 2005. MMWR Morb. Mortal. Wkly. Rep. 541201-1205. [PubMed] [Google Scholar]
- 8.Curry, S. R., J. W. Marsh, C. A. Muto, M. M. O'Leary, A. W. Pasculle, and L. H. Harrison. 2007. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J. Clin. Microbiol. 45215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaynes, R., D. Rimland, E. Killum, H. K. Lowery, T. M. Johnson, I. I. G. Kilgore, and F. C. Tenover. 2004. Outbreak of Clostridium difficile infection in a long-term care facility: association with gatifloxacin use. Clin. Infect. Dis. 38640-645. [DOI] [PubMed] [Google Scholar]
- 10.Geric, B., R. J. Carman, M. Rupnick, C. W. Genheimer, S. P. Sambol, D. M. Lyerly, D. N. Gerding, and S. Johnson. 2006. Binary toxin-producing, large clostridial toxin-negative Clostridium difficile strains are enterotoxic but do not cause disease in hamsters. J. Infect. Dis. 1931143-1150. [DOI] [PubMed] [Google Scholar]
- 11.Loo, V. G., L. Poirier, M. A. Miller, et al. 2005. A predominately clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 3532442-2449. [DOI] [PubMed] [Google Scholar]
- 12.McDonald, L. C., G. E. Killgore, A. Thompson, R. C. Owens, Jr., S. V. Kazakova, S. P. Sambol, S. Johnson, and D. N. Gerding. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 3532433-2441. [DOI] [PubMed] [Google Scholar]
- 13.McDonald, L. C., M. Owings, and D. B. Jernigan. 2006. Clostridium difficile infection in patients discharged from US short-stay hospitals, 1996-2003. Emerg. Infect. Dis. 3409-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEllison, M. C., R. J. Carman, D. N. Gerding, C. W. Genheimer, and L. Zheng. 2005. A hospital outbreak of Clostridium difficile disease associated with isolates carrying binary toxin genes. Clin. Infect. Dis. 40265-272. [DOI] [PubMed] [Google Scholar]
- 15.Musher, D. M., A. Manhas, P. Jain, F. Nuila, A. Waqar, N. Logan, B. Marino, and E. A. Gravis. 2007. Detection of Clostridium difficile toxin: comparison of enzyme immunoassay results with results obtained by cytotoxicity assay. J. Clin. Microbiol. 452737-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muto, C. A., M. Pokrywka, K. Shutt, A. B. Mendelsohn, K. Nouri, K. Posey, T. Roberts, K. Croyle, S. Krystofiak, S. Patel-Brown, A. W. Pasculle, D. L. Paterson, M. Saul, and L. H. Harrison. 2005. A large outbreak of Clostridium difficile-associated disease with an unexpected proportion of deaths and colectomies at a teaching hospital following increased fluoroquinolone use. Infect. Control Hosp. Epidemiol. 26273-280. [DOI] [PubMed] [Google Scholar]
- 17.Peterson, L. R., and P. J. Kelly. 1993. The role of the clinical microbiology laboratory in the management of Clostridium difficile-associated diarrhea. Infect. Dis. Clin. North Am. 7277-293. [PubMed] [Google Scholar]
- 18.Peterson, L. R., R. U. Manson, S. M. Paule, D. M. Hacek, A. Robicsek, R. B. Thomson, Jr., and K. L. Kaul. 2007. Detection of toxigenic Clostridium difficile in stool samples by real-time polymerase chain reaction for the diagnosis of C. difficile-associated diarrhea. Clin. Infect. Dis. 451152-1160. [DOI] [PubMed] [Google Scholar]
- 19.Peterson, L. R., M. M. Olson, C. J. Shanholtzer, and D. N. Gerding. 1988. Results of a prospective 18-month clinical evaluation of culture, cytotoxin testing and culturette brand (CTD) latex testing in the diagnosis of Clostridium difficile-associated diarrhea. Diagn. Microbiol. Infect. Dis. 1085-91. [DOI] [PubMed] [Google Scholar]
- 20.Redelings, M. D., F. Sorvillo, and L. Mascola. 2007. Increase in Clostridium difficile-related mortality rates, United States, 1999-2004. Emerg. Infect. Dis. 91417-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spigaglia, P., and P. Mastrantonio. 2002. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J. Clin. Microbiol. 403470-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terhes, G., E. Urban, J. Soki, K. A. Hamid, and E. Nagy. 2004. Community-acquired Clostridium difficile diarrhea caused by binary toxin, toxin A, and toxin B gene-positive isolates in Hungary. J. Clin. Microbiol. 424316-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ticehurst, J. R., D. Z. Aird, L. M. Dam, A. P. Borek, J. T. Hargrove, and K. C. Carroll. 2006. Effective detection of toxigenic Clostridium difficile by a two-step algorithm including tests for antigen and cytotoxin. J. Clin. Microbiol. 441145-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turgeon, D. K., T. J. Novicki, J. Quick, L. Carlson, P. Miller, B. Ulness, A. Cent, R. Ashley, A. Larson, M. Coyle, A. P. Limaye, B. T. Cookson, and T. R. Fritsche. 2003. Six rapid tests for direct detection of Clostridium difficile and its toxins in fecal samples compared with the fibroblast cytotoxicity assay. J. Clin. Microbiol. 41667-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Berg, R. J., N. Vaessen, H. P. Endtz, T. Schulin, E. R. van der Vorm, and E. J. Kuijper. 2007. Evaluation of real-time PCR and conventional diagnostic methods for the detection of Clostridium difficile-associated diarrhea in a prospective multicentre study. J. Med. Microbiol. 5636-42. [DOI] [PubMed] [Google Scholar]
- 26.Warny, M., J. Pepin, A. Fang, G. Killgore, A. Thompson, J. Brazier, E. Frost, and L. C. McDonald. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 3661079-1084. [DOI] [PubMed] [Google Scholar]
- 27.Wilkins, T. D., and D. M. Lyerly. 2003. Clostridium difficile testing: after 20 years, still challenging. J. Clin. Microbiol. 41531-534. [DOI] [PMC free article] [PubMed] [Google Scholar]