Abstract
In the present study, a novel broad-range real-time PCR was developed for the rapid detection of human pathogenic fungi. The assay targets a part of the 28S large-subunit ribosomal RNA (rDNA) gene. We investigated its application for the most important human pathogenic fungal genera, including Aspergillus, Candida, Cryptococcus, Mucor, Penicillium, Pichia, Microsporum, Trichophyton, and Scopulariopsis. Species were identified in PCR-positive reactions by direct DNA sequencing. A noncompetitive internal control was applied to prevent false-negative results due to PCR inhibition. The minimum detection limit for the PCR was determined to be one 28S rDNA copy per PCR, and the 95% detection limit was calculated to 15 copies per PCR. To assess the clinical applicability of the PCR method, intensive-care patients with artificial respiration and patients with infective endocarditis were investigated. For this purpose, 76 tracheal secretion samples and 70 heart valve tissues were analyzed in parallel by real-time PCR and cultivation. No discrepancies in results were observed between PCR analysis and cultivation methods. Furthermore, the application of the PCR method was investigated for other clinical specimens, including cervical swabs, nail and horny skin scrapings, and serum, blood, and urine samples. The combination of a broad-range real-time PCR and direct sequencing facilitates rapid screening for fungal infection in various clinical specimens.
Invasive fungal infections caused mainly by Candida and Aspergillus spp. have assumed an increasing importance over the last two decades, with a high mortality and morbidity among hospitalized and immunocompromised patients (36). Candida is a commensal of the human mucosa and has remained the fourth most important cause of nosocomial bloodstream infections. Over 200 species of Candida have been described, but 95% of all Candida infections are caused by five species: C. glabrata, C. parapsilosis, C. tropicalis, C. krusei, and C. albicans. C. albicans is the most important cause of candidemia worldwide (1, 6, 13, 33). Invasive aspergillosis is caused mainly by Aspergillus fumigatus and A. flavus; other species of Aspergillus associated with infection are A. terreus, A. niger, and A. nidulans (36). Fungal infections have also been considered in many superficial dermatological mycoses (22, 37) causing restricted infections of the corneocyte of skin, hair, and nails (27). Other important fungal pathogens that cause uncommon human diseases are Cryptococcus spp., Penicillium spp., Pichia spp., Fusarium spp., and Mucor spp. (19, 35, 36). The crucial factor in diagnosis and effective therapy is the identification of the etiologic agent. The standard method for the detection of fungal infections is mycological culture and direct microscopy of clinical specimen. Nevertheless, microscopy often lacks a satisfactory sensitivity, whereas diagnosis by mycological culture often require a long growth period. The application of serological tests for cryptococcus, aspergillus, and histoplasma antigens also lacks sufficient sensitivity (26). Furthermore, widely accepted antigen tests for Candida or dermatophytes are not available. In this context, rapid diagnosis of mycological infection by in vitro amplification and detection of fungal DNA is a common method used in clinical laboratories. A wide range of methods have been described, including restriction fragment length polymorphism (34), PCR amplification (18, 21), hybridization with species-specific probes, amplicon size differences (5, 14), and methods to identify unique DNA sequences (24, 28). Real-time PCR assays play an important role among molecular genetic screening methods because of the accelerated diagnostic outcome. A variety of real-time PCR assays for the detection of isolated fungal species have been described (3, 12, 16, 17, 30). Nevertheless, a broad-range real-time PCR assay targeting nearly all clinically relevant fungal species in one assay is not available. Furthermore, none of these assays include quality control patterns to identify false-negative results. The presence of amplification inhibitors, and thus the occurrence of false-negative results, is a common problem in PCR diagnostics (15).
The aim of this study was the development of a broad-range 28S rDNA real-time PCR assay for the rapid detection of fungal pathogens in various clinical specimens. The specific assay allows the simultaneous detection and discrimination of medically important fungal pathogens.
MATERIALS AND METHODS
Fungal and bacterial strains.
Fungal and bacterial strains were obtained from the American Type Culture Collection (ATCC) (LGC Promochem GmbH, Wesel, Germany) or the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ) (Braunschweig, Heidelberg) or were previously isolated from patients' specimens at our hospital. All clinical isolates used in this study were characterized in our microbiological laboratory by standard methods. Fungal and bacterial isolates were cultivated using standard cultivation methods.
Clinical samples.
Tracheal secretion (TS) samples (n = 76) from patients with artificial respiration and heart valve tissues (n = 70) from endocarditis patients were used for the evaluation of the sensitivity of the PCR assay in comparison to mycological cultivation. TS samples were examined for fungal infections by cultivation on Sabouraud agar (SGC 2; bioMérieux, Nürtingen, Germany) for 5 days at 30°C. Heart valve tissues were cultivated in tryptone soy broth and brain heart infusion bouillon (bioMérieux) for 21 days at 37°C. All isolates were identified using traditional morphology and the ID 32C and VITEK 2 ID-YST biochemical identification methods (bioMérieux). The remaining material was stored at −20°C until used for DNA extraction. Nail and horny skin scraping samples (n = 56), as well as cervical swabs (n = 98) from patients suspected of having fungal infections, were analyzed for the presence of fungal infections only by real-time PCR. Urine, EDTA-anticoagulated blood, and serum samples for spiking experiments were obtained from healthy volunteers.
Sample pretreatment and DNA extraction.
Samples were pretreated, depending on the clinical specimen. Total DNA was extracted from TS samples, heart valve tissues, cell suspensions, and nail and horny skin scraping samples by use of a QIAamp DNA blood kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. DNA was eluted from the QIAamp column with 50 μl sterile distilled water.
Cell suspensions of bacteria and fungi were prepared with 200 μl sterile distilled water. Bacteria and Candida spp. were lysed at 95°C for 10 min. Cell walls of Aspergillus spp., Mucor spp., and dermatophytes were disrupted mechanically in a RiboLyser (Hybaid, Teddington, England) using lysing matrix E (QBiogene, Heidelberg, Germany).
Viscous TS samples were diluted with an equal volume of sputasol (Oxoid, Poole, England) and liquefied at 37°C for 1 h. Thereafter, 400 μl of the sample was transferred to a tube containing lysing matrix E and mechanically disrupted in a RiboLyser. Subsequently, 200 μl of the sample was incubated with 200 μl buffer AL at 56°C for 10 min. Twenty milligrams of valvular tissue was treated with 180 μl buffer ATL and 20 μl proteinase K (30 U; Sigma-Aldrich, Taufkirchen, Germany) at 56°C until the tissue was completely lysed, followed by incubation with 200 μl buffer AL at 70°C for 10 min prior to DNA extraction. Skin and nail scraping samples were suspended in 200 μl sterile distilled water and transferred into a tube with lysing matrix E. After mechanical disruption, 180 μl of buffer ATL and 20 μl of proteinase K were added. Samples were incubated at 56°C overnight, followed by incubation with 200 μl of buffer AL at 70°C for 10 min.
Cervical swabs were rinsed in lysis buffer (Digene Hybrid Capture 2 CT/GC DNA kit; Digene, Dreieich, Germany). Total DNA was extracted from 200 μl cervical swab lysis solution, 200 μl EDTA-anticoagulated blood, 400 μl serum, and 400 μl urine using the NucliSens easyMAG (bioMérieux) automated DNA extraction system. DNA was eluted in 55 μl easyMAG elution buffer.
Analytical sensitivity and specificity.
Total nucleic acids from 38 different fungal strains were adjusted to a final concentration of 7 to 10 ng/nl and were analyzed with the 28S rDNA real-time PCR assay. The analytical sensitivity and the precision of the assay were determined using serially diluted copies of the 28S rDNA of Aspergillus fumigatus amplified with the NL1/NL4 (21) primer system as template for real-time PCR. The 95% detection limit was calculated by probit analysis using the SPSS 14.0 software (SPSS GmbH Software, version 14.0; SPSS, München, Germany). For the evaluation of the DNA extraction protocols and the determination of matrix effects of the individual specimen, 400-μl urine samples, 400-μl TS samples, 200-μl EDTA-coagulated blood samples, and 400-μl serum samples from healthy volunteers were spiked with serial dilutions of C. albicans of between 100 and 106 CFU/ml.
Primer design for amplification of fungal 28S rDNA.
The sequences of the probe and the PCR primers were selected from the sequences of fungal 28S (large-subunit) rRNA genes. DNA sequences of the following human pathogenic fungi were amplified with primers NL1 and NL4 targeting the D1 and D2 domain as described previously (21) (GenBank Accession numbers are in parentheses): Aspergillus flavus (EU071389), Aspergillus fumigatus (EU071392), Aspergillus niger (EU071393), Candida albicans (EU071395), Candida dubliniensis (EU071397), Candida krusei (EU071398), Candida parapsilosis (EU071400), Candida tropicalis (EU071401), Microsporum canis (EU071402), Mucor flavus (EU071390), Trichophyton rubrum (EU071403), Cryptococcus neoformans (EU071399), and Aureobasidium pullulans (EU071394). The alignment of the DNA sequences was performed using the DNasis software (Hitachi Software Engineering Co., version 2.06.006.001) with standard algorithms. A BLAST search was performed to check sequence specificity of the primers and probe used. The forward primer for the 28S rDNA was adapted from that described previously (21) (primer NL1), and the reverse primer was 5′-TTAGCTTTAGATGRARTTTACCACC-3′ (primer 260R). The TaqMan probe (5′-CGGCGAGTGAAGCGGSAARAGCTC-3′) was synthesized with the fluorescent reporter 6-carboxyfluorescein coupled to the 5′-end and a dark quencher coupled to the 3′ end (BHQ1). Sequences of the MS2 primers and probe for the internal control (IC) have been described previously (8); the TaqMan probe of the IC was labeled with Cy5 at the 5′ end and a dark quencher (BHQ2) at the 3′ end. The amplified fragments were 270 bp in length for the fungal 28S rDNA and 203 bp for the IC.
Real-time PCR.
DNA amplification was carried out in 0.2-ml tubes containing 40 μl reaction mix and 10 μl DNA extract. The reaction mix consisted of 1× Eurogentec Tth buffer, 6 mM MgCl2, 400 nM of each deoxynucleoside triphosphate, 200 nM of each 28S rDNA primer and probe, 200 nM of each IC primer, 100 nM of IC fluorescent probe, and 2.5 U of Eurogentec Tth DNA polymerase (Eurogentec Deutschland, Köln, Germany). PCR was performed on the Rotor-Gene 3000 system (Corbett Life Sciences, Sydney, Australia) with preliminary denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 35 s, and extension at 72°C for 20 s, with a single fluorescence acquisition step at the end of the annealing step. A 203-bp PCR product of the bacteriophage MS2 replicase gene was added to the reaction mixture as an exogenous IC sequence. To ensure that the IC sequence did not limit the amplification sensitivity of the 28S rDNA assay, low fungal DNA concentrations close to the assay's detection limit were tested with different concentrations of the IC. The detection of low target concentrations with and without IC coamplification was evaluated, confirming that the IC had no influence on the assay sensitivity (data not shown). DNA sequencing of real-time PCR products was performed as described previously (8).
RESULTS
Specificity, detection limit, and precision testing of the real-time PCR assay.
To prove the suitability of the generic probe and primer set for the detection of a broad range of human pathogenic fungi, total nucleic acids of 38 different fungal species, listed in Table 1, were extracted and assayed for 28S rDNA by real-time PCR. The specificity of the assay was evaluated with DNA extracts from 29 different bacterial strains and DNA extracts from 10 different human clinical specimens (data not shown). Cross-reactivity of the primers and probes was not detected, and the negative controls yielded no false-positive results. In order to evaluate the analytical sensitivity and the precision of the assay, serially diluted copies of the 28S rDNA of Aspergillus fumigatus were used as templates. The 95% detection limit was calculated as 15 copies per PCR (range, 10.8 to 25.9), whereas the lowest concentration detectable with this real-time PCR assay was one copy per PCR (Table 2). The reproducibility of the assay was demonstrated by analyzing the intra- and interassay variation for the crossing threshold (CT) values. The intra-assay variability was calculated from eight replicates, with CTs of 32.29 ± 0.41 for the 28S rDNA and 26.35 ± 0.48 for the IC. The interassay variability was determined from five independent PCR runs with four replicates per run. The CTs for the 28S rDNA and the IC were calculated as 32.55 ± 0.45 and 26.90 ± 0.57, respectively. Intra- and interassay variations, as well as standard deviations, are low for both targets.
TABLE 1.
Amplification of whole genomic fungal DNA (7 to 10 ng/μl) with the 28S rDNA real-time PCR assay using primers NL1 and 260R
| Fungal speciesa | Strain | CTb | Other species with identical sequence homologyc |
|---|---|---|---|
| Aspergillus flavus | RV2 | 22.72 | Aspergillus oryzae, Aspergillus caelatus, Aspergillus tamarii, Aspergillus parasiticus |
| Aspergillus fumigatus | 5226 | 22.69 | Neosartorya quadricincta, Neosartorya spinosa, Aspergillus phialisepticus, Neosartorya stramenia |
| Aspergillus niger | RV3 | 24.49 | Aspergillus carbonarius |
| Aspergillus niger | 1005 | 25.67 | Aspergillus carbonarius |
| Aspergillus penicilliodes | MA-1 | 23.57 | None |
| Aspergillus restrictus | MA-2 | 20.80 | Aspergillus caesiellus |
| Aspergillus versicolor | 237 | 24.17 | Davidiella tassiana, Aspergillus ustus, Aspergillus puniceus, Aspergillus pseudoeflectus, Aspergillus granulosus |
| Candida albicans | ATCC 10231 | 25.10 | Candida africana |
| Candida africana | RV1 | 25.69 | Candida albicans |
| Candida ciferrii | RV1 | 23.46 | None |
| Candida colliculosa | RV1 | 23.72 | None |
| Candida dubliniensis | RV4 | 22.85 | None |
| Candida glabrata | 5740 | 25.46 | None |
| Candida kefyr | RV1 | 25.90 | Candida sphaerica |
| Candida krusei | RV1 | 25.81 | None |
| Candida lipolytica | HAK-1 | 21.10 | None |
| Candida lusitaniae | 13898 | 21.73 | None |
| Candida parapsilosis | II30 | 25.81 | None |
| Candida intermedia | BCK-2417298 | 19.97 | None |
| Candida tropicalis | RV-1 | 22.74 | None |
| Candida zeylanoides | RV-1 | 22.83 | None |
| Dipodascus capitatus | RV-4 | 21.45 | None |
| Cryptococcus neoformans | 471 | 24.25 | None |
| Cryptococcus laurentii | RV-2 | 23.60 | None |
| Scopulariopsis brevicaulis | TV-1 | 25.88 | None |
| Trichophyton tonsurans | TV-1 | 23.10 | Trichophyton verrucosum, Trichophyton interdigitale |
| Trichophyton mentagrophytes | TV-1 | 21.01 | Trichophyton verrucosum, Trichophyten interdigitale, Trichophyton tonsurans |
| Trichophyton rubrum | TV-1 | 22.99 | None |
| Microsporum canis | TV-1 | 24.57 | Anthroderma otae |
| Mucor hiemalis | DSMZ 2656 | 26.19 | None |
| Mucor flavus | DSMZ 2184 | 26.42 | None |
| Pencillium expansum | 4786 | 24.79 | Miscellaneous Penicillium spp., no differentiation by 28S rDNA sequence |
| Penicillium polonicum | 4974 | 25.98 | Miscellaneous Penicillium spp., no differentiation by 28S rDNA sequence |
| Pichia anomala | VLB-2 | 20.34 | None |
| Pichia galeiformis | RV-1 | 21.26 | None |
| Pichia norvegensis | 129 | 19.92 | None |
| Schizosaccharomyces pombe | BCK-S50 | 20.11 | None |
| Saccharomyces cerevisiae | BCK-2345258 | 21.55 | None |
| Aureobasidium pullulans | 234 | 25.07 | None |
Species were identified by DNA sequence analysis.
The CT (crossing threshold) values describe the point where the amplification curve exceeds the noise band.
Results of sequencing analysis of the PCR products obtained with 28S rDNA real-time PCR.
TABLE 2.
Evaluation of analytical sensitivity with serial dilutions of 28S rDNA of Aspergillus fumigatus
| Copies/μla | No. PCR-positive samples/total | % Positiveb |
|---|---|---|
| 110 | 24/24 | 100 |
| 55 | 24/24 | 100 |
| 28 | 23/24 | 99 |
| 14 | 22/24 | 94 |
| 7 | 19/24 | 76 |
| 4 | 17/24 | 64 |
| 2 | 14/24 | 55 |
| 1 | 10/24 | 50 |
Serial dilution of 28S rDNA copies of Aspergillus fumigatus amplified with the NL1/NL4 primer system (21).
Probability of a positive PCR signal calculated by probit analysis.
Sensitivity of the real-time PCR assay.
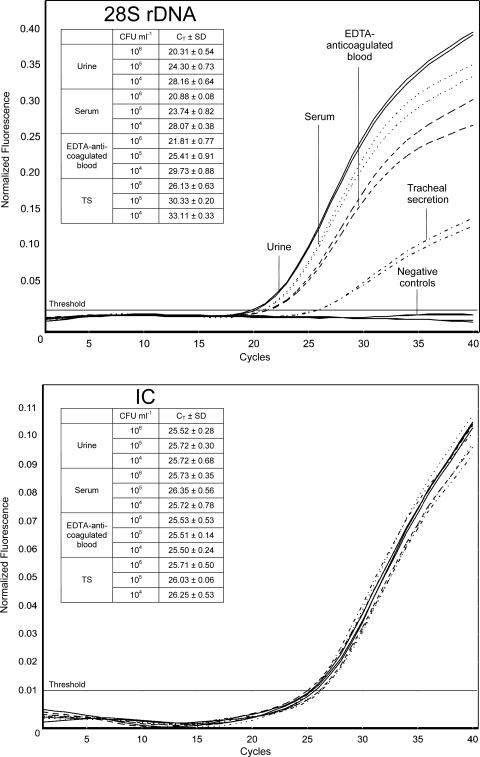
The sensitivity of a PCR test is also highly influenced by the DNA extraction protocol and the specimen itself. Nucleic acids were extracted from urine, serum, EDTA-anticoagulated blood, and TS samples inoculated with 100 to 106 CFU/ml of C. albicans (ATCC 10231) and analyzed in triplicate by 28S rDNA real-time PCR. The lowest concentrations obtaining a positive PCR result after DNA extraction were 10 CFU/400 μl of urine or serum, 50 CFU/400 μl TS sample, and 10 CFU/200 μl of EDTA-anticoagulated blood. Furthermore, the mean CT values of urine, serum, EDTA-anticoagulated blood, and TS samples inoculated with 104 to 106 CFU/ml C. albicans were compared with regard to matrix effects. The mean CT values are displayed along with examples of amplification plots for specimens inoculated with 106 CFU/ml in Fig. 1. All samples demonstrate decreasing mean CT values for the 28S rDNA, corresponding to the different concentrations of fungal DNA. Urine and serum samples show the lowest mean CT values for all concentrations, with almost equal values. The mean CT values in EDTA-anticoagulated blood were slightly lower, whereas the mean CT values for the TS samples demonstrate a subsequent fluorescence signal with a distance of approximately 5 CT compared to urine and serum samples.
FIG. 1.
Amplification plots for various specimens inoculated with 106 CFU/ml of C. albicans. Nucleic acids were extracted from urine (thin solid lines), serum (dotted lines), EDTA-anticoagulated blood (dashed lines), and TS (dashed-dotted lines) samples inoculated with 104 to 106 CFU/ml of C. albicans (ATCC 10231) as described in Materials and Methods. Nucleic acids were analyzed in triplicate by 28S rDNA real-time PCR. Amplification plots for specimens inoculated with 106 CFU/ml are shown, and the CT for 28S rDNA (upper panel) and the IC (lower panel) are arranged tabularly. Thick solid lines indicate negative controls (noninoculated specimen).
Identification specificity of the 28S rDNA PCR products.
Species identification of real-time PCR products was performed by sequencing analysis as described previously (8). A BLAST search was performed to determine the specificity of species identification. The PCR products of all 38 fungal species investigated had 98 to 100% sequence identity. Seventeen isolates revealed identical sequence homologies to other species (Table 1), which limits characterization to the genus level. For intraspecies differentiation, isolated DNA was amplified with PCR assay targeting the internal transcribed spacer 1 (ITS 1) and parts of the 5.8S rDNA (25).
Detection of fungal species in clinical samples.
To evaluate the assay sensitivity for the detection of fungi in clinical samples, the results of PCR examination and microbiological culture were compared for TS samples and heart valve tissues. TS samples were obtained routinely from intensive care patients with artificial respiration and thus an increased risk to develop an atypical pneumonia. In 20 TS samples (26%), fungi were detected by real-time PCR as well as by microbiological cultivation (Table 3). The remaining 54 TS samples were negative for fungal pathogens by both methods. The comparison of positive real-time PCR and cultivation results showed concordant findings for 18 TS samples. In sample 5, C. albicans and C. parapsilosis were detected by cultivation, whereas sequencing analysis of the real-time PCR products exclusively identified C. parapsilosis. In samples 8 and 12, mycological culture identified Candida spp. without intraspecies differentiation. Sequencing analysis of real-time PCR products identified, with a similar homology, C. albicans/C. africana in sample 8 and C. albicans/C. africana/C. dublienensis in sample 12. Subsequent amplification and sequencing analysis of the ITS region characterized the pathogens in sample 8 and sample 12 as C. albicans. Screening of 70 heart valves obtained from patients with definite infectious endocarditis by real-time PCR and mycological culture obtained one positive sample (sample 26). The remaining 69 samples were negative for fungal pathogens by both methods.
TABLE 3.
Comparison of mycological culture and real-time PCR results for TS samples and heart valve tissues
| Patient | Sex | Age (yr) | Clinical specimen | Culture result | Result of sequencing analysis of real-time PCR products |
|---|---|---|---|---|---|
| 1 | Female | 53 | TS | C. albicans | C. albicans |
| 2 | Female | 87 | TS | C. tropicalis | C. tropicalis |
| 3 | Male | 70 | TS | C. albicans | C. albicans or C. africana |
| 4 | Female | 53 | TS | C. albicans | C. albicans |
| 5 | Female | 65 | TS | C. albicans, C. parapsilosis | C. parapsilosis |
| 6 | Male | 46 | TS | P. aeruginosa,aC. parapsilosis | C. parapsilosis |
| 7 | Female | 53 | TS | C. albicans, Enterococcus spp.a | C. albicans |
| 8 | Male | 79 | TS | Candida spp. | C. albicans or C. africana |
| 9 | Male | 83 | TS | C. glabrata, Enterobacter chloaceaa | C. glabrata |
| 10 | Male | 79 | TS | C. albicans | C. albicans |
| 11 | Male | 79 | TS | C. albicans | C. albicans |
| 12 | Female | 42 | TS | Candida spp. | C. albicans, C. africana, C. dublienensis |
| 13 | Male | 50 | TS | Enterococcus spp.,aC. albicans | C. albicans |
| 14 | Female | 1 | TS | C. albicans | C. albicans |
| 15 | Male | 50 | TS | C. glabrata | C. glabrata |
| 16 | Male | 67 | TS | C. albicans | C. albicans |
| 17 | Male | 70 | TS | C. albicans | C. albicans |
| 18 | Male | 79 | TS | C. albicans, Enterococcus spp.a | C. albicans |
| 19 | Male | 60 | TS | C. albicans | C. albicans |
| 20 | Female | 69 | TS | C. albicans | C. albicans |
| 21 | Male | 74 | Heart valve tissue | C. albicans, Corynebacterium spp.a | C. albicans |
Molecular genetic screening for bacterial DNA was not performed.
Ninety-eight cervical swabs were routinely examined by real-time PCR for the presence of fungal DNA. Fungi were detected in 12 samples (12%), and subsequent sequencing analysis identified C. albicans.
Additionally, 56 nail and horny skin clinical samples derived from patients with possible skin mycoses were assayed by real-time PCR. The presence of fungal DNA was confirmed in 15 samples (27%). Sequencing analysis identified the following pathogens with similar DNA homology: (i) Trichophyton rubrum (60%), (ii) T. verrucosum/T. interdigitale (20%), and (iii) T. tonsurans/T. verrucosum/T. interdigitale (20%). Subsequent sequencing analysis of the ITS region characterized pathogens ii and iii as T. verrucosum.
DISCUSSION
The detection of fungal DNA by PCR has been described as an important tool in the early diagnosis of fungal infection. Mycological cultivation methods often require long growth periods, especially in the diagnosis of dermatological diseases. Real-time PCR amplification offers a rapid diagnostic tool with the advantage of the detection of viable/nonviable cells, as well as circulating free fungal DNA (4). The aim of this present study was the development of a broad-range real-time PCR assay as a rapid screening method for the presence of fungal DNA in various clinical samples. This test is primarily intended to be used as a rapid diagnostic screening tool for infection control in addition to the sample analysis currently performed in the standard microbiology laboratory. In particular, rapid detection of fungi is frequently required for hematological or long-term intensive care patients. Therefore, we investigated the clinical applicability for TS samples due to our cohort consisting of intensive care patients with artificial respiration. In addition, the clinical applicability was investigated for heart valve tissue samples from endocarditis patients, referring to broad-range screening methods for bacterial infection as a routine diagnostic tool (23, 29). Furthermore, the detection of DNA from serum, urine, and EDTA-anticoagulated blood, the most common materials from patients with invasive disease, was evaluated with spiking experiments, due to the absence of a corresponding patient cohort. The detection of fungal DNA from various clinical samples has been shown to be highly sensitive. Moreover, the combination of real-time PCR and direct sequencing of positive PCR products improves the diagnostic turnaround time. The total time required for the detection of fungal pathogens in cervical swabs and TS samples, including DNA extraction, real-time PCR, and sequencing, is less than 9 h. The time-limiting step is the sample pretreatment prior to DNA extraction. In comparison, the earliest time that mycological cultivation results can be obtained is 24 h after sample processing (e.g., for Candida spp. and Penicillium spp.). Slower-growing fungi such as Aspergillus spp. require at least up to 48 h incubation, whereas cultivation of fastidious fungi such as dermatophytes lasts up to a week or longer. In summary, the detection of fungal DNA by real-time PCR is more than 2 to 15 times faster than standard cultivation methods, depending on the pathogen. Although the material and reagent costs for cultivation are lower than those for PCR, the faster pathogen identification by real-time PCR enables an earlier onset of antibiotic treatment and, therefore, reduced hospitalization, including lower subsequent health care costs. So far, a variety of real-time PCR assays based on species-specific TaqMan or hybridization probes for the detection and identification of Candida and Aspergillus species targeting the 18S rDNA (9, 20), the mitochondrial cytochrome b (31), the ITS between 18S and 28S rDNA (16), the CaMP65 (65-kDa mannoprotein) gene (2), the RNase P RNA gene (17), and the ITS 2 region (30) have been described. All assays demonstrated high specificity and sensitivity for the detection and quantification of medically important Candida and Aspergillus species. Nevertheless, a broad-range real-time PCR assay targeting clinical relevant fungal pathogens besides Candida and Aspergillus in one assay is currently not available. In comparison, the application of broad-range PCR assays for the detection and identification of bacteria is a common method used in the clinical laboratory (7, 29). We investigated our assay using the Rotorgene 3000 system (Corbett Life Sciences, Sydney, Australia). However, this assay is easily adaptable to all common real-time platforms available in most clinical laboratories.
Assays based on species-specific probes were designed to identify common fungal pathogens, but rare or infrequent fungi were unaccounted for (17, 30). The numbers of species-specific probes in such assays are terminated by biochemical limitations, and consequently, the number of identifiable pathogens is always limited. Targeting the 28S rDNA offers the possibility of developing a broad-range PCR assay covering a wide range of potential and infrequent fungal pathogens. Our assay is able to detect a broad range of clinical relevant fungi with one set of primers and probe. The subsequent sequence analysis enables an unlimited species identification spectrum. The complete sequences of the 28S rDNA and a large number of partial 28S rDNA sequences for several fungal species have been published in GenBank. Nevertheless, some species share identical sequence homologies within the 28S rRNA gene region. For example, many closely related species of Aspergillus and Penicillium are not able to be discriminated by the 28S rDNA sequence, and sequencing analysis initially ascertains only the genus of the causative pathogen. However, this should be not of major concern for the beginning of an adequate antibiotic treatment, which is primarily based on the genus and the localization of the pathogen. A positive screening further legitimates the beginning of an antibiotic treatment with all known adverse effects. Certainly, the accurate determination of the species is necessary for the derivation of species-specific antibiotic resistance, for example, the fluconazole resistance of many C. glabrata or C. krusei strains or the amphotericin B resistance of some Aspergillus strains. In cases of ambiguous sequencing results, we performed a second PCR using a primer/probe set specific for ITS 1 (25). The ITS 1 and ITS 2 regions offer sufficient intraspecies diversity and were shown to be suitable for the discrimination of medically important fungi (5, 10, 11, 14). The obvious application of a broad-range real-time PCR screening based on the ITS 1 and ITS 2 region is problematic due to sequence variability and the absence of a generic probe sequence. However, with inclusion of the second identification step, which is infrequently essential, molecular genetic identification by real-time PCR and sequencing still show a significantly reduced diagnostic turnaround time, particularly for the diagnosis of slow-growing filamentous fungi.
The detection limit of our real-time PCR assay is one 28S rDNA copy per PCR. The sensitivity of the assay was also determined with regard to divergent clinical matrices. Inhibitory effects on real-time PCR by clinical sample compounds, and consequently reduced sensitivity or false-negative results, are a well-known problem for the clinical reliability of real-time PCR assays. It was shown that divergent matrices had influences on the sensitivity of the assay. Urine and serum samples had at least an equal effect on the CT values, whereas EDTA-anticoagulated blood and TS samples had stronger effects. The delayed CT values of EDTA-anticoagulated blood samples is mainly influenced by the impact of background human DNA, whereas the viscous sample character primarily affects the CT values of TS samples. The application of an IC markedly improves the reliability by dramatically decreasing the risk of false-negative results due to PCR inhibition.
The primer specificity for the 28S rDNA of human pathogen fungi was determined by the exclusion of cross-amplification with 29 different bacterial or human DNAs. The novel PCR assay offers a high specificity and minimizes the risk of false-positive results. Nevertheless, the potential for rare cross-reactivity with organisms not included in our specificity panel could not be ruled out.
To assess the clinical applicability of the PCR assay, TS and heart valve tissue samples were analyzed in parallel by real-time PCR and microbiological cultivation. In the interpretation of positive results, attention should be paid to the importance of distinguishing between colonization and acute disease. In general, PCR methods do not have the potential to afford this distinction comparably to mycological cultivation. A differentiation between colonization and acute disease is feasible only by careful simultaneous consideration of clinical symptoms. A positive PCR result also indicates the presence of nucleic acids without prediction of pathogen viability. Nevertheless, the aim of this study was the evaluation of the sensitivity of our 28S rDNA real-time PCR assay in comparison to mycological cultivation. The assay was developed to facilitate a rapid screening method for the presence of fungal DNA, independently from the patient's disease pattern. Twenty-two TS samples (29%) and one heart valve tissue were positive for fungal pathogens by real-time PCR, with 100% concordance to culture detection. The sensitivity of the PCR assay examined with the limited number of TS and heart valve tissue samples demonstrated a sensitivity level similar to that of microbiological cultivation. Evaluation of the sensitivity of the 28S real-time PCR assay in comparison to mycological cultivation for the screening of dermatological and cervical swab samples is currently under investigation. Furthermore, the 28S rDNA broad-range real-time PCR assay is applicable for molecular sterility testing of cell therapeutic products in combination with broad-range bacterial screening (32). In the future we will investigate the suitability of a multiplex real-time PCR assay combining screening for bacterial, fungal, and mycoplasmal contamination of platelet concentrates and cell culture therapeutic products.
In summary, our novel broad-range PCR assay offers a rapid, highly specific, and cost-effective screening method for fungal pathogens in different clinical specimens with the common advantages of real-time PCR. Moreover, the application of an IC provides a quality control and further improves the assay's diagnostic reliability. Our results demonstrate a sensitivity similar to that of cultivation methods, with a markedly reduced diagnostic turnaround time. Considering the utility of this real-time PCR assay for routine sample processing currently performed in the microbiological laboratory, our method offers the possibility of faster detection and identification of fungi in various clinical samples. Nevertheless, the application of real-time PCR has to be evaluated as a supplementary method to mycological cultivation. Cultivation methods are mandatory to measure the antibiotic resistance and, in particular, the viability of a pathogen. Thus, broad-range molecular genetic methods can be an appropriate addition to mycological cultivation, and the combination of both methods offers a powerful diagnostic tool for the detection and identification of fungal pathogens.
Acknowledgments
We thank Christiane Szliska (Bethesda Hospital, Freudenberg, Germany) for the provision of patients' skin and nail scraping samples, as well as Arndt Gröning, (Labor WagnerStibbe, Hannover, Germany) for the provision of patients' cervical swabs. We also thank Sarah Kirkby for linguistic advice.
Footnotes
Published ahead of print on 2 April 2008.
REFERENCES
- 1.Almirante, B., D. Rodríguez, B. J. Park, M. Cuenca-Estrella, A. M. Planes, M. Almela, J. Mensa, F. Sanchez, J. Ayats, M. Gimenez, P. Saballs, S. K. Fridkin, J. Morgan, J. L. Rodriguez-Tudela, D. W. Warnock, and A. Pahissa. 2005. Epidemiology and predictors of mortality in cases of Candida bloodstream infection: results from population-based surveillance, Barcelona, Spain, from 2002 to 2003. J. Clin. Microbiol. 431829-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arancia, S., A. Carattoli, R. La Valle, A. Cassone, and F. De Bernardis. 2006. Use of 65 kDa mannoprotein gene primers in real time PCR identification of Candida albicans in biological samples. Mol. Cell Probes. 20263-268. [DOI] [PubMed] [Google Scholar]
- 3.Binnicker, M. J., S. P. Buckwalter, J. J. Eisberner, R. A. Stewart, A. E. McCullough, S. L. Wohlfiel, and N. L. Wengenack. 2007. Detection of Coccidioides species in clinical specimens by real-time PCR. J. Clin. Microbiol. 45173-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bretagne, S., and J. M. Costa. 2005. Towards a molecular diagnosis of invasive aspergillosis and disseminated candidosis. FEMS Immunol. Med. Microbiol. 45361-368. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. Lafe, U. Bui, A. P. Limaye, and B. T. Cookson. 2001. Polymorphic internal transcribed spacer region 1 DNA sequences identify medically important yeasts. J. Clin. Microbiol. 394042-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colombo, A. L., M. Nucci, B. J. Park, S. A. Nouér, B. Arthington-Skaggs, D. A. da Matta, D. Warnock, and J. Morgan. 2006. Epidemiology of candidemia in Brazil: a nationwide sentinel surveillance of candidemia in 11 medical centers. J. Clin. Microbiol. 442816-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreier, J., M. Störmer, and K. Kleesiek. 2007. Real-time polymerase chain reaction in transfusion medicine: applications for detection of bacterial contamination in blood products. Transfus. Med. Rev. 21237-254. [DOI] [PubMed] [Google Scholar]
- 8.Dreier, J., M. Störmer, and K. Kleesiek. 2005. Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. J. Clin. Microbiol. 434551-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez-Lopez, A., M. T. Martin-Gomez, P. Martin-Davila, P. Lopez-Onrubia, J. Gavalda, J. Fortun, A. Pahissa, J. L. Rodriguez-Tudela, and M. Cuenca-Estrella. 2006. Detection of fungal DNA by real-time polymerase chain reaction: evaluation of 2 methodologies in experimental pulmonary aspergillosis. Diagn. Microbiol. Infect. Dis. 56387-393. [DOI] [PubMed] [Google Scholar]
- 10.Gräser, Y., M. El Fari, R. Vilgalys, A. F. Kuijpers, G. S. De Hoog, W. Presber, and H. Tietz. 1999. Phylogeny and taxonomy of the family Arthrodermataceae (dermatophytes) using sequence analysis of the ribosomal ITS region. Med. Mycol. 37105-114. [PubMed] [Google Scholar]
- 11.Gräser, Y., A. F. Kuijpers, M. El Fari, W. Presber, and G. S. de Hoog. 2000. Molecular and conventional taxonomy of the Microsporum canis complex. Med. Mycol. 38143-153. [DOI] [PubMed] [Google Scholar]
- 12.Gutzmer, R., S. Mommert, U. Küttler, T. Werfel, and A. Kapp. 2004. Rapid identification and differentiation of fungal DNA in dermatological specimens by LightCycler PCR. J. Med. Microbiol. 531207-1214. [DOI] [PubMed] [Google Scholar]
- 13.Hajjeh, R. A., A. N. Sofair, L. H. Harrison, G. M. Lyon, B. A. Arthington-Skaggs, S. A. Mirza, M. Phelan, J. Morgan, W. Lee-Yang, M. A. Ciblak, L. E. Benjamin, L. T. Sanza, S. Huie, S. F. Yeo, M. E. Brandt, and D. W. Warnock. 2004. Incidence of bloodstream infections due to Candida species and in vitro susceptibilities of isolates collected from 1998 to 2000 in a population-based active surveillance program. J. Clin. Microbiol. 421519-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henry, T., P. C. Iwen, and S. H. Hinrichs. 2000. Identification of Aspergillus species using internal transcribed spacer regions 1 and 2. J. Clin. Microbiol. 381510-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoorfar, J., B. Malorny, A. Abdulmawjood, N. Cook, M. Wagner, and P. Fach. 2004. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 421863-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu, M. C., K. W. Chen, H. J. Lo, Y. C. Chen, M. H. Liao, Y. H. Lin, and S. Y. Li. 2003. Species identification of medically important fungi by use of real-time LightCycler PCR. J. Med. Microbiol. 521071-1076. [DOI] [PubMed] [Google Scholar]
- 17.Innings, A., M. Ullberg, A. Johansson, C. J. Rubin, N. Noreus, M. Isaksson, and B. Herrmann. 2007. Multiplex real-time PCR targeting the RNase P RNA gene for detection and identification of Candida species in blood. J. Clin. Microbiol. 45874-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaeger, E. E., N. M. Carroll, S. Choudhury, A. A. Dunlop, H. M. Towler, M. M. Matheson, P. Adamson, N. Okhravi, and S. Lightman. 2000. Rapid detection and identification of Candida, Aspergillus, and Fusarium species in ocular samples using nested PCR. J. Clin. Microbiol. 382902-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein, A. S., G. T. Tortora, R. Malowitz, and W. H. Greene. 1988. Hansenula anomala: a new fungal pathogen. Two case reports and a review of the literature. Arch. Intern. Med. 1481210-1213. [DOI] [PubMed] [Google Scholar]
- 20.Klingspor, L., and S. Jalal. 2006. Molecular detection and identification of Candida and Aspergillus spp. from clinical samples using real-time PCR. Clin. Microbiol. Infect. 12745-753. [DOI] [PubMed] [Google Scholar]
- 21.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 351216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwong-Chung, K. J., and J. E. Bennet. 1992. Medical mycology. Lea & Febiger, Philadelphia, PA.
- 23.Marin, M., P. Munoz, M. Sanchez, M. del Rosal, L. Alcala, M. Rodriguez-Creixems, and E. Bouza. 2007. Molecular diagnosis of infective endocarditis by real-time broad-range polymerase chain reaction (PCR) and sequencing directly from heart valve tissue. Medicine (Baltimore) 86195-202. [DOI] [PubMed] [Google Scholar]
- 24.Martin, C., D. Roberts, M. van Der Weide, R. Rossau, G. Jannes, T. Smith, and M. Maher. 2000. Development of a PCR-based line probe assay for identification of fungal pathogens. J. Clin. Microbiol. 383735-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCullough, M. J., K. V. Clemons, J. H. McCusker, and D. A. Stevens. 1998. Intergenic transcribed spacer PCR ribotyping for differentiation of Saccharomyces species and interspecific hybrids. J. Clin. Microbiol. 361035-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrison, C. J., and M. D. Lindsley. 2001. Serological approaches to the diagnosis of invasive fungal infections, p. 667-716. In R. Calderone and R. Cihlar (ed.), Fungal pathogenesis: principles and practice. Marcel Dekker, Inc., New York, NY.
- 27.Ninet, B., I. Jan, O. Bontems, B. Léchenne, O. Jousson, R. Panizzon, D. Lew, and M. Monod. 2003. Identification of dermatophyte species by 28S ribosomal DNA sequencing with a commercial kit. J. Clin. Microbiol. 41826-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Playford, E. G., F. Kong, Y. Sun, H. Wang, C. Halliday, and T. C. Sorrell. 2006. Simultaneous detection and identification of Candida, Aspergillus, and Cryptococcus species by reverse line blot hybridization. J. Clin. Microbiol. 44876-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rovery, C., G. Greub, H. Lepidi, J. P. Casalta, G. Habib, F. Collart, and D. Raoult. 2005. PCR detection of bacteria on cardiac valves of patients with treated bacterial endocarditis. J. Clin. Microbiol. 43163-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schabereiter-Gurtner, C., B. Selitsch, M. L. Rotter, A. M. Hirschl, and B. Willinger. 2007. Development of novel real-time PCR assays for detection and differentiation of 11 medically important Aspergillus and Candida species in clinical specimens. J. Clin. Microbiol. 45906-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spiess, B., D. Buchheidt, C. Baust, H. Skladny, W. Seifarth, U. Zeilfelder, C. Leib-Mösch, H. Mörz, and R. Hehlmann. 2003. Development of a LightCycler PCR assay for detection and quantification of Aspergillus fumigatus DNA in clinical samples from neutropenic patients. J. Clin. Microbiol. 411811-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Störmer, M., K. Kleesiek, and J. Dreier. 2007. High-volume extraction of nucleic acids by magnetic bead technology for ultrasensitive detection of bacteria in blood components. Clin. Chem. 53104-110. [DOI] [PubMed] [Google Scholar]
- 33.Takakura, S., N. Fujihara, T. Saito, T. Kudo, Y. Iinuma, and S. Ichiyama. 2004. National surveillance of species distribution in blood isolates of Candida species in Japan and their susceptibility to six antifungal agents including voriconazole and micafungin. J. Antimicrob. Chemother. 53283-289. [DOI] [PubMed] [Google Scholar]
- 34.Tintelnot, K., G. S. De Hoog, E. Antweiler, H. Losert, M. Seibold, M. A. Brandt, A. H. Van Den Ende, and M. C. Fisher. 2007. Taxonomic and diagnostic markers for identification of Coccidioides immitis and Coccidioides posadasii. Med. Mycol. 45385-393. [DOI] [PubMed] [Google Scholar]
- 35.Walsh, T. J., A. Groll, J. Hiemenz, R. Fleming, E. Roilides, and E. Anaissie. 2004. Infections due to emerging and uncommon medically important fungal pathogens. Clin. Microbiol. Infect. 10(Suppl. 1)48-66. [DOI] [PubMed] [Google Scholar]
- 36.Warnock, D. W. 2007. Trends in the epidemiology of invasive fungal infections. Nippon Ishinkin Gakkai Zasshi 481-12. [DOI] [PubMed] [Google Scholar]
- 37.Weitzman, I., and R. C. Summerbell. 1995. The dermatophytes. Clin. Microbiol. Rev. 8240-259. [DOI] [PMC free article] [PubMed] [Google Scholar]