Abstract
We describe a Klebsiella oxytoca infection outbreak in a renal transplant unit that involved seven patients. All strains belonged to a single pulsed-field gel electrophoresis pattern and were resistant to amoxicillin-clavulanate, cefuroxime, piperacillin-tazobactam, and aztreonam but susceptible to ceftriaxone, ceftazidime, cefepime, and imipenem. Chromosomal β-lactamase hyperproduction was caused by a point mutation in the blaOXY-2 gene promoter region.
Klebsiella oxytoca is an opportunistic pathogen responsible for causing health care-associated infections (14, 15, 17). These species possess chromosomal genes encoding β-lactamases that are constitutively expressed at low levels and that confer resistance to amino- and carboxypenicillins but not to other β-lactams (4). K. oxytoca β-lactamases were initially divided into the two main groups OXY-1 and OXY-2, which possessed distinct β-lactam hydrolytic profiles (11, 12). Recently, other OXY-type β-lactamases (OXY-3 to OXY-6) have been reported among K. oxytoca isolates (6). Distinct point mutations in the −35 and −10 promoter regions of these β-lactamase genes have been pointed out as a reason for OXY hyperproduction in 10 to 20% of K. oxytoca isolates and led to a broader spectrum of β-lactam resistance (10, 13).
Susceptibility to bacterial infection in renal transplantation recipients is related directly to the level and duration of the pharmacological immunosuppression. Bacterial urinary tract infections are frequently associated with early onset chronic rejection and may lead to reduced transplantation survival (8).
K. oxytoca infection outbreaks have been documented in multiple settings (4, 15, 18, 19, 21). However, K. oxytoca infection outbreaks in transplantation units have not yet been reported. The aim of this study was to evaluate the antimicrobial susceptibility profiles, the genetic relatedness, and the mechanisms of β-lactam resistance among clinical isolates of K. oxytoca that caused health care-associated infections in a renal transplantation unit of a teaching hospital located in Buenos Aires, Argentina.
Seven K. oxytoca strains were isolated from the urine, peritoneal fluid, and central venous catheters of renal and renal-pancreas transplantation patients hospitalized at the transplantation unit of the University Hospital, CEMIC, between March and August of 2005. According to an epidemiological investigation, the index case was a renal transplantation patient who developed a urinary tract infection caused by this strain during the hospitalization period. The outbreak of infection involved a total of seven patients (one isolate per patient). Horizontal transmission was suspected because at that time, the transplantation unit was located in a shared facility without individual rooms, and all patients were attended by a common group of health care workers.
The isolates were associated with infection and were identified by conventional methods (5). Antimicrobial susceptibility testing was performed by using the Clinical and Laboratory Standards Institute (CLSI) broth microdilution method and interpreted according to CLSI breakpoints (CLSI, 2006). Quality control was performed by testing Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853. All quality control results were within published MIC ranges (CLSI, 2006) (2, 3). Pulsed-field gel electrophoresis (PFGE) was performed using the restriction endonuclease SpeI as previously described (16). Analysis of PFGE patterns was performed by visual inspection of photographs of ethidium bromide-stained gels. The isolates were classified according to the criteria described by Tenover et al. (20).
Detection of the blaOXY group genes and promoter regions was carried out by PCR, followed by DNA sequencing. PCR was performed under standard conditions, using the primers OXY-F (5′-GATTTCACAAAGCGCTCGGC-3′) and OXY-R (5′-CCTGCTGCGGCTGGGTAAAA-3′), designed based on the nucleotide sequences of the blaOXY-1 and blaOXY-2 genes available at GenBank, under accession numbers Z30177 and Z49084, respectively. PCR products were analyzed by electrophoresis in 1.0% agarose gels and were sequenced on both strands by using an ABI Prism 377 sequencer unit. The nucleotide sequences and deduced amino acid sequences were analyzed by using Lasergene software (DNAStar, Madison, WI). The sequences obtained were compared to sequences available at http://www.ebi.ac.uk/fasta33/.
The outer membrane proteins of the isolates were studied according to the method described by Filip et al. (7). Wild-type K. oxytoca strains susceptible to penicillins and broad-spectrum cephalosporins were included as control strains.
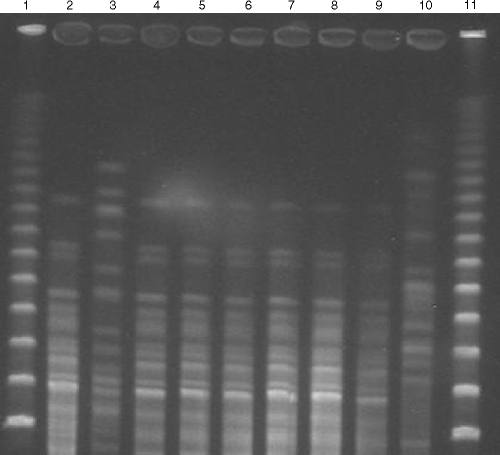
The antimicrobial susceptibility profiles of the K. oxytoca strains studied are presented in Table 1. The seven K. oxytoca strains were resistant to piperacillin, piperacillin-tazobactam, amoxicillin-clavulanic acid, cephalothin, cefuroxime, and aztreonam but were susceptible to ceftriaxone, ceftazidime, cefepime, and imipenem. A single isolate was resistant to cefoxitin (MIC, 32 μg/ml). No plasmids were found by phenotypic and molecular analyses. Seven K. oxytoca strains showed a unique PFGE pattern (pattern A), as shown in Fig. 1. The presence of the blaOXY-2 gene was detected in all strains. The blaOXY-2 promoter sequence region of clone A was P (−35[TTGTCA]; −10[GATAAT]), which differed by one base (underlined) from the weak promoter, P (−35[TTGTCA]; −10[GATAGT]), present in the K. oxytoca wild type carrying blaOXY-2. The outer membrane profiles were identical among the K. oxytoca isolates studied, except for the cefoxitin-resistant strain, which exhibited reduced expression of the 36-kDa outer membrane protein on a sodium dodecyl sulfate-polyacrylamide gel.
TABLE 1.
Antimicrobial susceptibility profiles of wild-type K. oxytoca and K. oxytoca OXY-2 hyperproducer strains evaluated in this study
| Isolate | MIC (μg/ml) of the indicated drugs to K. oxytoca isolatesa
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK | GEN | AMP | SAM | AMC | FEP | CRO | CAZ | CXM | CEF | CIP | MEZ | TZP | PIP | TIM | CTT | SXT | ATM | IPM | MER | |
| 7 | 16 | >8 | >16 | >16/8 | >16/8 | 1 | 1 | 2 | >16 | >16 | 2 | >64 | >64/2 | >64 | >64/2 | 4 | >2/38 | >256 | 0.25 | 0.25 |
| 6 | 8 | 4 | >16 | >16/8 | >16/8 | 1 | 2 | 2 | >16 | >16 | 2 | >64 | >64/4 | >64 | >64/2 | 8 | >2/38 | >256 | 0.5 | 0.25 |
| 5 | 16 | >8 | >16 | >16/8 | >16/8 | 2 | 8 | 2 | >16 | >16 | 8 | >64 | >64/4 | >64 | >64/4 | 32 | >2/38 | >256 | 0.25 | 0.25 |
| 4 | 16 | 1 | >16 | >16/8 | >16/8 | 1 | 2 | 1 | >16 | >16 | 2 | >64 | >64/4 | >64 | >64/4 | 4 | >2/38 | >256 | 0.5 | 0.25 |
| 3 | 2 | 4 | >16 | >16/8 | >16/8 | 1 | 2 | 1 | >16 | >16 | 2 | >64 | >64/4 | >64 | >64/4 | 4 | >2/38 | >256 | 0.5 | 0.25 |
| 2 | 16 | 4 | >16 | >16/8 | >16/8 | 1 | 1 | 2 | >16 | >16 | 2 | >64 | >64/4 | >64 | >64/4 | 4 | >2/38 | >256 | 0.5 | 0.25 |
| 1 | 8 | 2 | >16 | >16/8 | >16/8 | 1 | 1 | 2 | >16 | >16 | 2 | >64 | >64/4 | >64 | >64/4 | 4 | >2/38 | >256 | 0.5 | 0.25 |
MIC values were determined by broth microdilution technique (3). AMK, amikacin; AMC, amoxicillin-clavulanic acid; SAM, ampicillin-sulbactam; AMP, ampicillin; FEP, cefepime; CRO, ceftriaxone; CAZ, ceftazidime; CXM, cefuroxime; CEF, cephalothin; CIP, ciprofloxacin; GEN, gentamicin; IPM, imipenem; MER, meropenem; ATM, aztreonam; MEZ, mezlocillin; TZP, piperacillin-tazobactam; PIP, piperacillin; TIM, ticarcillin-clavulanic acid; CTT, cefoxitin; SXT, trimethoprim-sulfamethoxazole.
FIG. 1.
PFGE patterns of SpeI-digested chromosomal DNA restriction fragments from K. oxytoca isolates resolved in 1% SeaKem Gold agarose. Lanes 1 and 11, molecular weight standards of lambda ladder. Lane 2, strain 1 (PFGE pattern A); lane 3, strain X (quality control strain); lane 4, strain 2 (PFGE pattern A); lane 5, strain 3 (PFGE pattern A); lane 6, strain 4 (PFGE pattern A); lane 7, strain 5 (PFGE pattern A); lane 8, strain 6 (PFGE pattern A); lane 9, strain 7 (PFGE pattern A); and lane 10, strain U (quality control strain).
We describe an outbreak of K. oxytoca infection among transplantation patients (hospitalized in a single institution), caused by an isolate that overproduces OXY-2 due to a mutation in the blaOXY-2 promoter region. OXY β-lactamases are chromosomally encoded and are usually synthesized at low levels, conferring resistance to amino- and carboxypenicillins. Overproduction of such enzymes results from a mutation in the β-lactamase gene promoter, enhancing the hydrolytic substrate profile and conferring resistance to penicillins and some extended-spectrum β-lactams, especially aztreonam (9). In contrast, these strains are susceptible to ceftriaxone, ceftazidime, and cefepime, as observed with our study. This feature helps to distinguish OXY-type overproducers from K. oxytoca isolates that harbor plasmid-encoded extended-spectrum β-lactamases (4, 6). In spite of being encoded by chromosomal DNA, this group of enzymes has been grouped with the extended-spectrum β-lactamases in the 2be class of the classification by Bush et al., due to the substrate profiles and inhibition patterns found by clavulanic acid (1).
OXY-2-producing K. oxytoca strains are more commonly (53 to 74%) isolated from clinical specimens and are usually more resistant than OXY-1 producers. The substitution of guanine with adenine in the −10 blaOXY-2 promoter region, as observed with this study, has already been described to enhance OXY-2 expression about 13-fold in K. oxytoca strain SL911 (9, 13). This is the probable reason for the β-lactam susceptibility profile displayed by clone A.
The outbreak of infection was controlled by an enhancement of standard biosafety precautions (i.e., hand-washing practice) through an educational program and contact isolation procedures. All the outbreak control procedures were supervised directly by the infection control department of the institution.
In summary, a unique clone of K. oxytoca was responsible for causing infections in a renal transplantation unit, suggesting that the patients might have acquired this clone from a common source. These isolates displayed enhanced resistance to β-lactams, including cefuroxime and aztreonam, which was attributed to a single base change in the −10 consensus sequence of the promoter. In addition, it is possible that cefoxitin resistance in one of these strains was caused by the diminished expression of the 36-kDa outer membrane protein. Epidemiologic surveillance of transplantation units is of major importance to the prevention and early detection of outbreaks caused by multidrug-resistant bacteria.
Acknowledgments
We thank the personnel of ALERTA, LEMC, and CEMIC for their contributions during the performance of this study and especially Jussimara Monteiro for performing the PFGE technique.
A.C.G. is partially funded by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (process no. 307714/2006-3).
Footnotes
Published ahead of print on 16 April 2008.
REFERENCES
- 1.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 391211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial disk susceptibility tests, 9th ed. Approved standard M2-A9. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed. Approved standard M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 4.Decre, D., B. Burghoffer, V. Gautier, J. C. Petit, and G. Arlet. 2004. Outbreak of multi-resistant Klebsiella oxytoca involving strains with extended-spectrum beta-lactamases and strains with extended-spectrum activity of the chromosomal beta-lactamase. J. Antimicrob. Chemother. 54881-888. [DOI] [PubMed] [Google Scholar]
- 5.Farmer, J., III. 2003. Enterobacteriaceae: introduction and identification, p. 636-653. In P. R. Murray, E. J. Baron, M. A. Pfaller, J. H. Jorgensen, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 6.Fevre, C., M. Jbel, V. Passet, F. X. Weill, P. A. Grimont, and S. Brisse. 2005. Six groups of the OXY β-lactamase evolved over millions of years in Klebsiella oxytoca. Antimicrob. Agents Chemother. 493453-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filip, C., G. Fletcher, J. L. Wulf, and C. F. Earhart. 1973. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J. Bacteriol. 115717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fishman, J. A. 2007. Infection in renal transplant recipients. Semin. Nephrol. 27445-461. [DOI] [PubMed] [Google Scholar]
- 9.Fournier, B., P. H. Lagrange, and A. Philippon. 1996. β-Lactamase gene promoters of 71 clinical strains of Klebsiella oxytoca. Antimicrob. Agents Chemother. 40460-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fournier, B., A. Gravel, D. C. Hooper, and P. Roy. 1999. Strength and regulation of the different promoters for chromosomal β-lactamase of Klebsiella oxytoca. Antimicrob. Agents Chemother. 43850-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fournier, B., P. H. Roy, P. H. Lagrange, and A. Philippon. 1996. Chromosomal β-lactamase genes of Klebsiella oxytoca are divided into two main groups: blaOXY-1 and blaOXY-2. Antimicrob. Agents Chemother. 40454-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fournier, B., and P. H. Roy. 1997. Variability of chromosomally encoded β-lactamases from Klebsiella oxytoca. Antimicrob. Agents Chemother. 411641-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fournier, B., C. Y. Liu, P. H. Lagrange, R. Krishnamoorthy, and A. Philippon. 1995. Point mutation in the Pribnow box, the molecular basis of β-lactamase overproduction in Klebsiella oxytoca. Antimicrob. Agents Chemother. 391365-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogenauer, C., C. Langner, E. Beubler, I. T. Lippe, R. Schicho, G. Gorkiewicz, R. Krause, N. Gerstgrasser, G. J. Krejs, and T. A. Hinterleitner. 2006. Klebsiella oxytoca as a causative organism of antibiotic-associated hemorrhagic colitis. N. Engl. J. Med. 3552418-2426. [DOI] [PubMed] [Google Scholar]
- 15.Jeong, S. H., W. M. Kim, C. L. Chang, J. M. Kim, K. Lee, Y. Chong, H. Y. Hwang, Y. W. Baek, H. K. Chung, I. G. Woo, and J. Y. Ku. 2001. Neonatal intensive care unit outbreak caused by a strain of Klebsiella oxytoca resistant to aztreonam due to overproduction of chromosomal b-lactamase. J. Hosp. Infect. 48281-288. [DOI] [PubMed] [Google Scholar]
- 16.Maslow, J. N., M. E. Mulligan, and R. D. Arbeit. 1993. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin. Infect. Dis. 17153-162. [DOI] [PubMed] [Google Scholar]
- 17.Podschun, R., and U. Ullmann. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11589-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reiss, I., A. Borkhardt, R. Füssle, A. Sziegoleit, and L. Gortner. 2000. Disinfectant contaminated with Klebsiella oxytoca as a source of sepsis in babies. Lancet 356310. [DOI] [PubMed] [Google Scholar]
- 19.Sardan, Y. C., P. Zarakolu, A. Altun, A.Yildirim, G. Yildirim, G. Hascelik, and O. Uzun. 2004. A cluster of nosocomial Klebsiella oxytoca bloodstream infections in a university hospital. Infect. Control Hosp. Epidemiol. 25878-882. [DOI] [PubMed] [Google Scholar]
- 20.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson, J. T., R. C. Jones, A. M. Siston, J. R. Fernandez, K. Martin, E. Beck, S. Sokalski, B. J. Jensen, M. J. Arduino, A. Srinivasan, and S. I. Gerber. 2005. Outbreak of catheter-associated Klebsiella oxytoca and Enterobacter cloacae bloodstream infections in an oncology chemotherapy center. Arch. Intern. Med. 1652639-2643. [DOI] [PubMed] [Google Scholar]