Abstract
Quantitative real-time PCR (qPCR) may improve the detection of fungal pathogens. Extraction of DNA from fungal pathogens is fundamental to optimization of qPCR; however, the loss of fungal DNA during the extraction process is a major limitation to molecular diagnostic tools for pathogenic fungi. We therefore studied representative automated and manual extraction methods for Aspergillus fumigatus and Rhizopus oryzae. Both were analyzed by qPCR for their ability to extract DNA from propagules and germinated hyphal elements (GHE). The limit of detection of A. fumigatus and R. oryzae GHE in bronchoalveolar lavage (BAL) fluid with either extraction method was 1 GHE/ml. Both methods efficiently extracted DNA from A. fumigatus, with a limit of detection of 1 × 102 conidia. Extraction of R. oryzae by the manual method resulted in a limit of detection of 1 × 103 sporangiospores. However, extraction with the automated method resulted in a limit of detection of 1 × 101 sporangiospores. The amount of time to process 24 samples by the automated method was 2.5 h prior to transferring for automation, 1.3 h of automation, and 10 min postautomation, resulting in a total time of 4 h. The total time required for the manual method was 5.25 h. The automated and manual methods were similar in sensitivity for DNA extraction from A. fumigatus conidia and GHE. For R. oryzae, the automated method was more sensitive for DNA extraction of sporangiospores, while the manual method was more sensitive for GHE in BAL fluid.
The detection and identification of medically important filamentous fungi in immunocompromised and diabetic patients may be limited by low sensitivity and time-consuming methods. Invasive pulmonary aspergillosis is an important cause of morbidity and mortality in patients with hematological malignancy and transplantation (1, 3, 7, 14, 23, 24, 28, 29). The most common cause of invasive infection by an Aspergillus species is infection by Aspergillus fumigatus (31, 38). When the organism is inhaled by an immunocompromised host, uninhibited germination of conidia into hyphae may result in pulmonary tissue hemorrhage and infarction (35). However, the diagnostic yield of bronchoalveolar lavage (BAL) fluid for the diagnosis of invasive pulmonary aspergillosis using conventional microbiological methods is relatively low (33, 36).
The number of cases of zygomycosis has increased over the last six decades, making diagnosis of these infections a necessity (20, 32, 34). In the immunocompromised host, rapid progression of pneumonia and dissemination are frequently due to the inhalation of sporangiospores, which contributes to the high mortality rate (76%), underscoring the urgency of making a rapid and accurate diagnosis of pulmonary zygomycosis (16, 17, 27, 32). Even when both culture and histopathologic analysis of BAL fluid are performed, many suspected infections are not confirmed. Roden et al. found that Rhizopus spp. were the most commonly recovered organisms among 218 microbiologically defined infections (32). Given the increase in the number of these infections in recent years, a molecular approach to detection of zygomycete molds may increase sensitivity and rapid diagnosis, resulting in earlier therapy.
Thus, there is a need for the development of more sensitive and more rapid techniques that would aid in the early diagnosis of patients with these life-threatening infections and improve clinical outcomes. Currently, the use of real-time PCR is a standard method accepted for the detection of nucleic acids from many microorganisms in clinical samples. Although widely used for detection of many viruses and mycobacteria, quantitative real-time PCR (qPCR) is not yet similarly accepted in clinical mycology laboratories. A lack of standardized methods for diagnostic PCR of medically important fungi has led to divergent results (4). Nevertheless, the application of diagnostic PCR to BAL fluid in immunocompromised patients with suspected fungal pneumonia appears to be promising (5, 15, 19).
The use of an efficient, rapid, standardized method of DNA extraction from the pathogen is a fundamental component for the optimization and reproducibility of qPCR assays (15, 18). The sensitivity of any PCR assay for the detection of fungal pathogens ultimately depends on efficient lysis of fungal cells from biological samples and purification of DNA that is free of inhibitors (11). Filamentous fungi have complex cell walls consisting of chitin, (1→3)-β-d-glucan, (1→6)-β-glucan, lipids, and peptides that are difficult to disrupt, thus requiring rigorous extraction methods. These methods are time-consuming and therefore reduce the ability for rapid diagnosis. The efficiency of extraction of fungal DNA may vary considerably depending on the method chosen (9, 11, 13, 15, 25). Thus, the extraction method chosen may often represent a compromise between efficiency, lack of exogenous contamination, and the ability to be adapted by routine high-throughput laboratories (15).
The development and availability of automated techniques for DNA extraction and product detection may facilitate fungal DNA detection in clinical diagnostic laboratories (4, 15). Thus, given the importance of developing optimal DNA extraction methods for diagnostic PCR assays, we investigated both automated and manual methods for their ability to extract DNA from germinated hyphal elements (GHE) of A. fumigatus and Rhizopus oryzae in normal rabbit BAL fluid. BAL fluid was selected because it is a common clinical specimen submitted for detection of fungi causing lower respiratory tract infections and in which the organism may exist as GHE and spores. DNA also was extracted from conidia and sporangiospores for quantitation from fungal propagules. The analytical yield and sensitivity of each extraction method were determined by qPCR.
MATERIALS AND METHODS
Organism.
The organisms, A. fumigatus (NCI 4215, ATCC MYA-1163) and R. oryzae (NCI 98), were subcultured from frozen slants (stored at −70°C) onto Sabouraud dextrose agar slants (K-D Medical, Inc., Columbia, MD) and incubated for 24 h at 37°C. The slants were then incubated at room temperature for an additional 5 days before harvesting. Conidia and sporangiospores were harvested under a laminar airflow hood with a solution of 0.025% Tween 20 (Fisher Scientific, Fair Lawn, NJ) in normal saline (K-D Medical, Inc., Columbia, MD), filtered, washed, and counted on a hemacytometer.
DNA extraction of GHE in normal rabbit BAL fluid.
BAL fluid was obtained as previously described (30). A. fumigatus and R. oryzae samples were set up in triplicate using a 24-well flat-bottom plate (Corning, Corning, NY). The following were added to each well: 800 μl normal rabbit BAL fluid, 200 μl yeast nitrogen broth (KD Medical, Columbia, MD), gentamicin (20 μg/ml; Hospira, Inc., Lake Forest, IL), vancomycin (20 μg/ml; Hospira, Inc., Lake Forest, IL), and 100 μl of conidia or sporangiospores (104, 103, 102, 101, or 100). Normal rabbit BAL fluid samples, without any organism, were set up as negative controls using the same growth medium as described above. Samples were incubated at 37°C for 24 h. Germination was confirmed by visual inspection using an inverted microscope. Following incubation, the contents were harvested from individual wells and placed into Fast Prep Lysing Matrix D (LMD) tubes (Q·BIOgene/MP Biomedical, Morgan Irvine, CA) containing no lysing matrix (LM). Each well was subsequently rinsed with 200 μl phosphate-buffered saline (PBS) (Quality Biological, Inc., Gaithersburg, MD), and the well contents were added to corresponding samples for DNA extraction. DNA extraction was performed immediately after harvesting in an AirClean PCR work station (AirClean Systems, Raleigh, NC).
Automated DNA extraction method.
The MagNA Pure LC system can purify DNA from different biological samples by incorporating cell disruption and protein digestion, DNA binding to magnetic glass particles, removal of cellular debris by extensive washing, magnetic separation of the bead-DNA complex, and DNA elution.
BAL fluid samples were centrifuged for 10 min at 16,000 × g, and supernatants were discarded. An aliquot of 150 μl of spheroplast buffer (1.0 M sorbitol [Sigma S-1876, St. Louis, MO], 50.0 mM sodium phosphate monobasic [Sigma S-0751], 0.1% 2-mercaptoethanol [Sigma M-3148], 10 mg/ml lyticase [Sigma L-2524]), 10 μl lysing enzymes (Novozyme [20 mg/ml; Sigma L-1412]), and LM were added to each specimen. Samples were briefly vortexed and incubated at 30°C for 5 min at 1,200 rpm in an Eppendorf thermomixer (Eppendorf, Westbury, NY). Mixing was terminated and sample incubation continued for 25 min. Samples were processed using a Fast Prep instrument (Q·BIOgene/MP Biomedical, Morgan Irvine, CA) at speed 5 for 30 s and placed on ice for 5 min; this process was performed a total of three times. Samples were equilibrated to room temperature and centrifuged for 1 min at 1,000 × g. The samples were then processed with a MagNA Pure LC instrument using a MagNA Pure LC DNA isolation kit III (bacteria, fungi) (Roche Applied Science, Indianapolis, IN) as recommended by the manufacturer. Samples were eluted in 100 μl of kit elution buffer.
Manual DNA extraction method.
The DNeasy Plant minikit is a spin column procedure that incorporates sample lysis, removal of RNA, removal of proteins and polysaccharides, DNA precipitation, and binding to the spin column membrane. Multiple washes are performed to remove contaminants, and DNA is then eluted from the membrane.
BAL fluid samples were centrifuged for 10 min at 16,000 × g, and supernatants were discarded. The samples were gently resuspended in 100 μl spheroplast buffer plus 10 μl of lysing enzymes and incubated at 30°C in an Eppendorf thermomixer for 45 min at 1,200 rpm. After centrifugation for 20 min at 400 × g, the spheroplast-BAL fluid pellets were resuspended in 400 μl AP1 buffer (DNeasy Plant minikit, Qiagen, Valencia, CA). The samples were added to LMD tubes, processed using a Fast Prep instrument at speed 5 for 30 s, and placed on ice for 5 min (25). This process was performed a total of three times. Samples were centrifuged at 16,000 × g for 60 s and then gently vortexed. The specimens (approximately 300 μl) were transferred to new tubes. The beads in the LMD tubes were rinsed with 100 μl AP1 buffer, and this wash was added to each corresponding sample (resulting in a 400-μl final volume). Four microliters of RNase A (100 mg/ml) was added to each sample, vortexed vigorously, and incubated for 10 min at 65°C in an Eppendorf thermomixer at 1,200 rpm. The samples were further processed according to the DNeasy Plant minikit (Qiagen, Valencia, CA) protocol with the following modification: after 200 μl preheated (65°C) AE buffer was applied to the column, the entire apparatus (column and collection tube) was heated at 65°C in the Eppendorf thermomixer for 5 min (10, 26).
DNA extraction from conidia and sporangiospores.
All DNA samples were extracted in an AirClean PCR work station. Genomic DNA was extracted from 10-fold serial dilutions (104, 103, 102, 101, and 100) of A. fumigatus conidia or R. oryzae sporangiospores suspended in PBS. An aliquot of 100 μl of each conidial or spore dilution was placed in LMD tubes without LM and centrifuged for 10 min at 16,000 × g. The supernatant was gently removed from each sample, and samples were further processed by either the automated or manual method as described above.
qPCR assays.
When extracting with the automated method, the final eluate frequently may have residual cellular debris and a slight red color related to the magnetic particles [Roche Applied Science, MagNA Pure LC DNA isolation kit III (bacteria, fungi) user manual]. Therefore, prior to qPCR, all samples extracted by either method were briefly vortexed and then centrifuged for 45 s at 4,500 × g to pellet any particulates that might be present. If samples were not centrifuged prior to qPCR, inhibition was often observed. An aliquot of 5 μl was drawn from the surface of each DNA specimen and added to 15 μl of master mix for qPCR. The master mixes were prepared in a biosafety cabinet located in a room different from where DNA extractions were performed. LightCycler carousel loading was performed in a room separate from where the PCR master mix was prepared.
Aspergillus fumigatus PCR assay.
Both methods were analyzed for efficiency of DNA extraction from A. fumigatus conidia and GHE using the A. fumigatus-specific qPCR assay described previously (10, 26). Briefly, the PCR master mix consisted of 0.5 μM of each of the primers (Cap positive sense, 5′CGAAGACCCCAACATG3′; Cap negative sense, 5′TGAGGGCAGCAATGAC3′), 5 mM MgCl2, 0.025% bovine serum albumin (Sigma), 0.025 U/ml Platinum Taq DNA polymerase (Invitrogen Corp., Carlsbad, CA), 10× PCR buffer (Invitrogen Corp., Carlsbad, CA), 0.2 mM PCR Nucleotide Mix Plus (1 dATP, dCTP, dGTP, and 3 dUTP in proportionate ratios; Roche Applied Science, Indianapolis, IN), and 0.1 μM each of the fluorescein (5′AGTATGCAGTCTGAGTTGATTATCG3′) and LC Red-640 (5′ATCAGTTAAAACTTTCAACAACGGA3′) probes. To prevent potential amplicon carryover, each reaction mixture also contained HK-UNG thermostable uracil N-glycosylase (Epicenter, Madison, WI) as recommended by the manufacturer. Each reaction mixture contained a 5-μl aliquot of extracted specimen, together with 15 μl of the master mix. The LightCycler 2.0 instrument (Roche Applied Science) was used with the following cycling conditions: uracil activation at 37°C for 180 s and uracil heat inactivation at 95°C for 60 s for 1 cycle; amplification cycles of denaturation at 95°C for 0 s (slope, 20°C/s), annealing at 58°C for 3 s (slope, 10°C/s), extension at 72°C for 15 s (slope, 3°C/s), and cool down at 40°C for 120 s. The total number of cycles was 45. Quantitation standards (10-fold serial dilutions of A. fumigatus genomic DNA ranging from 1 × 105 fg to 1 × 103 fg) and a set of negative controls were run in conjunction with each set of samples. The amplicon generated was 253 bp in size. A crossover value of ≤36 cycles was considered positive (10).
Rhizopus species PCR assay.
Primers were chosen which anneal to the 28S rRNA gene sequences within the genera Rhizopus, Mucor, and Rhizomucor but not to those in unrelated fungi, such as Penicillium, Aspergillus, or Candida, that might also be present in clinical samples (S. M. Harrington, M. Kasai, A. Francesconi, R. Petraitiene, V. Petraitis, and T. J. Walsh, presented at the 107th General Meeting of the American Society for Microbiology, Toronto, Canada, 21 to 25 May 2007). Primers were designed using software available through the Whitehead Institute for Biomedical Research, MIT (http://frodo.wi.mit.edu/primer3/input.htm). Primer sequences were as follows: Zygo-F1, 5′TTCAAAGAGTCAGGTTGTTTGG3′, and Zygo-R1, 5′CAGTCTGGCTCCAAACGGTTC3′ (Midland Certified Reagent Co., Midland, TX). Hybridization fluorescence resonance energy transfer probes were chosen using Oligo software (Molecular Biology Insights, Cascade, CO) and synthesized by Operon Biotechnologies, Inc. (Huntsville, AL). Probe sequences for the zygomycete PCR were 5′GGCGAGAAACCGATAGCGAAC-fluorescein isothiocyanate3′ and 5′RD640-GTACCGTGAG-GGAAAGATGAAAAGAACTTTGAAA3′.
Real-time PCR was performed with a LightCycler 2.0 instrument. The PCR master mix consisted of 0.025% bovine serum albumin, 3 mM MgCl2, 0.025 U Platinum Taq DNA polymerase, 0.2× PCR buffer, 0.2 mM PCR Nucleotide Mix Plus, 0.002 U/μl HK-UNG, 0.25 μM of each primer, and 0.1 μM of each RD640- and fluorescein isothiocyanate-labeled probe. To 15 μl of master mix, 5 μl of extracted specimen was added. Uracil was released by incubating at 37°C for 900 s, and then enzyme was inactivated at 95°C for 180 s. Touchdown PCR cycling was performed as follows: 95°C denaturation for 0 s (20°C/s), followed by annealing in 1°C steps between 68°C and 54°C for 5 s (10°C/s), each with a 72°C extension of 15 s (3°C/s) for each cycle. Touchdown cycling was followed by 35 cycles of 95°C for 0 s (20°C/s), 54°C for 5 s (10°C/s), and 72°C for 15 s (3°C/s). A postamplification melt analysis was performed by cooling from 96°C to 40°C for 30 s (20°C/s), followed by a gradual increase in temperature (2°C/s) to 75°C for 0 s (0.2°C/s). Quantitation standards (10-fold serial dilutions of R. oryzae genomic DNA ranging from 1 × 103 fg to 1 × 101 fg) were run in conjunction with each set of samples to assess assay sensitivity and linearity and qPCR results. The amplicon generated was 180 bp in length. A crossover value of ≤22 cycles was considered positive.
Inhibition studies.
Separate PCRs were performed on all samples to test for any inhibitors of PCR. A master mix consisting of the A. fumigatus primers/probes, A. fumigatus genomic DNA, and R. oryzae sample DNA was used as described above to test for inhibitors in the R. oryzae samples. Conversely, the master mix consisting of the R. oryzae primers/probes, R. oryzae genomic DNA, and A. fumigatus sample DNA was used as described above to test for inhibitors in the A. fumigatus samples. Presence of inhibitors was determined by comparing the amplification efficiency of the spiked genomic DNA in the same reaction with the extracted experimental DNA samples against reaction mixtures containing just water. The presence of inhibition would result in a higher crossover value than those of water samples. No inhibition was observed in any of the samples.
Statistical analysis.
Data are expressed as means and standard errors of the means. Sensitivity was assessed using categorical variables in two-by-two tables. Differences in proportions were determined by Fisher's exact test. Yields of DNA from the two methods were assessed by differences in continuous variables measured by the Mann-Whitney U test. A P value of ≤0.05 was considered significant.
RESULTS
Extraction of DNA from Aspergillus fumigatus conidia and GHE.
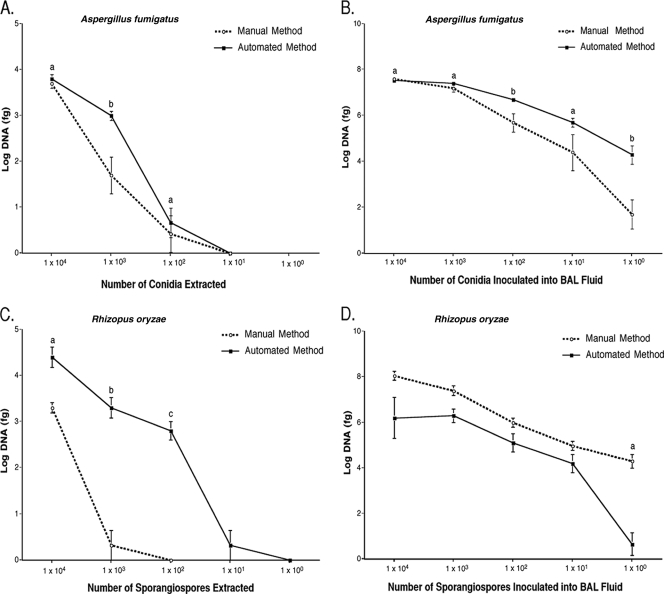
Both methods extracted DNA from A. fumigatus conidia in PBS, resulting in a lower limit of detection of 1 × 102 conidia. There was no significant difference in the amount of DNA amplified at this level of detection (Fig. 1A). When DNA was extracted from A. fumigatus GHE in BAL fluid, both extraction methods resulted in a level of detection of 1 × 100 GHE/ml (Fig. 1B). At this level of detection, the automated method demonstrated a trend toward greater sensitivity in percent yield of positive samples (P = 0.08) (Table 1). At a given concentration of 1 GHE/ml, the automated method extracted more DNA than did the manual method (P = 0.008) (Fig. 1B).
FIG. 1.
Amounts of DNA extracted by the automated and manual methods of DNA extraction. (A) Extraction of 10-fold serial dilutions of Aspergillus fumigatus. a, not significant; b, P = 0.0003. (B) Extraction of Aspergillus fumigatus GHE in normal rabbit BAL fluid. a, not significant; b, P ≤ 0.008. (C) Extraction of 10-fold serial dilutions of Rhizopus oryzae sporangiospores. a, b, and c, P ≤ 0.008. (D) Extraction of Rhizopus oryzae GHE in normal rabbit BAL fluid. The manual method extracted significantly more DNA (a, P = 0.0006) at a level of detection of 1 × 100 GHE than the automated method at the same level.
TABLE 1.
Sensitivity of DNA extraction methods for Aspergillus fumigatus and Rhizopus oryzae
| Isolate and amt | % Sensitivity (na)
|
|
|---|---|---|
| Automated method | Manual method | |
| A. fumigatus | ||
| Conidia | ||
| 1 × 104 | 100 (8) | 100 (7) |
| 1 × 103 | 100 (8) | 86 (7) |
| 1 × 102 | 38b (8) | 17b (6) |
| 1 × 101 | 0 (8) | 0 (6) |
| 1 × 100 | 0 (8) | 0 (6) |
| GHE | ||
| 1 × 104 | 100 (9) | 100 (9) |
| 1 × 103 | 100 (9) | 100 (9) |
| 1 × 102 | 100 (9) | 100 (9) |
| 1 × 101 | 100 (9) | 89 (9) |
| 1 × 100 | 100c (9) | 56c (9) |
| R. oryzae | ||
| Sporangiospores | ||
| 1 × 104 | 100 (6) | 100 (6) |
| 1 × 103 | 100e (7) | 17e (6) |
| 1 × 102 | 100d (6) | 0d (6) |
| 1 × 101 | 17 (6) | 0 (6) |
| 1 × 100 | 0 (6) | 0 (6) |
| GHE | ||
| 1 × 104 | 100 (7) | 100 (7) |
| 1 × 103 | 100 (7) | 100 (7) |
| 1 × 102 | 100 (7) | 100 (7) |
| 1 × 101 | 100 (7) | 100 (7) |
| 1 × 100 | 14e (7) | 100e (7) |
No. of samples processed.
P = 0.58.
P = 0.08.
P = 0.002.
P = 0.005.
Extraction of DNA from Rhizopus oryzae sporangiospores and GHE.
Extraction of DNA from R. oryzae sporangiospores in PBS by the manual method resulted in a level of detection of 1 × 103 sporangiospores (Table 1), whereas extraction by the automated method resulted in a level of detection of 1 × 101 sporangiospores (Table 1). In addition, significantly more DNA (P ≤ 0.008) was amplified at sporangiospore levels of 1 × 104,1 × 103, and 1 × 102 with the automated method (Fig. 1C). When DNA was extracted from R. oryzae GHE, both extraction methods resulted in a level of detection of 1 × 100 GHE/ml (Table 1). At this level of detection, however, the manual method demonstrated greater sensitivity in percent yield of positive samples (P = 0.005) and recovered significantly more DNA (P = 0.0006) (Fig. 1D).
Sample processing time.
The amount of time to process 24 samples by the automated method was 2.5 h for sample collection, enzymatic pretreatment, and mechanical disruption prior to transferring for automation. The time of automation was 1.3 h by the MagNA Pure LC instrument followed by 10 min postautomation, resulting in a total time of 4 h. The total time required for the manual method was 5.25 h, which included sample collection, enzymatic pretreatment, mechanical disruption, and further processing with the DNeasy Plant minikit.
DISCUSSION
The methods for DNA extraction from fungi for clinical detection have long been ignored. Yet, the loss of as much as 99.9% of fungal DNA during the extraction process is a major limitation to the analytical sensitivity of diagnostic PCR for invasive apergillosis and other life-threatening mycoses.
Due to the limitations of the techniques currently used to diagnose invasive pulmonary aspergillosis and invasive pulmonary zygomycosis, there is a need for more sensitive, non-culture-based techniques such as PCR. One parameter, which influences the clinical usefulness of PCR, is the ability to efficiently isolate DNA from a clinical sample. This paper describes two different DNA extraction methods that can be used for A. fumigatus, the most common Aspergillus species causing invasive pulmonary aspergillosis, and R. oryzae, an increasingly important cause of zygomycosis. The data from this study demonstrate that both methods are comparable. However, the automated method is faster and can be used for high-throughput clinical laboratories. To our knowledge, this is the first study to investigate these two medically important fungi for the extraction of DNA from BAL fluid using automated and manual methods. Understanding these methods and their comparative yields improves the use of these systems in clinical laboratories.
To date, there is no standardized PCR method for the detection of fungal pathogens. The lack of standardization is due in part to the uncertainty about the optimal sample (e.g., blood, serum, plasma, tissue, or BAL fluid) and inconsistency between DNA extraction methods (6). There is a need for more time-efficient automated DNA extraction methods for fungi that may be standardized and that may decrease work burden (6, 18). The ability to detect fungal pathogens in clinical samples by qPCR requires extraction methods that can efficiently lyse their cell walls.
The structure of the fungal cell wall is highly complex compared to the structures of mammalian cell membranes and bacterial cell walls (18). The fungal cell wall consists of α- and β-(1→3)-glucans, β-(1→6)-glucans, chitin, galactomannan, mannans, mannoproteins, lipids, and peptides. Due to the complexity of the fungal cell wall, conventional methods employed for extracting DNA from viruses and bacteria may not be suitable for the extraction of DNA from these complex organisms.
No single DNA extraction method is optimal for the efficient extraction of all fungi. Therefore, in addition to some type of enzymatic and or mechanical pretreatment, modifications of kit protocols may be necessary (11, 18, 26). The manual DNA extraction methods described in the literature for filamentous fungi tend to be labor-intensive and time-consuming, rendering them unsuitable for high-throughput diagnostic laboratories (8, 9, 11, 13, 18, 21, 22, 25, 26). When small numbers of samples need to be processed, our laboratory uses the manual DNA extraction method incorporating enzymatic pretreatment and mechanical disruption with a modified version of the DNeasy Plant minikit protocol (25, 26). The DNeasy Plant minikit is a spin column procedure that incorporates sample lysis, removal of RNA, removal of proteins and polysaccharides, DNA precipitation, and binding to the spin column membrane. Multiple washes are performed to remove contaminants, and DNA is then eluted from the membrane.
The automated or manual method may be suitable for DNA extraction from BAL fluid containing mixed fungal elements of A. fumigatus or R. oryzae. Our data demonstrate that the manual extraction method shows good correlation with the automated method. These data are in agreement with results presented by White et al. for the extraction of DNA from water or EDTA-treated whole blood spiked with A. fumigatus conidia (37).
The evolution of molecular diagnostics for mycology is advancing with the development of fully automated platforms which have the ability to extract DNA from fungi (8, 13, 19, 21). We investigated the MagNA Pure LC instrument, a widely used fully automated closed system, for DNA extraction. This device can purify DNA from different biological samples utilizing magnetic bead technology. Although MagNA Pure technology has been applied to blood and tissue for the detection of yeasts and filamentous fungi, little has been known about the comparative application of automated and manual technologies for detection of conidia and GHE of Aspergillus spp. and Rhizopus spp. from lower respiratory tract specimens, particularly BAL fluid. BAL fluid was used in these studies because it is a common specimen chosen for analysis by clinical microbiology laboratories for the detection of organisms in patients with fungal pneumonia.
The automated and manual DNA extraction methods described in this study effectively extracted DNA from A. fumigatus and R. oryzae fungal propagules and GHE in BAL fluid. Depending on the cellular stage of growth of the organism, BAL fluid samples may contain a combination of GHE and spores (2, 12). Our data demonstrate that both DNA extraction methods can efficiently extract DNA from A. fumigatus conidia at the same level of detection with no significant difference in sensitivity. In addition, no significant difference in sensitivity was shown between DNA extraction methods when extracting DNA from A. fumigatus GHE. Therefore, when extracting BAL fluid samples that may contain diverse forms of A. fumigatus, either method could be implemented. When extracting DNA from R. oryzae sporangiospores, the automated method demonstrated greater sensitivity in percent yield of positive samples. The extraction of BAL fluid samples containing at least 10 GHE of R. oryzae with both the automated and manual methods showed similar sensitivity. However, at the lower limit of detection (1 GHE), the manual method showed better sensitivity. Thus, either the automated or manual method may be suitable for DNA extraction from BAL fluid containing mixed fungal elements of R. oryzae.
In analyzing the data from these studies, a distinction was made between sample sensitivity and absolute yield of DNA per sample. The automated method resulted in similar or greater quantities of DNA extracted for Aspergillus and Rhizopus propagules and Aspergillus GHE. Greater quantities of DNA may be extracted from these types of samples using the automated method because unlike the manual method, following enzymatic treatment, the automated method does not incorporate a centrifugation step and subsequent removal of supernatant. Hence, the entire sample is carried through the automated extraction method. The centrifugation step and removal of supernatant in the manual method may result in the loss of organism and therefore the isolation of less DNA using the manual method.
The manual method resulted in similar or greater quantities of DNA extracted for Rhizopus GHE. There may be several factors contributing to these differences. By visual inspection with an inverted microscope, the hyphal elements of A. fumigatus were narrow, septate, and dichotomously branched with uniform diameter, whereas the R. oryzae hyphal elements were broad, bulky, and sparsely septate with nonparallel irregular branching. Therefore, when R. oryzae GHE were processed, there likely would have been denser cellular debris in these samples. The manual method incorporates a filtration and homogenization unit designed to help remove cellular debris and precipitates, which may interfere with efficient DNA isolation from zygomycetes such as Rhizopus spp. By comparison, the automated method does not include a filtration and homogenization unit for the removal of cellular debris and precipitates. Therefore, any cellular debris not removed by the repeated washing of the magnetic glass particles used in the automated method may reduce the efficiency of isolation. At the lower level of detection there may be such a small amount of DNA extracted that any cellular debris may have affected the ability of the DNA to bind to the magnetic glass particles, resulting in the isolation of less DNA by the automated method. Conversely, at the higher cellular concentrations there may be so much DNA that the cellular debris has a minimal effect on the amount of DNA obtained.
Even with the implementation of the enzymatic pretreatment and mechanical disruption, the automated method was still 1.25 h faster than the manual method when extracting 24 samples. For this reason, the automated method is an encouraging option for high-throughput laboratories with a need to extract DNA from filamentous fungi in multiple clinical samples. The manual method remains a useful option when small numbers of samples need to be processed.
The MagNA Pure technology reduces the number of manual steps needed for DNA extraction from various types of samples. However, when using it to extract DNA from fungi, additional manual steps need to be incorporated. This instrument can extract up to 32 samples at one time, therefore making it more time-efficient than manual methods. The automated method has been implemented in our laboratory for high-throughput extraction of in vivo samples. As we transfer our PCR technology for detection of Aspergillus and zygomycetes to the clinical laboratory, the manual method of extraction will be incorporated for the relatively small number of BAL fluid samples submitted daily.
These data provide a foundation for the use of either an automated or manual method of DNA extraction of BAL fluid samples from immunocompromised patients for whom a diagnosis of pulmonary aspergillosis or zygomycosis is being considered. The automated method is more time-efficient when extracting 24 samples and demonstrates equal or greater levels of detection. The automated DNA extraction method may provide a favorable option for high-throughput clinical laboratories. The manual method is useful if small numbers of samples need to be processed.
Footnotes
Published ahead of print on 19 March 2008.
REFERENCES
- 1.Alangaden, G. J., M. Wahiduzzaman, and P. H. Chandrasekar. 2002. Aspergillosis: the most common community-acquired pneumonia with gram-negative Bacilli as copathogens in stem cell transplant recipients with graft-versus-host disease. Clin. Infect. Dis. 35659-664. [DOI] [PubMed] [Google Scholar]
- 2.Bauters, T. G., and H. J. Nelis. 2000. Rapid and sensitive plate method for detection of Aspergillus fumigatus. J. Clin. Microbiol. 383796-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin, D. K., Jr., W. C. Miller, S. Bayliff, L. Martel, K. A. Alexander, and P. L. Martin. 2002. Infections diagnosed in the first year after pediatric stem cell transplantation. Pediatr. Infect. Dis. J. 21227-234. [DOI] [PubMed] [Google Scholar]
- 4.Bretagne, S., and J. M. Costa. 2005. Towards a molecular diagnosis of invasive aspergillosis and disseminated candidosis. FEMS Immunol. Med. Microbiol. 45361-368. [DOI] [PubMed] [Google Scholar]
- 5.Buchheidt, D., C. Baust, H. Skladny, J. Ritter, T. Suedhoff, M. Baldus, W. Seifarth, C. Leib-Moesch, and R. Hehlmann. 2001. Detection of Aspergillus species in blood and bronchoalveolar lavage samples from immunocompromised patients by means of 2-step polymerase chain reaction: clinical results. Clin. Infect. Dis. 33428-435. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S. C., C. L. Halliday, and W. Meyer. 2002. A review of nucleic acid-based diagnostic tests for systemic mycoses with an emphasis on polymerase chain reaction-based assays. Med. Mycol. 40333-357. [DOI] [PubMed] [Google Scholar]
- 7.Cornet, M., L. Fleury, C. Maslo, J. F. Bernard, and G. Brucker. 2002. Epidemiology of invasive aspergillosis in France: a six-year multicentric survey in the Greater Paris area. J. Hosp. Infect. 51288-296. [DOI] [PubMed] [Google Scholar]
- 8.Costa, C., J. M. Costa, C. Desterke, F. Botterel, C. Cordonnier, and S. Bretagne. 2002. Real-time PCR coupled with automated DNA extraction and detection of galactomannan antigen in serum by enzyme-linked immunosorbent assay for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 402224-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faggi, E., G. Pini, and E. Campisi. 2005. Use of magnetic beads to extract fungal DNA. Mycoses 483-7. [DOI] [PubMed] [Google Scholar]
- 10.Francesconi, A., M. Kasai, R. Petraitiene, V. Petraitis, A. M. Kelaher, R. Schaufele, W. W. Hope, Y. R. Shea, J. Bacher, and T. J. Walsh. 2006. Characterization and comparison of galactomannan enzyme immunoassay and quantitative real-time PCR assay for detection of Aspergillus fumigatus in bronchoalveolar lavage fluid from experimental invasive pulmonary aspergillosis. J. Clin. Microbiol. 442475-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fredricks, D. N., C. Smith, and A. Meier. 2005. Comparison of six DNA extraction methods for recovery of fungal DNA as assessed by quantitative PCR. J. Clin. Microbiol. 435122-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glazer, M., S. Nusair, R. Breuer, J. Lafair, Y. Sherman, and N. Berkman. 2000. The role of BAL in the diagnosis of pulmonary mucormycosis. Chest 117279-282. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths, L. J., M. Anyim, S. R. Doffman, M. Wilks, M. R. Millar, and S. G. Agrawal. 2006. Comparison of DNA extraction methods for Aspergillus fumigatus using real-time PCR. J. Med. Microbiol. 551187-1191. [DOI] [PubMed] [Google Scholar]
- 14.Grow, W. B., J. S. Moreb, D. Roque, K. Manion, H. Leather, V. Reddy, S. A. Khan, K. J. Finiewicz, H. Nguyen, C. J. Clancy, P. S. Mehta, and J. R. Wingard. 2002. Late onset of invasive aspergillus infection in bone marrow transplant patients at a university hospital. Bone Marrow Transplant. 2915-19. [DOI] [PubMed] [Google Scholar]
- 15.Hope, W. W., T. J. Walsh, and D. W. Denning. 2005. Laboratory diagnosis of invasive aspergillosis. Lancet Infect. Dis. 5609-622. [DOI] [PubMed] [Google Scholar]
- 16.Horger, M., H. Hebart, H. Schimmel, M. Vogel, H. Brodoefel, K. Oechsle, U. Hahn, M. Mittelbronn, W. Bethge, and C. D. Claussen. 2006. Disseminated mucormycosis in haematological patients: CT and MRI findings with pathological correlation. Br. J. Radiol. 79e88-95. [DOI] [PubMed] [Google Scholar]
- 17.Husain, S., B. D. Alexander, P. Munoz, R. K. Avery, S. Houston, T. Pruett, R. Jacobs, E. A. Dominguez, J. G. Tollemar, K. Baumgarten, C. M. Yu, M. M. Wagener, P. Linden, S. Kusne, and N. Singh. 2003. Opportunistic mycelial fungal infections in organ transplant recipients: emerging importance of non-Aspergillus mycelial fungi. Clin. Infect. Dis. 37221-229. [DOI] [PubMed] [Google Scholar]
- 18.Karakousis, A., L. Tan, D. Ellis, H. Alexiou, and P. J. Wormald. 2006. An assessment of the efficiency of fungal DNA extraction methods for maximizing the detection of medically important fungi using PCR. J. Microbiol. Methods 6538-48. [DOI] [PubMed] [Google Scholar]
- 19.Klingspor, L., and S. Jalal. 2006. Molecular detection and identification of Candida and Aspergillus spp. from clinical samples using real-time PCR. Clin. Microbiol. Infect. 12745-753. [DOI] [PubMed] [Google Scholar]
- 20.Kontoyiannis, D. P., M. S. Lionakis, R. E. Lewis, G. Chamilos, M. Healy, C. Perego, A. Safdar, H. Kantarjian, R. Champlin, T. J. Walsh, and Raad, I. I. 2005. Zygomycosis in a tertiary-care cancer center in the era of Aspergillus-active antifungal therapy: a case-control observational study of 27 recent cases. J. Infect. Dis. 1911350-1360. [DOI] [PubMed] [Google Scholar]
- 21.Loeffler, J., K. Schmidt, H. Hebart, U. Schumacher, and H. Einsele. 2002. Automated extraction of genomic DNA from medically important yeast species and filamentous fungi by using the MagNA Pure LC system. J. Clin. Microbiol. 402240-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loffler, J., H. Hebart, U. Schumacher, H. Reitze, and H. Einsele. 1997. Comparison of different methods for extraction of DNA of fungal pathogens from cultures and blood. J. Clin. Microbiol. 353311-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marr, K. A., R. A. Carter, M. Boeckh, P. Martin, and L. Corey. 2002. Invasive aspergillosis in allogeneic stem cell transplant recipients: changes in epidemiology and risk factors. Blood 1004358-4366. [DOI] [PubMed] [Google Scholar]
- 24.Morgan, J., K. A. Wannemuehler, K. A. Marr, S. Hadley, D. P. Kontoyiannis, T. J. Walsh, S. K. Fridkin, P. G. Pappas, and D. W. Warnock. 2005. Incidence of invasive aspergillosis following hematopoietic stem cell and solid organ transplantation: interim results of a prospective multicenter surveillance program. Med. Mycol. 43(Suppl. 1)S49-S58. [DOI] [PubMed] [Google Scholar]
- 25.Muller, F. M., K. E. Werner, M. Kasai, A. Francesconi, S. J. Chanock, and T. J. Walsh. 1998. Rapid extraction of genomic DNA from medically important yeasts and filamentous fungi by high-speed cell disruption. J. Clin. Microbiol. 361625-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Sullivan, C. E., M. Kasai, A. Francesconi, V. Petraitis, R. Petraitiene, A. M. Kelaher, A. A. Sarafandi, and T. J. Walsh. 2003. Development and validation of a quantitative real-time PCR assay using fluorescence resonance energy transfer technology for detection of Aspergillus fumigatus in experimental invasive pulmonary aspergillosis. J. Clin. Microbiol. 415676-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pagano, L., P. Ricci, A. Tonso, A. Nosari, L. Cudillo, M. Montillo, A. Cenacchi, L. Pacilli, F. Fabbiano, A. Del Favero, et al. 1997. Mucormycosis in patients with haematological malignancies: a retrospective clinical study of 37 cases. Br. J. Haematol. 99331-336. [DOI] [PubMed] [Google Scholar]
- 28.Paterson, D. L., and N. Singh. 1999. Invasive aspergillosis in transplant recipients. Medicine (Baltimore) 78123-138. [DOI] [PubMed] [Google Scholar]
- 29.Patterson, T. F., W. R. Kirkpatrick, M. White, J. W. Hiemenz, J. R. Wingard, B. Dupont, M. G. Rinaldi, D. A. Stevens, J. R. Graybill, et al. 2000. Invasive aspergillosis. Disease spectrum, treatment practices, and outcomes. Medicine (Baltimore) 79250-260. [DOI] [PubMed] [Google Scholar]
- 30.Petraitiene, R., V. Petraitis, A. H. Groll, T. Sein, R. L. Schaufele, A. Francesconi, J. Bacher, N. A. Avila, and T. J. Walsh. 2002. Antifungal efficacy of caspofungin (MK-0991) in experimental pulmonary aspergillosis in persistently neutropenic rabbits: pharmacokinetics, drug disposition, and relationship to galactomannan antigenemia. Antimicrob. Agents Chemother. 4612-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rementeria, A., N. Lopez-Molina, A. Ludwig, A. B. Vivanco, J. Bikandi, J. Ponton, and J. Garaizar. 2005. Genes and molecules involved in Aspergillus fumigatus virulence. Rev. Iberoam Micol. 221-23. [DOI] [PubMed] [Google Scholar]
- 32.Roden, M. M., T. E. Zaoutis, W. L. Buchanan, T. A. Knudsen, T. A. Sarkisova, R. L. Schaufele, M. Sein, T. Sein, C. C. Chiou, J. H. Chu, D. P. Kontoyiannis, and T. J. Walsh. 2005. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin. Infect. Dis. 41634-653. [DOI] [PubMed] [Google Scholar]
- 33.Saito, H., E. J. Anaissie, R. C. Morice, R. Dekmezian, and G. P. Bodey. 1988. Bronchoalveolar lavage in the diagnosis of pulmonary infiltrates in patients with acute leukemia. Chest 94745-749. [DOI] [PubMed] [Google Scholar]
- 34.Siwek, G. T., K. J. Dodgson, M. de Magalhaes-Silverman, L. A. Bartelt, S. B. Kilborn, P. L. Hoth, D. J. Diekema, and M. A. Pfaller. 2004. Invasive zygomycosis in hematopoietic stem cell transplant recipients receiving voriconazole prophylaxis. Clin. Infect. Dis. 39584-587. [DOI] [PubMed] [Google Scholar]
- 35.Stergiopoulou, T., J. Meletiadis, E. Roilides, D. E. Kleiner, R. Schaufele, M. Roden, S. Harrington, L. Dad, B. Segal, and T. J. Walsh. 2007. Host-dependent patterns of tissue injury in invasive pulmonary aspergillosis. Am. J. Clin. Pathol. 127349-355. [DOI] [PubMed] [Google Scholar]
- 36.Tarrand, J. J., M. Lichterfeld, I. Warraich, M. Luna, X. Y. Han, G. S. May, and D. P. Kontoyiannis. 2003. Diagnosis of invasive septate mold infections. A correlation of microbiological culture and histologic or cytologic examination. Am. J. Clin. Pathol. 119854-858. [DOI] [PubMed] [Google Scholar]
- 37.White, P. L., R. Barton, M. Guiver, C. J. Linton, S. Wilson, M. Smith, B. L. Gomez, M. J. Carr, P. T. Kimmitt, S. Seaton, K. Rajakumar, T. Holyoake, C. C. Kibbler, E. Johnson, R. P. Hobson, B. Jones, and R. A. Barnes. 2006. A consensus on fungal polymerase chain reaction diagnosis? A United Kingdom-Ireland evaluation of polymerase chain reaction methods for detection of systemic fungal infections. J. Mol. Diagn. 8376-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zmeili, O. S., and A. O. Soubani. 2007. Pulmonary aspergillosis: a clinical update. Q. J. Med. 100317-334. [DOI] [PubMed] [Google Scholar]