Abstract
We have isolated a number of vanB-containing Enterococcus faecium isolates on bile esculin screening agar containing 6 mg/liter vancomycin, which on subsequent susceptibility testing using Etest have repeatedly demonstrated vancomycin MICs of ≤4 mg/liter. To investigate this genotype-phenotype incongruence of “low-MIC vancomycin-resistant enterococci” (LM-VRE), we examined the molecular characteristics of these isolates, including the presence of the vanB operon, using PCR amplification and DNA sequencing. All LM-VRE isolates contained vanB associated with the transposon Tn1549 and were polyclonal. Sequencing of the vanB ligase gene showed no differences from the prototype vanB2. In addition, we examined supplemented media to improve phenotypic detection of these isolates. Etest detection of LM-VRE improved when Mueller-Hinton agar (MHA) and brain heart infusion agar (BHIA) were supplemented with 10 g/liter oxgall (MHA-Oxg and BHIA-Oxg, respectively). We assessed the sensitivity and specificity of these media to detect vancomycin resistance using vancomycin-resistant vanB-containing E. faecium (n = 11), vancomycin-susceptible (van negative) E. faecium (n = 11), vancomycin-susceptible (van negative) E. faecalis (n = 11), and our LM-VRE (n = 23) isolates. After 48 h of incubation, both MHA-Oxg and BHIA-Oxg were 100% (34/34) sensitive and 100% (22/22) specific in the identification of vancomycin resistance. These findings suggest that supplementation of MHA or BHIA with 10 g/liter oxgall should be considered in laboratories where VRE detection protocols rely primarily on strain phenotype rather than early vanB gene detection by PCR.
Vancomycin-resistant enterococci (VRE) are significant nosocomial pathogens worldwide, and both vanA and vanB strains of VRE may result in serious infections, including bacteremia (15, 23). In Australia, unlike the United States and Europe, vanB-containing Enterococcus faecium predominates (7, 18). vanB-containing isolates show variable phenotypic resistance to vancomycin (MIC, 4 to 1,024 mg/liter) and can therefore be more difficult to detect than vanA-containing VRE that readily demonstrate high-level resistance to vancomycin (9, 10, 14, 20).
Both the vanA and vanB operons are associated with transposons, and transfer of resistance among enterococci appears to be mediated via acquisition of these elements (22). The vanA operon is located on the transposon Tn1546, while three transposons, Tn1547, Tn5382, and Tn1549, have been described which contain the vanB operon; sequencing data suggest Tn1549 is essentially identical to Tn5382 (22).
Using Enterococcosel agar (BBL, Sparks, MD) containing 6 mg/liter vancomycin (EVA) for VRE screening, we have identified a number of vanB-containing E. faecium isolates that on subsequent susceptibility testing using the standard Etest (AB Biodisk, Solna, Sweden) method (inoculum of 0.5 McFarland standard [0.5 McF] on Mueller-Hinton agar [MHA]) have repeatedly demonstrated vancomycin MICs of ≤4 mg/liter. This difficulty in phenotypic identification of “low-MIC VRE” (LM-VRE) may result in misidentification, potentially with important clinical and infection control consequences (9).
We present here a description of the resistance genotype-phenotype incongruence of these variants of VRE, including molecular characteristics. These LM-VRE strains can be detected on EVA but not on MHA or brain heart infusion agar (BHIA) using Etest, and either of the manufacturer's two recommended methods (standard susceptibility testing for enterococci [MHA, inoculum of 0.5 McF] and VRE screening [BHIA, inoculum of 2 McF]). We therefore assessed the performance of these agars when supplemented with various components of EVA that are not found in MHA or BHIA. Using this approach, we aimed to develop a simple modification to MHA and BHIA that could be readily used for accurate screening for VRE.
(This work was presented in part at the 45th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, 16 to 19 December 2005.)
MATERIALS AND METHODS
Isolates.
We identified 23 E. faecium isolates from fecal or rectal swab screening specimens following culture on EVA (containing 6 mg/liter vancomycin). These isolates were subsequently cultured on horse blood agar (Columbia blood base; Oxoid, Basingstoke, United Kingdom) and found to be vancomycin susceptible (MIC, ≤4 mg/liter) using the Etest according to the manufacturer's standard methods (MHA; inoculum of 0.5 McF), with CLSI criteria for breakpoints. However, they contained vanB when assessed by PCR (1, 8, 12, 13). We defined these isolates as LM-VRE.
All 23 LM-VRE isolates were compared to 11 vancomycin-resistant (MIC, ≥8 mg/liter) vanB-containing E. faecium strains (VRE), 11 vancomycin-susceptible (MIC, ≤4 mg/liter) van-negative E. faecium strains (VSEfm), 11 vancomycin-susceptible (MIC, ≤4 mg/liter) van-negative E. faecalis strains (VSEfs), and 5 ATCC control strains (vanB1-containing E. faecalis [ATCC 51299], vancomycin-susceptible E. faecalis [ATCC 29212], E. faecium [ATCC 35667], E. gallinarum [ATCC 700425], and E. casseliflavus [ATCC 700327]). All non-ATCC isolates were obtained from fecal or rectal swab specimens collected from patients being screened for VRE colonization or from clinical specimens of urine, blood, and/or biliary fluid.
Studies of antimicrobial susceptibility using various medium supplements.
In a preliminary assessment of the LM-VRE isolates, susceptibility to vancomycin was assessed using an inoculum of 0.5 McF with Etest (AB Biodisk, Solna, Sweden) on MHA and BHIA in which the media were supplemented with either 10 g/liter oxgall (MHA-Oxg and BHIA-Oxg; dehydrated bile [BBL]), 0.05 g/liter ammonium ferric citrate (AFC) (MHA-AFC and BHIA-AFC; BDH, Poole, United Kingdom), or 1 g/liter esculin (MHA-E or BHIA-E; BDH) since these compounds are present in EVA but not in MHA and BHIA. All cultures were incubated at 35°C and assessed after 24 h and 48 h (1, 12). The following definitions were used for vancomycin and teicoplanin susceptibility: vancomycin susceptible, MIC of ≤4 mg/liter, and vancomycin intermediate or resistant, MIC of ≥8 mg/liter; and teicoplanin susceptible, MIC of ≤8 mg/liter, and teicoplanin intermediate or resistant, MIC of ≥16 mg/liter.
Subsequently, the susceptibilities to vancomycin and teicoplanin of all isolates (LM-VRE, n = 23; VSEfm, n = 11; VSEfs, n = 11; and VRE, n = 11) were tested in duplicate by Etest, agar dilution, and automated Vitek I procedures (BioMérieux; Durham, NC). Because the media, inoculum sizes, and incubation periods specified by AB Biodisk for standard and VRE screening methods for enterococcal glycopeptide susceptibility testing vary, we assessed susceptibility using Etest after 24 and 48 h of incubation on MHA and MHA-Oxg using an inoculum of 0.5 McF and BHIA and BHIA-Oxg using an inoculum of 2 McF (1, 2). Using agar dilution and CLSI guidelines (19), susceptibility to vancomycin and teicoplanin (inoculum of 104 CFU, serial doubling dilutions of 1 to 64 mg/liter for both agents) were assessed on MHA, MHA-Oxg, BHIA containing 6 mg/liter vancomycin (BHIV6-AD), and EVA. In addition 10-μl aliquots of the inoculum of 0.5 McF were inoculated onto BHIA containing 6 mg/liter vancomycin (BHIV6-S) as per CLSI recommendations for screening for vancomycin resistance (19). Susceptibility was also tested using Vitek I (software version R09.01) and the GPS-431 card (BioMérieux).
PFGE.
All 56 clinical isolates were assessed by pulsed-field gel electrophoresis (PFGE) using SmaI DNA digestion, with gels analyzed by GelCompar II (Applied Maths, Sint-Martens-Latem, Belgium) using the Dice coefficient and unweighted pair group method using average linkages with settings for tolerance and optimization of 1% and 0.75%, respectively (6, 17). Isolates were considered to be in the same PFGE group if band-based similarity was greater than 80% (17).
PCR amplification, DNA sequencing, and analysis.
Species identification of all 56 clinical isolates was confirmed by multiplex PCR for the d-Ala-d-Ala ligase gene (13). The glycopeptide resistance genotype was determined by multiplex PCR for vanA, vanB, and vanC (8). Total genomic DNA was extracted using the Qiagen Dneasy tissue kit according to the manufacturer's instructions (Qiagen Pty., Ltd., Victoria, Australia). The resistance genotype of the LM-VRE isolates was confirmed by amplification of the Tn5382/1549-vanB operon by long-range PCR as previously described (6). This PCR utilized the primers 16924 and 16925 and comprised the entire vanB operon with flanking DNA derived from Tn5382/1549 to give an amplicon size of 6,729 bp (6). The Tn5382/1549-vanB operon amplicon was also used as a template for sequencing of the vanB ligase gene from a representative isolate of each PFGE group as described previously (5).
RESULTS
Description of LM-VRE isolates.
LM-VRE isolates were confirmed as vanB-positive E. faecium by multiplex PCR and grew on nonsupplemented horse blood agar and EVA. The results of vancomycin susceptibility testing are shown in Table 1. The vancomycin MICs of LM-VRE ranged from 1.5 mg/liter to 6 mg/liter (MIC50, 2 mg/liter; MIC90, 3 mg/liter), and teicoplanin MICs ranged from 0.5 mg/liter to 1.5 mg/liter after 24 h of incubation on MHA. This vancomycin-susceptible phenotype was confirmed on repeat testing.
TABLE 1.
Comparison of vancomycin susceptibilities of isolates tested using Etest and agar dilution with 24- and 48-h incubations
| Isolate group (n) | MIC90 (MIC50) in mg/liter by:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Etest (inoculum of 0.5 McF)
|
Etest (inoculum of 2 McF)
|
Agar dilution
|
||||||||||
| MHA
|
MHA-Oxg
|
BHIA
|
BHIA-Oxg
|
MHA
|
MHA-Oxg
|
|||||||
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |
| LM-VRE (23) | 3 (2) | 8 (2) | 32 (8) | 64 (32) | 16 (6) | 32 (16) | 32 (24) | 48 (32) | 2 (1) | 2 (2) | 16 (4) | 16 (4) |
| VSEfm (11) | 1.5 (1) | 1.5 (1) | 3 (2) | 3 (2) | 3 (3) | 3 (3) | 3 (3) | 4 (3) | 1 (1) | 1 (1) | 1 (1) | 2 (2) |
| VSEfs (11) | 3 (2) | 3 (2) | 3 (3) | 4 (3) | 6 (4) | 8 (4) | 6 (4) | 8 (4) | 2 (1) | 2 (1) | 4 (2) | 4 (2) |
| VRE (11) | 96 (32) | ≥256 (≥256) | ≥256 (≥256) | ≥256 (≥256) | ≥256 (96) | ≥256 (≥256) | ≥256 (≥256) | ≥256 (≥256) | 32 (16) | 64 (32) | 64 (64) | ≥128 (≥128) |
LM-VRE isolates belonged to seven different PFGE groups, suggesting the LM-VRE isolates were polyclonal. Some of these PFGE groups also contained VRE isolates.
Studies of antimicrobial susceptibility using various media supplements. (i) Preliminary medium studies.
Etest detected only 6/23 and 4/23 LM-VRE isolates using nonsupplemented MHA and BHIA, respectively, after 48 h of incubation. MHA or BHIA supplemented with AFC or esculin did not improve detection of LM-VRE (data not shown). However, the vancomycin Etest using MHA-Oxg accurately identified resistance in 14/23 and 23/23 LM-VRE isolates after 24 and 48 h of incubation, respectively. Similarly, Etest assessment on BHIA-Oxg identified vancomycin resistance in 14/23 and 21/23 LM-VRE isolates after 24 and 48 h, respectively.
(ii) Studies using media with and without oxgall supplementation.
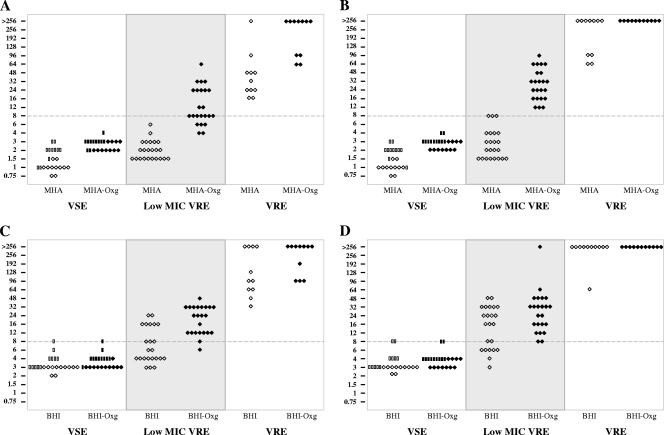
The MIC data from both Etest methods for all patient isolates are presented in Fig. 1 and summarized with the agar dilution MIC data as MIC90 and MIC50 in Table 1. As expected, the Etest on nonsupplemented media performed poorly in identifying LM-VRE on MHA (0/23 and 3/23 isolates after 24 and 48 h incubation, respectively) and BHIA (10/23 and 16/23 isolates after 24 and 48 h of incubation, respectively). In contrast, after 48 h of incubation, the sensitivity and specificity of MHA-Oxg were excellent (100% and 100%, respectively) and those of BHIA-Oxg (100% and 91%, respectively) were very good.
FIG. 1.
Vancomycin MIC by Etest on selected media with and without 10-g/liter oxgall supplementation. Open symbols represent E. faecium (diamonds) and E. faecalis (rectangles) isolates tested on medium without oxgall supplementation. Solid symbols represent E. faecium (diamonds) and E. faecalis (rectangles) isolates tested on oxgall-supplemented media. (A) MHA and MHA-Oxg at 24 h of incubation. (B) MHA and MHA-Oxg at 48 h of incubation. (C) BHIA and BHIA-Oxg at 24 h of incubation. (D) BHIA and BHIA-Oxg at 48 h of incubation.
Using agar dilution, after 48 h of incubation, vancomycin MICs for LM-VRE isolates were 1 to 4 mg/liter (MHA) and 2 to 16 mg/liter (MHA-Oxg). All 11 VSEfm and 11 VSEfs strains were vancomycin susceptible (MHA MICs of 1 to 2 mg/liter and MHA-Oxg MICs of 1 to 4 mg/liter), while all 11 VRE isolates were vancomycin intermediate or resistant (MHA MICs of 16 to 64 mg/liter and MHA-Oxg MICs of 64 to ≥128 mg/liter). Therefore, for detection of vancomycin resistance by agar dilution using MHA-Oxg, sensitivity was limited (21/34 [62%]), but specificity was good (22/22 [100%]).
Only 4/23 LM-VRE isolates grew on BHIV6-AD after 48 h of incubation. All VRE isolates but none of the VSE isolates grew on BHIV6-AD and EVA screening plates after 48 h. Sensitivity and specificity were 44% (15/34) and 100% (22/22), respectively, for BHIV6-AD. Similar results were noted on the BHIV6 with the higher inoculum (BHIV6-S): sensitivity and specificity were 41% (14/34) and 100% (22/22), respectively. In contrast, EVA was 100% (34/34) sensitive and 100% (22/22) specific for detection of vancomycin resistance. Similarly, Vitek I susceptibility testing was 100% sensitive and 100% specific.
All patient isolates were teicoplanin susceptible on unsupplemented media. However, oxgall-supplemented media substantially altered the observed susceptibility to teicoplanin of all isolates using both Etest and agar dilution. Use of Etest on MHA-Oxg, demonstrated teicoplanin MICs of 24 to 48 mg/liter, 8 to 32 mg/liter, 16 to 64 mg/liter, and 32 to 128 mg/liter for VSEfm, VSEfs, LM-VRE, and VRE, respectively, after 24 h of incubation, with similar findings noted for BHIA-Oxg (data not shown). Elevated teicoplanin MICs were also observed using agar dilution with MHA-Oxg: MIC range, 8 to 32 mg/liter. All patient isolates were teicoplanin susceptible by Vitek I analysis.
PCR amplification, DNA sequencing, and analysis.
All LM-VRE isolates demonstrated an ∼7-kb Tn5382/1549-vanB amplicon by PCR, confirming that all isolates contained vancomycin resistance associated with Tn1549, with no evidence for major insertions or deletions based on the product size obtained. Using the Tn5382/1549-vanB amplicon as a template, the sequences of the vanB ligase gene in LM-VRE were demonstrated to be 99 to 100% identical to the sequence of prototype vanB2 in Tn1549 (GenBank accession no. AF192329).
DISCUSSION
This is the first report of a large polyclonal series of vancomycin-resistant E. faecium isolates that are phenotypically susceptible to vancomycin (as determined by Etest) but carry a vanB2 ligase gene associated with a Tn1549-like element. They are not vancomycin dependent as all isolates grew well on nonsupplemented horse blood agar (16).
We found that oxgall supplementation of both MHA and BHIA substantially improved Etest detection of LM-VRE (100% sensitivity and 100% specificity) in this collection of 23 isolates. In comparison, both supplementation of MHA with oxgall and use of the BHIV6-AD and BHIV6-S in agar dilution were less sensitive for detection of vancomycin resistance (62% versus 44% and 41%, respectively) but had a specificity of 100% for each. The Vitek I method readily discriminated LM-VRE from VSE isolates (sensitivity, 100%; specificity, 100%). Our study has some potential limitations. Notably, oxgall appears to induce resistance to teicoplanin, with all 56 strains that were initially teicoplanin susceptible using nonsupplemented media being observed as teicoplanin resistant using Etest on MHA-Oxg or BHIA-Oxg. Oxgall-supplemented medium is not recommended for teicoplanin testing, and the explanation for this observation is unknown.
We found that LM-VRE isolates contained vanB2 associated with a Tn5382/Tn1549 element. The ligase gene was identical to that of VRE, with no gross structural changes in the vanB operon based on the size of the PCR product obtained. Subtle sequence or functional changes in the operon, particularly in the vanS/vanR regulatory system, may account for the vancomycin-susceptible phenotype observed with LM-VRE (4), as might operon copy number and the presence of remnant fragments within the genome of these isolates (3, 11, 21).
Subsequent to this study, we have found that LM-VRE is a significant problem in our institution, representing up to 25% of new VRE isolates. Oxgall supplementation of MHA or BHIA improves detection of these LM-VRE by Etest and therefore may be useful in laboratories where VRE detection protocols are reliant on strain phenotype or prioritize phenotype detection ahead of van genotyping by PCR.
Footnotes
Published ahead of print on 23 April 2008.
REFERENCES
- 1.AB Biodisk. 2000. Etest technical guide M0000199. Detection of glycopeptide resistance. AB Biodisk, Solna, Sweden.
- 2.AB Biodisk. 2007. Etest application guide M0000187 MH0245. AB Biodisk, Solna, Sweden.
- 3.Arthur, M., F. Depardieu, and P. Courvalin. 1999. Regulated interactions between partner and non-partner sensors and response regulators that control glycopeptide resistance gene expression in enterococci. Microbiology 1451849-1858. [DOI] [PubMed] [Google Scholar]
- 4.Arthur, M., and R. Quintiliani, Jr. 2001. Regulation of VanA- and VanB-type glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 45375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballard, S. A., E. A. Grabsch, P. D. R. Johnson, and M. L. Grayson. 2005. Comparison of three PCR primer sets for the identification of vanB gene carriage in feces and correlation with carriage of vancomycin-resistant enterococci: interference by vanB-containing anaerobic bacilli. Antimicrob. Agents Chemother. 4977-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballard, S. A., K. K. Pertile, M. Lim, P. D. R. Johnson, and M. L. Grayson. 2005b. Molecular characterization of vanB elements in naturally occurring gut anaerobes. Antimicrob. Agents Chemother. 491688-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell, J., J. Turnidge, G. Coombs, and F. O'Brien. 1998. Emergence and epidemiology of vancomycin-resistant enterococci in Australia. Commun. Dis. Intell. 22249-252. [PubMed] [Google Scholar]
- 8.Bell, J. M., J. C. Paton, and J. Turnidge. 1998. Emergence of vancomycin-resistant enterococci in Australia: phenotypic and genotypic characteristics of isolates. J. Clin. Microbiol. 362187-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cetinkaya, Y., P. Falk, and C. G. Mayhall. 2000. Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13686-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chavers, L. S., S. A. Moser, W. H. Benjamin, Jr., S. E. Banks, J. R. Steinhauer, A. M. Smith, C. N. Johnson, E. Funkhouser, L. P. Chavers, A. M. Stamm, and K. B. Waites. 2003. Vancomycin-resistant enterococci: 15 years and counting. J. Hosp. Infect. 53159-171. [DOI] [PubMed] [Google Scholar]
- 11.Choi, J. Y., K. H. Chang, K. S. Lee, Y. G. Song, and J. M. Kim. 2004. Increasing vancomycin susceptibility in vancomycin resistant enterococci by vanH promoter and ddl transformation. J. Infect. 48314-319. [DOI] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing: fifteenth informational supplement. M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 13.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 3324-27. (Erratum, 33:1434.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gold, H. S., and R. C. Moellering, Jr. 1996. Antimicrobial-drug resistance. N. Engl. J. Med. 3351445-1453. [DOI] [PubMed] [Google Scholar]
- 15.Goossens, H., D. Jabes, R. Rossi, C. Lammens, G. Privitera, and P. Courvalin. 2003. European survey of vancomycin-resistant enterococci in at-risk hospital wards and in vitro susceptibility testing of ramoplanin against these isolates. J. Antimicrob. Chemother. 51(Suppl. 3)iii5-iii12. [DOI] [PubMed] [Google Scholar]
- 16.Green, M., J. H. Shlaes, K. Barbadora, and D. M. Shlaes. 1995. Bacteremia due to vancomycin-dependent Enterococcus faecium. Clin. Infect. Dis. 20712-714. [DOI] [PubMed] [Google Scholar]
- 17.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains J. Clin. Microbiol. 411574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mutnick, A. H., D. J. Biedenbach, and R. N. Jones. 2003. Geographic variations and trends in antimicrobial resistance among Enterococcus faecalis and Enterococcus faecium in the SENTRY Antimicrobial Surveillance Program (1997-2000). Diagn. Microbiol. Infect. Dis. 4663-68. [DOI] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptiblity tests for bacteria that grow aerobically, 6th ed. M7-A6. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 20.Teixeira, L. M., and R. R. Facklam. 2003. Enterococcus, p. 422-433. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. F. Yolken (ed.), Manual of clinical microbiology, 8th ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 21.Torres Viera, C., S. Tsiodras, H. S. Gold, E. P. G. Coakley, C. Wennersten, G. M. Eliopoulos, R. C. Mollering, Jr., and R. T. Inouye. 2001. Restoration of vancomycin susceptibility in Enterococcus faecalis by antiresistance determinant gene transfer. Antimicrob. Agents Chemother. 45973-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weaver, K. E., L. B. Rice, and G. Churchward. 2002. Plasmids and transposons, p. 219-263, In D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (ed.), The enterococci, ASM Press, Washington, DC.
- 23.Worth, L. J., K. A. Thursky, J. F. Seymour, and M. A. Slavin. 2007. Vancomycin-resistant Enterococcus faecium infection in patients with hematologic malignancy: patients with acute myeloid leukemia are at high-risk. Eur. J. Haematol. 79226-233. [DOI] [PubMed] [Google Scholar]