Abstract
We examined the ability of 206 clinical isolates of Staphylococcus aureus to lyse T cells and found differences between Agr groups. We found that the beta and delta hemolysins are involved and that methicillin-resistant S. aureus strains are less toxic than methicillin-susceptible S. aureus strains.
Staphylococcus aureus is a major human pathogen, and its ability to secrete toxins significantly contributes to host damage (3). Studies linking disease characteristics with specific genes in S. aureus are often hampered by the difficulty with the genetic manipulation of clinical isolates (18), the fact that laboratory strains are no longer a faithful representation of those causing disease today (5), and the fact that S. aureus often has multiple genes whose activities can be substituted for by other genes (4). In this study, we adopted an alternative approach, in that we took a large S. aureus strain collection and assayed a specific characteristic of each isolate. Because we had a large amount of genetic information (e.g., clonality by multilocus sequence type [MLST] [2] and the presence of 33 adhesins and toxins [11, 13]) relating to this strain collection, we sought to make associations between these factors and toxicity.
T cells are an important component of vertebrate immunity, although their role in protection against S. aureus infections is unclear. Using an immortalized T-cell line (T2 cells [15]), we developed an assay in which washed bacterial cells (grown in brain heart infusion medium overnight at 37°C shaking) were incubated with T cells (grown in RPMI [Gibco] supplemented with 10% fetal bovine serum [Gibco], 1 mM l-glutamine, 200 units/ml penicillin, and 0.1 mg/ml streptomycin [Sigma] at 37°C with 10% CO2) for 30 min. The number of T cells lysed by the de novo production of proteins by S. aureus was measured by using trypan blue (Sigma) in a Fast-Read counting chamber (Immune Systems, United Kingdom).
Table 1 shows the factors found to be associated with T-cell toxicity. The data could not be normalized, and so nonparametric tests were performed to identify associations between T-cell toxicity and a specific genetic factor(s). The first factor that was found to be associated with T-cell toxicity was Agr (accessory gene regulator) function. The Agr proteins are responsible for the down-regulation of surface proteins and the up-regulation of exoproteins when they reach a quorum (1) and are measured by determination of the level of expression of the delta toxin (11). The mean T-cell survival achieved when the T cells were incubated with strains expressing delta toxin was 59.2%, whereas 72.4% survival was achieved when the T cells were incubated with strains not expressing the delta toxin (P < 0.001, Mann-Whitney U test). As the delta toxin is itself a pore-forming protein, it is possible that our observation could be explained, at least in part, by the loss of this protein's expression alone and not as a result of the loss of other Agr-regulated proteins.
TABLE 1.
Bacterial factors significantly associated with T-cell toxicity
| Bacterial phenotype | Mean (median) % T-cell survivala | No. of strains |
|---|---|---|
| Beta toxin positive | 59.4 (61.9) | 163 |
| Beta toxin negative | 74.5 (77.1) | 42 |
| Delta toxin positive | 59.2 (61.7) | 51 |
| Delta toxin negative | 72.4 (77.7) | 154 |
| TSST-1 positive | 69.1 (72.4) | 53 |
| TSST-1 negative | 60.4 (63.6) | 151 |
| MRSA | 69.8 (77.2) | 23 |
| MSSA | 61.6 (66.1) | 183 |
| Agr type 1 | 54.2 (56.6) | 77 |
| Agr type 2 | 66.3 (70.6) | 47 |
| Agr type 3 | 68.4 (72.1) | 75 |
| CC1, CC12, CC15, CC22, CC30, CC39, CC51 | 67.69 (67.9) | 110b |
| CC5, CC8, CC9, CC16, CC25, CC45 | 55.57 (57.8) | 93c |
For beta toxin positive versus beta toxin negative, P < 0.001; for delta toxin positive versus delta toxin negative, P < 0.001; for TSST-1 positive versus TSST-1 negative, P = 0.009; for MRSA versus MSSA, P = 0.045; for Agr type 1 versus Agr type 2, P = 0.005; for Agr type 1 versus Agr type 3, P < 0.001; for Agr type 2 versus Agr type 3, P > 0.1; and for CC1, CC12, CC15, CC22, CC30, CC39, and CC51 versus CC5, CC8, CC9, CC16, CC25, and CC45, P < 0.001. All P values were determined by the Mann-Whitney U test.
CC1, n = 9; CC12, n = 6; CC15, n = 19; CC30, n = 57; CC39, n = 13; CC51, n = 6.
CC5, n = 12; CC8, n = 10; CC9, n = 4; CC16, n = 12; CC22, n = 17; CC25, n = 19; CC45, n = 22.
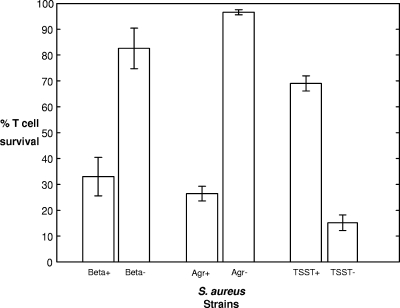
We next examined whether the presence of 16 other toxins (encoded by sea, seb, sec, sed, see, seg, she, sei, sej, eta, etb, pvl, hlg, hla, hlb, and tst) was associated with T-cell toxicity. Note that the alpha toxin was expressed by all strains tested, and as such, alpha toxin expression was not informative. We found that T-cell survival was significantly lower when the T cells were incubated with bacteria expressing the beta toxin (P < 0.001, Mann-Whitney U test). The beta toxin is sphingomyelinase, the activity of which is assayed by the hot-cold lysis of sheep erythrocytes (note that even though beta toxin is regulated by Agr, beta toxin expression is detectable in Agr null strains [14]). There was a close association between beta toxin and delta toxin expression, in which of the 51 isolates that did not express the delta toxin (i.e., they had dysfunctional Agr systems), 35 were also beta toxin negative, whereas of the 154 isolates that expressed the delta toxin, only 7 were beta toxin negative. As such, we needed to examine whether the beta toxin has a specific role in T-cell lysis or whether this association is a result of a linkage between beta toxin expression and Agr dysfunction. To test for this, we assayed the toxicities of some isogenic pairs of strains in which specific genes were inactivated (Fig. 1). As expected, the inactivation of the Agr locus (strains RN6911 [9] and RN6390B [12]) had a significant effect on T-cell toxicity; however, the effect of beta toxin alone (strains 8325-4 [8] and DU5719 [10]) was also significant, suggesting that it has a direct role in T-cell lysis as well as other Agr-regulated factors.
FIG. 1.
Comparison of isogenic strains with specific gene inactivation. For the beta toxin-positive strains (n = 6) and the beta toxin-negative strains (n = 6), the mean T-cell survival rates were 33.00% and 82.55%, respectively (P < 0.01). For the Agr-positive strains (n = 6) and the Agr-negative strains (n = 6), the mean T-cell survival rates were 26.42% and 96.55%, respectively (P < 0.01). For the TSST-1-positive strains (n = 7) and the TSST-1-knockout strains (n = 6), the mean T-cell survival rates were 69.02% and 15.17%, respectively (P < 0.01).
The third toxin associated with T-cell toxicity was toxic shock syndrome toxin 1 (TSST-1). Rather than contributing positively to T-cell toxicity, we found that the presence of the tst gene resulted in a decrease in the ability of a strain to lyse T cells. This could be explained by the findings of another study showing that TSST-1 has a global regulatory effect on exoprotein expression (17). To verify that TSST-1 and not some other factor in association with the gene for this protein was responsible for this, we compared the activity of a TSST-1-producing strain (strain RN4282 [7]) and an isogenic TSST-1 mutant (strain RN6938 [16]) (Fig. 1) and found the observed effect to be specific to the toxin, verifying that the regulatory effect of this gene is widespread in clinical populations of S. aureus.
We found that the methicillin-resistant S. aureus (MRSA) strains in our collection (all of which belonged to MLST-derived clonal complex 22 [CC22] and CC30 and contained the type II staphylococcal chromosomal cassette mec element) were significantly less toxic than the methicillin-susceptible S. aureus (MSSA) strains (P = 0.045). Upon examination of the distribution of toxin expression among these isolates, we observed that while 20% of the MSSA population of isolates were delta toxin negative, 72.7% of the MRSA isolates were delta toxin negative. It would therefore seem to be more likely that the observed decrease in toxicity is as a result of an association with Agr dysfunction rather than with the staphylococcal chromosomal cassette mec element per se.
As Agr was unquestionably a significant factor in this study, we examined whether there was a difference between the cross-inhibiting Agr types. We found that strains of Agr type 1 were significantly more toxic than those of Agr type 2 or 3 (P < 0.01 in both cases). We found no significant difference between Agr types 2 and 3. The Agr type of a strain is strongly associated with clonality (6), and so we examined the toxicity of our CCs (CCs 1 to 51). We found that toxicity resulted in T-cell survival rates that varied from 45.5% (CC16) to 72.2% (CC1). Using the median value (62.3%) to dichotomize the data into high and low toxicity, we found that CCs 1, 12, 15, 22, 30, 39, and 51 were all in the low-toxicity range, with T-cell survival rates of 72.7, 72.1, 67.8, 62.4, 68.2, 62.7, and 67.9%, respectively. CCs 5, 8, 9, 16, 25, and 45 were all in the high-toxicity range, with T-cell survival rates of 59.1, 58.9, 60.4, 45.5, 52.9, and 56.6%, respectively. Only one CC of Agr type 1 fell into the lower-toxicity range, and that was CC22, the toxicity of which fell only just above the point of dichotomization of the strains. All other Agr type 1 CCs were in the high-toxicity range. All MRSA strains were found in the low-toxicity group of strains. Delta toxin was expressed by only 65.4% of strains in the low-toxicity group but was expressed by 87.7% of the strains in the high-toxicity group (P < 0.001). Beta toxin was expressed by only 70.9% of the strains in the low-toxicity group but was expressed by 91% of the strains in the high-toxicity group (P < 0.001), and the toxic shock syndrome gene was found in 38% of the low-toxicity strains but only 5.3% of the high-toxicity strains (P < 0.001). No single factor was indicative of either high or low toxicity, and other factors associated with the genetic backgrounds of the strains may also contribute.
In this study, we have examined the toxicity of a genetically diverse population of clinical S. aureus isolates. We found that a functional Agr locus is required and that the beta toxin and possibly the delta toxin are directly involved. We also found that transcriptional repression by TSST-1 is widespread among clinical isolates and that certain Agr and CC types are more toxic than others. Our findings suggest that these activities are unlikely to be specific to T cells, as many other host cell types will be affected by the toxins found to be important here and their regulators. Other factors are clearly involved but have yet to be identified. Nevertheless, we found this to be an informative alternative to the usual approaches of studying pathogenicity traits, as it shows us how the truly wild-type bacteria are behaving and allows the identification of multiple contributing factors in a single assay.
Acknowledgments
This work was funded by Stiefel.
Footnotes
Published ahead of print on 16 April 2008.
REFERENCES
- 1.Bronner, S., H. Monteil, and G. Prevost. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28183-200. [DOI] [PubMed] [Google Scholar]
- 2.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. J. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 1853307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferry, T., T. Perpoint, F. Vandenesch, and J. Etienne. 2005. Virulence determinants in Staphylococcus aureus and their involvement in clinical syndromes. Curr. Infect. Dis. Rep. 7420-428. [DOI] [PubMed] [Google Scholar]
- 4.Greene, C., D. McDevitt, P. Francois, P. E. Vaudaux, D. P. Lew, and T. J. Foster. 1995. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 171143-1152. [DOI] [PubMed] [Google Scholar]
- 5.Grundmeier, M., M. Hussain, P. Becker, C. Heilmann, G. Peters, and B. Sinha. 2004. Truncation of fibronectin-binding proteins in Staphylococcus aureus strain Newman leads to deficient adherence and host cell invasion due to loss of the cell wall anchor function. Infect. Immun. 727155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindsay, J. A., A. Ruzin, H. F. Ross, N. Kurepina, and R. P. Novick. 1998. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29527-543. [DOI] [PubMed] [Google Scholar]
- 8.Novick, R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33155-166. [DOI] [PubMed] [Google Scholar]
- 9.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 123967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel, A. H., P. Nowlan, E. D. Weavers, and T. Foster. 1987. Virulence of protein A-deficient and alpha-toxin-deficient mutants of Staphylococcus aureus isolated by allele replacement. Infect. Immun. 553103-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peacock, S. J., C. E. Moore, A. Justice, M. Kantzanou, L. Story, K. Mackie, G. O'Neill, and N. P. J. Day. 2002. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 704987-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng, H. L., R. P. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 1704365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roche, F. M., R. Massey, S. J. Peacock, N. P. J. Day, L. Visai, P. Speziale, A. Lam, M. Pallen, and T. J. Foster. 2003. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology 149643-654. [DOI] [PubMed] [Google Scholar]
- 14.Sakoulas, G., G. M. Eliopoulos, R. C. Moellering, Jr., C. Wennersten, L. Venkataraman, R. P. Novick, and H. S. Gold. 2002. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 461492-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salter, R. D., and P. Cresswell. 1986. Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell hybrid. EMBO. 5943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sloane, R., J. C. S. De Azavedo, J. P. Arbuthnott, P. J. Hartigan, B. Kreiswirth, R. Novick, and T. J. Foster. 1991. A toxic shock syndrome toxin mutant of Staphylococcus aureus isolated by allelic replacement lacks virulence in a rabbit uterine model. FEMS Microbiol. Lett. 78239-244. [DOI] [PubMed] [Google Scholar]
- 17.Vojtov, N., H. F. Ross, and R. P. Novick. 2002. Global repression of exotoxin synthesis by staphylococcal superantigens. Proc. Natl. Acad. Sci. USA 9910102-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waldron, D. E., and J. A. Lindsay. 2006. Sau1: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 1885578-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]