Abstract
A new commercial real-time human cytomegalovirus (HCMV) PCR kit was evaluated after automated DNA extraction of 153 amniotic fluids in parallel with an in-house real-time PCR assay. The commercial kit displayed 100% sensitivity/specificity compared to the “in-house” assay and was suitable for prenatal diagnosis of HCMV congenital infection.
The diagnosis of fetal human cytomegalovirus (HCMV) infection is usually made by amplification of HCMV genome in amniotic fluid (AF) sampled by amniocentesis. The overall sensitivity of HCMV DNA detection for prenatal diagnosis in AF by PCR ranges from 70 to 100% (13). However, the sensitivity of HCMV PCR in AF is close to 100% when using a PCR test and appropriate timing for amniocentesis (e.g., after 20 weeks of gestation and at least 6 weeks after maternal infection) (4). Nevertheless, false-negative HCMV PCR results have been reported in AF samples even under these optimal diagnostic conditions (5, 7, 13). These false-negative results were related to DNA amplification inhibition by inhibitory properties of AF (2). Moreover, although 100% specificity was reported for HCMV detection by PCR in AF (1, 3, 11, 13), rare cases of false-positive prenatal diagnosis have also been published (5, 7, 9). In these cases, although AF tested PCR positive, the AF culture was negative and the child was not infected at birth. False-positive results could be due to contamination occurring during PCR testing. This risk is higher with nested PCR, a technology that is very sensitive but exposed to contamination. Generalization of semiautomated real-time PCR methodology might help to overcome the risk of contamination and to achieve absolute specificity for HCMV prenatal diagnosis.
In this study, we evaluated the performance of an automated DNA extraction system from AF samples to avoid cross-contamination between samples and to remove PCR inhibitors. We also compared the sensitivity, specificity, and quantification performance for amplification of HCMV in AF of a commercial real-time HCMV PCR assay including a PCR inhibitor detection system with our in-house real-time HCMV PCR assay (10).
We tested 153 AF samples obtained from 153 women who presented with HCMV seroconversion in pregnancy and/or whose fetuses had ultrasound features compatible with HCMV infection. The samples were collected between 2002 and 2007 in two prenatal diagnosis centers in Poissy Hospital and in Necker Hospital. Among these 153 samples, 115 tested HCMV DNA negative and 38 tested HCMV DNA positive with our in-house HCMV PCR assay (10). According to French law, all women gave written consent for CMV detection by PCR in their AF.
In this retrospective study, DNA was extracted from 200 μl of the 153 AF samples on the Magna Pure LC Instrument (RocheMolecular Biochemicals, Meylan, France) using the Total NA serum-plasma kit (Roche Diagnostic), with extracted DNA eluted in 100 μl of the kit elution buffer. An extraction control was included in each batch. Each known positive sample was extracted between two negative samples to check for potential cross-contamination during extraction. DNA extracts from known positive samples were amplified undiluted and 1:10 diluted.
CMV DNA was amplified from these extracted DNA samples by two methods. For method 1, 5 μl of extracted DNA was amplified using an “in-house” CMV PCR test targeting the UL123 gene in 25 μl of mixture containing 1× Platinum qPCR superMix-UDG (Invitrogen, Cergy-Pontoise, France), 400 nM reverse and forward primers, and 200 nM probe (10). Amplification was performed in a DNA Engine Opticon (Bio-Rad, Marne La Coquette, France). Quantification was achieved with a commercial standard diluted to obtain a standard curve of 1,000 to 1,000,000 copies/ml (AD 169 DNA; Tebu-Bio, Perray-en-Yvelynes, France). The 95% sensitivity of this in-house quantitative CMV PCR test was 500 copies/ml (10). For method 2, 10 μl of DNA was amplified in 25 μl PCR mixture with the CMV R-gene kit according to the manufacturer (Argene, Varilhes, France). This kit was based on a duplex PCR which allowed in a single tube amplification of CMV DNA in the UL83 gene with a 6-carboxyfluorescein-labeled probe and of an inhibitor control with a VIC fluorochrome-labeled probe. Amplification was performed in an ABI PRISM 7300 (Applied Biosystems, Courtaboeuf, France). Four quantification standards (2,500 to 2,500,000 copies/ml) were supplied in the kit, and the 95% sensitivity of the kit was 150 copies/ml. The recommendations of the manufacturer for validation of the test results were as follows. (i) When CMV DNA amplification was negative and the value of the cycle threshold (CT) of the internal control (IC) obtained in the sample was less than 2 CT over the CT value obtained when the internal control was amplified alone, the sample was validated as CMV negative. (ii) When CMV DNA amplification was negative and the IC value incorrect, the sample had to be retested. (iii) When CMV DNA amplification was positive and the value of the IC was correct, the sample was validated as CMV positive and the viral load was considered as accurate. (iv) When CMV DNA amplification was positive and the IC value incorrect, the sample was validated as CMV positive but the viral load was considered as inaccurate and reamplification of a 1:100 or 1:1,000 dilution of the extracted DNA was recommended to obtain an accurate quantification of the viral load.
All 115 CMV-negative AF samples were found negative by both the in-house test and the commercial kit. The CT values of the internal controls obtained with the negative AF samples were within the expected range (see above), except for one sample. In this DNA extract, the IC control was >34; however, when the AF sample was reextracted the IC had the correct value. CMV DNA was detected by the two PCR assays in 38 samples, and these samples were validated as positive with both tests. The CT values of the IC were in the normal range (32 ± 2) for three positive AF samples (7.9%) when the extracted DNAs were amplified undiluted (the viral loads in those three samples were less than 105 copies/ml) and for nine AF samples (24%) when the 1:10-diluted extracted DNAs were amplified. (The viral loads of those nine AF samples were less than 106 copies/ml.) In the 26 other AF samples, the CT value of the IC was either undetectable or >34 when both 1:10-diluted and undiluted DNA extracts were amplified: in all of these AF samples, the viral load was over 106 copies/ml. Among these 26 samples, 19 were submitted to a second extraction and amplification using 1:100- and 1:1,000-diluted extracts. The IC values were correct for 15 samples when the 1:100 DNA extracts were used, and a 1:1,000 dilution was necessary to obtain a correct IC value in the further 4 samples.
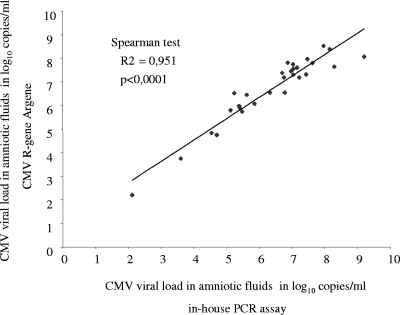
The viral loads obtained with the in-house technique and with the commercial kit were compared. We used the viral load value obtained with the smaller DNA extract dilution in which a correct amplification was obtained (e.g., CT value of the IC of 32 ± 2). The viral loads obtained for each sample with both tests were highly correlated when analyzed by the Spearman's rank test (r2 = 0.951, P < 0.0001) (Fig. 1). The median viral loads obtained with the commercial kit and with the “in-house” assay were 7.18 (range, 2.20 to 8.52) log10 copies/ml and 6.76 (range, 2.12 to 8.29) log10 copies/ml, respectively, with a median difference between viral load values of 0.42 (0.04 to 1.28) log10 copies/ml.
FIG. 1.
Correlation between HCMV viral loads in 31 of the 38 positive amniotic fluid samples after quantification by the in-house PCR assay and the CMV R-gene (Argene).
The new real-time PCR commercial assay had 100% sensitivity and 100% specificity for prenatal diagnosis of HCMV congenital infection in AF compared to an in-house HCMV real-time PCR. In this evaluation, DNA extraction was automated with no cross-contamination between samples. PCR inhibitors were detected in only one negative sample (1/115 [0.86%]) and did not persist when the sample was reanalyzed. As viral loads are generally very high in AF, extracted DNA had to be diluted to 1:100, or even to 1:1,000 for some samples, to obtain a correct IC value and therefore an accurate quantification of CMV DNA loads. Quantification of DNA loads in AF was highly correlated with both quantitative assays used. Quantification of HCMV DNA in AF could be of interest as it was shown in four studies that the median levels of HCMV DNA in AF were higher in symptomatic fetuses than in fetuses with subclinical infection, even if this difference did not always reach significance (6, 8, 12, 14). To our knowledge, no evaluation of a CMV commercial quantitative real-time PCR assay in AF has ever been reported before. We have demonstrated that the CMV R-gene is suitable for standardized prenatal diagnosis of CMV congenital infection.
Footnotes
Published ahead of print on 16 April 2008.
REFERENCES
- 1.Antsaklis, A. J., G. J. Daskalakis, S. A. Mesogitis, P. T. Koutra, and S. S. Michalas. 2000. Prenatal diagnosis of fetal primary cytomegalovirus infection. BJOG 10784-88. [DOI] [PubMed] [Google Scholar]
- 2.Avidor, B., G. Efrat, M. Weinberg, Z. Kra-Oz, J. Satinger, S. Mitrani-Rosenbaum, Y. Yaron, L. Shulman, M. Tepperberg-Oikawa, D. Wolf, S. A. Berger, S. Lipitz, E. Mendelson, and M. Giladi. 2004. Insight into the intrinsic sensitivity of the PCR assay used to detect CMV infection in amniotic fluid specimens. J. Clin. Virol. 29260-270. [DOI] [PubMed] [Google Scholar]
- 3.Azam, A. Z., Y. Vial, C. L. Fawer, J. Zufferey, and P. Hohlfeld. 2001. Prenatal diagnosis of congenital cytomegalovirus infection. Obstet. Gynecol. 97443-448. [DOI] [PubMed] [Google Scholar]
- 4.Bodeus, M., C. Hubinont, P. Bernard, A. Bouckaert, K. Thomas, and P. Goubau. 1999. Prenatal diagnosis of human cytomegalovirus by culture and polymerase chain reaction: 98 pregnancies leading to congenital infection. Prenat. Diagn. 19314-317. [DOI] [PubMed] [Google Scholar]
- 5.Enders, G., U. Bader, L. Lindemann, G. Schalasta, and A. Daiminger. 2001. Prenatal diagnosis of congenital cytomegalovirus infection in 189 pregnancies with known outcome. Prenat. Diagn. 21362-377. [DOI] [PubMed] [Google Scholar]
- 6.Gouarin, S., E. Gault, A. Vabret, D. Cointe, F. Rozenberg, L. Grangeot-Keros, P. Barjot, A. Garbarg-Chenon, P. Lebon, and F. Freymuth. 2002. Real-time PCR quantification of human cytomegalovirus DNA in amniotic fluid samples from mothers with primary infection. J. Clin. Microbiol. 401767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gouarin, S., P. Palmer, D. Cointe, S. Rogez, A. Vabret, F. Rozenberg, F. Denis, F. Freymuth, P. Lebon, and L. Grangeot-Keros. 2001. Congenital HCMV infection: a collaborative and comparative study of virus detection in amniotic fluid by culture and by PCR. J. Clin. Virol. 2147-55. [DOI] [PubMed] [Google Scholar]
- 8.Guerra, B., T. Lazzarotto, S. Quarta, M. Lanari, L. Bovicelli, A. Nicolosi, and M. P. Landini. 2000. Prenatal diagnosis of symptomatic congenital cytomegalovirus infection. Am. J. Obstet. Gynecol. 183476-482. [DOI] [PubMed] [Google Scholar]
- 9.Lazzarotto, T., S. Varani, B. Guerra, A. Nicolosi, M. Lanari, and M. P. Landini. 2000. Prenatal indicators of congenital cytomegalovirus infection. J. Pediatr. 13790-95. [DOI] [PubMed] [Google Scholar]
- 10.Leruez-Ville, M., M. Ouachée, R. Delarue, A.-S. Sauget, S. Blanche, A. Buzyn, and C. Rouzioux. 2003. Monitoring cytomegalovirus infection in adult and pediatric bone marrow transplant recipients by a real-time PCR assay performed with blood plasma. J. Clin. Microbiol. 412040-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liesnard, C., C. Donner, F. Brancart, F. Gosselin, M. L. Delforge, and F. Rodesch. 2000. Prenatal diagnosis of congenital cytomegalovirus infection: prospective study of 237 pregnancies at risk. Obstet. Gynecol. 95881-888. [DOI] [PubMed] [Google Scholar]
- 12.Picone, O., J. M. Costa, M. Leruez-Ville, P. Ernault, M. Olivi, and Y. Ville. 2004. Cytomegalovirus (CMV) glycoprotein B genotype and CMV DNA load in the amniotic fluid of infected fetuses. Prenat. Diagn. 241001-1006. [DOI] [PubMed] [Google Scholar]
- 13.Revello, M. G., and G. Gerna. 2002. Diagnosis and management of human cytomegalovirus infection in the mother, fetus, and newborn infant. Clin. Microbiol. Rev. 15680-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Revello, M. G., M. Zavattoni, M. Furione, F. Baldanti, and G. Gerna. 1999. Quantification of human cytomegalovirus DNA in amniotic fluid of mothers of congenitally infected fetuses. J. Clin. Microbiol. 373350-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]