Abstract
The Klebsiella pneumoniae carbapenemase (KPC) was detected in carbapenem-resistant isolates of Citrobacter freundii and Klebsiella oxytoca recovered from different patients in a Michigan hospital. Restriction analysis and hybridization with a KPC-specific probe showed the blaKPC-2 genes of these two genera of the family Enterobacteriaceae are carried on a common plasmid.
The numbers of carbapenem-hydrolyzing β-lactamases in members of the family Enterobacteriaceae are increasing in the United States (4, 11, 19). The most frequently encountered are the plasmid-encoded Ambler class A Klebsiella pneumoniae carbapenemase (KPC)-type enzymes found in isolates predominantly from the eastern United States (2, 5, 11, 15, 24), particularly from the New York City region (4). More recently, the geographical distribution of KPC-producing isolates within the United States has widened to include Pennsylvania, Ohio, and Arkansas (11, 18) and Georgia, Colorado, New Mexico, Arizona, and California (CDC, unpublished data). KPC-producing Escherichia coli and K. pneumoniae isolates that are thought to have originated outside of the United States have been reported in Israel (17), Colombia (21), Greece (10), and China (23).
KPC was first identified in a K. pneumoniae isolate from North Carolina (24), and the enzyme has been found the most frequently in K. pneumoniae (2, 5, 6). In addition, KPC enzymes have been detected in multiple genera and species of the Enterobacteriaceae, including Salmonella enterica serotype Cubana (14), K. oxytoca (2, 11, 25), Enterobacter spp. (3, 11, 13), Citrobacter freundii, E. coli, and Serratia marcescens (11, 12). A recent report from Colombia also describes KPC-producing isolates of Pseudomonas aeruginosa (22).
In this report, we describe the characterization of two carbapenem-resistant isolates, a C. freundii isolate and a K. oxytoca isolate, obtained from two different patients in a Michigan hospital and found to produce the KPC type 2 (KPC-2) enzyme encoded by a common plasmid.
In November 2004, C. freundii 13692 was isolated from the urine of a patient hospitalized for complications of chronic liver disease. This isolate was found, by the reference broth microdilution method (7), to be susceptible to amikacin but resistant to all other antibiotics tested, including meropenem (MIC = 16 μg/ml). The patient was treated with intravenous amikacin, and subsequent urine cultures were negative for C. freundii. Due to underlying disease and multisystem organ failure, the patient died on hospital day 75. Prior to culture, this patient had received multiple antibacterial agents for treatment and prophylaxis, including piperacillin-tazobactam, ampicillin-sulbactam, cefepime, ceftriaxone, levofloxacin, vancomycin, metronidazole, and aminoglycosides. In August 2005, K. oxytoca 15002 was isolated in the same hospital from the sputum of a second patient who had undergone cardiac transplantation and who developed pneumonia. This isolate was tested by disk diffusion (8) and was found to be susceptible to gentamicin, tobramycin, amikacin, and trimethoprim-sulfamethoxazole; intermediate to ciprofloxacin and levofloxacin; and resistant to all other antibiotics tested, including meropenem. The patient was treated with inhaled colistin (25 days), intravenous gentamicin (18 days), and intravenous tigecycline (18 days); and the pneumonia resolved. Subsequent sputum cultures showed meropenem-susceptible K. oxytoca on one occasion without associated clinical respiratory disease. The patient was discharged from the hospital on hospital day 104. Prior to the collection of samples for culture, the patient was exposed to the following antibacterials: piperacillin-tazobactam, cefepime, cefuroxime, cefazolin, aztreonam, levofloxacin, linezolid, metronidazole, clindamycin, rifampin, and dapsone. Both patients had multiple previous hospitalizations, and their hospital stays overlapped during 2 weeks in October 2004. Both patients developed renal failure requiring hemodialysis; the first patient received continuous renal replacement therapy (CRRT) administered in the hospital intensive care unit (ICU) room from 21 October through 13 December 2004, and the second patient received intermittent hemodialysis (HD), which began on 24 July 2005 and which was administered in an ICU different from that where the first patient was located. The patients shared no equipment or personnel, as different machines and dialysates are used for CRRT and HD. No other common procedures, wards, personnel, or epidemiological links were identified. Neither patient was treated with carbapenems.
Antimicrobial susceptibility testing at the CDC by the CLSI reference broth microdilution method (7, 9) confirmed that C. freundii 13692 was highly resistant to all three carbapenems tested (MICs ≥ 16 μg/ml) (Table 1) and that K. oxytoca 15002 was resistant to ertapenem and meropenem (MICs > 16 μg/ml) but intermediate to imipenem (MIC = 8 μg/ml). Both isolates were also resistant to all other β-lactams and fluoroquinolones tested; but the two isolates demonstrated different susceptibilities to aminoglycosides, chloramphenicol, and trimethoprim-sulfamethoxazole. Both isolates were susceptible to colistin and tigecycline.
TABLE 1.
Antimicrobial susceptibilities of clinical isolates and transformants
| Antimicrobial agent | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|
| C. freundii 13692 | E. coli TF/13692 | K. oxytoca 15002 | E. coli TF/15002 | E. colia | |
| Ertapenem | >16 | >16 | >16 | >16 | ≤0.5 |
| Imipenem | 16 | >16 | 8 | 32 | ≤1 |
| Meropenem | 16 | >16 | >16 | >16 | ≤0.25 |
| Amikacin | 2 | 2 | ≤1 | 2 | 2 |
| Amoxicillin-clavulanate | >32/16 | >32/16 | >32/16 | >32/16 | 4/2 |
| Ampicillin | >64 | >32 | >64 | >32 | 4 |
| Aztreonam | >64 | >64 | >64 | >64 | ≤1 |
| Cefepime | >32 | >32 | >32 | >32 | ≤0.5 |
| Cefotaxime | >256 | 256 | 256 | 256 | 2 |
| Cefotaxime-clavulanate | 128/4 | 32/4 | 2/4 | 32/4 | |
| Cefoxitin | >32 | >32 | 32 | >32 | 4 |
| Cefpodoxime | >16 | >16 | >16 | >16 | 1 |
| Ceftazidime | >256 | 64 | 32 | 128 | ≤4 |
| Ceftazidime-clavulanate | 256/4 | 64/4 | 16/4 | 64/4 | ≤2/4 |
| Ceftriaxone | >64 | >64 | >64 | >64 | ≤1 |
| Chloramphenicol | >16 | ≤2 | 8 | ≤2 | ≤2 |
| Ciprofloxacin | >8 | ≤0.25 | 8 | ≤0.12 | ≤0.12 |
| Colistin | 1 | 0.5 | 1 | 2 | 1 |
| Gentamicin | 16 | 0.5 | ≤0.25 | 0.5 | 1 |
| Levofloxacin | >8 | ≤0.25 | 8 | ≤0.25 | ≤0.25 |
| Piperacillin-tazobactam | >128/4 | >128/4 | >64/4 | >128/4 | 2/4 |
| Polymyxin B | 1 | 0.5 | 1 | ≤0.5 | 1 |
| Tigecycline | 1 | 0.12 | 1 | 0.12 | 0.12 |
| Tobramycin | 16 | 0.5 | ≤0.25 | 0.5 | 0.5 |
| Trimethoprim-sulfamethoxazole | >8/152 | ≤0.25/4.75 | ≤0.25/4.75 | ≤0.25/4.75 | ≤0.25/4.75 |
E. coli EP-Max 10B competent cells were used for transformation by electroporation.
The β-lactamases of both isolates were characterized by isoelectric focusing (20) of cell extracts. C. freundii 13692 produced two β-lactamases with isoelectric points (pIs) of >8.4 and 6.9, consistent with AmpC and KPC-type enzymes, respectively. K. oxytoca 15002 produced only one detectable enzyme with a pI of 6.9. The presence of the blaKPC gene in both isolates was confirmed by amplification of a 1,011-bp PCR product by using forward primer 5′-TGT CAC TGT ATC GCC GTC-3′ and reverse primer 5′-GTC AGT GCT CTA CAG AAA ACC-3′. The DNA sequences of both strands of the coding region of the blaKPC gene were determined from a 989-bp PCR product amplified with forward primer 5′-GCT ACA CCT AGC TCC ACC TTC-3′ and reverse primer 5′-ACA GTG GTT GGT AAT CCA TGC-3′. The DNA sequence, determined from independent amplification reactions with previously described oligonucleotide primers (24), confirmed the presence of the blaKPC-2 gene in both C. freundii 13692 and K. oxytoca 15002.
Plasmid analysis revealed that C. freundii 13692 contained three plasmids with molecular sizes of approximately 165, 145, and 95 kb, as well as an additional one of <2.1 kb (data not shown); K. oxytoca 15002 contained two plasmids of approximately 145 and 95 kb. To isolate the KPC-producing plasmid, E. coli EP-Max 10B cells (Bio-Rad, Hercules, CA) were transformed with plasmid DNA from C. freundii 13692 and K. oxytoca 15002 by electroporation (Gene Pulser Xcell; Bio-Rad). Transformants of both C. freundii 13692 (TF/13692) and K. oxytoca 15002 (TF/15002) were selected on LB agar containing 4 μg/ml meropenem. Transformants TF/15002 and TF/13692 each produced only one β-lactamase with a pI of 6.9, consistent with that of the KPC enzyme, and were positive for blaKPC by PCR. Both also contained the ca. 95-kb plasmid seen in the clinical isolates, but the latter also contained the smallest plasmid found in C. freundii 13692, which was <2.1 kb. Only β-lactam resistance was transferred in each transformation, including resistance to imipenem, meropenem, and ertapenem (MICs ≥ 16 μg/ml) (Table 1).
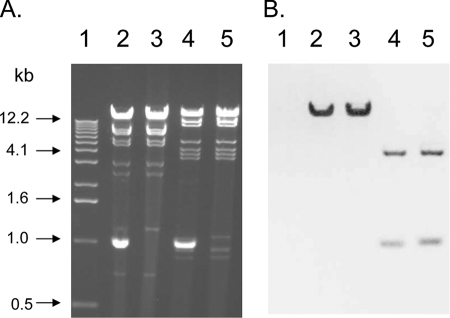
Plasmid DNA from both transformants was compared by restriction digestion with EcoRI and HindIII (Fig. 1A). The <2.1-kb plasmid from TF/13692 was not cut by either enzyme (Fig. 1A, lanes 2 and 4). The restriction profiles of the ca. 95-kb plasmid DNA prepared from TF/13692 and TF/15002 were identical (Fig. 1A, lanes 2 and 3, respectively [EcoRI], and lanes 4 and 5 [HindIII], respectively). To localize the blaKPC gene, digested plasmid DNA from transformants TF/13692 and TF/15002 was hybridized with a blaKPC probe in a Southern blot analysis. Briefly, by using the Genius nonradioactive nucleic acid labeling and detection system (Roche, Indianapolis, IN), a 552-bp digoxigenin-labeled blaKPC fragment generated with forward primer 5′-CACACCCATCCGTTACGG-3′ and reverse primer 5′-GCCTCGCTGTGCTTGTCA-3′ was hybridized overnight at 65°C to EcoRI- or HindIII-digested plasmid DNA which had been transferred to a nylon membrane (Zeta-Probe; Bio-Rad). The blaKPC probe hybridized with a single fragment larger than 12.2 kb in the EcoRI-restricted plasmid DNA from both TF/13692 and TF/15002 (Fig. 1B, lanes 2 and 3, respectively). The probe also hybridized with two fragments of approximately 4.1 kb and 1.1 kb in the HindIII-restricted DNA (Fig. 1B, lanes 4 and 5, respectively). These results are consistent with the restriction profile of the blaKPC-2 gene and flanking sequences reported from S. enterica serotype Cubana (GenBank accession number AF481906) (14).
FIG. 1.
(A) Restriction digests of plasmid DNA from E. coli transformants of C. freundii 13692 and K. oxytoca 15002 obtained with EcoRI (lanes 2 and 3, respectively) and HindIII (lanes 4 and 5, respectively). Lane 1 contains a 1-kb ladder size standard. (B) Southern blot hybridized with a blaKPC-specific probe.
In this report we described a ca. 95-kb, KPC-producing plasmid that was common to two different genera of Enterobacteriaceae from two different patients in a single hospital. These findings demonstrate the potential for the horizontal transfer of carbapenemase-producing plasmids between clinically relevant gram-negative bacilli. We were not able to conjugate this resistance plasmid to E. coli in the laboratory, as has been demonstrated for other KPC plasmids (13-16, 25). This means that plasmid transfer occurred either by transformation or by a conjugative event that could not be duplicated in vitro. No other KPC-producing isolates have been identified at this institution, so these two isolates could be considered a “warning signal” that more resistance could emerge. There was no clear epidemiological link between the two patients described in this report, which makes the identification of prevention strategies difficult. However, neither of the patients described in this report was treated with a carbapenem. This suggests that restricting carbapenem use may have little impact on limiting the dissemination of this carbapenemase. In any case, it is important to recognize that the KPC enzyme can occur in Enterobacteriaceae other than K. pneumoniae and that these isolates have the potential to transmit the carbapenemase.
Hospital laboratories should suspect carpbapenemase (e.g., KPC) production in an isolate of Enterobacteriaceae when it tests nonsusceptible to a carbapenem or when the carbapenem MIC is elevated but the isolate remains susceptible (e.g., a meropenem or imipenem MIC of >1 μg/ml) (9). Carbapenemase production can be confirmed by performing a phenotypic carbapenemase test, such as the modified Hodge test, and/or by detection of the carbapenemase gene by PCR (1). Clinical microbiology laboratories should promptly report any isolates with a carbapenemase phenotype to the hospital infection control department so that the use of appropriate contact isolation precautions may be considered.
Acknowledgments
We thank Arjun Srinivasan, Linda Weigel, and Brandon Kitchel for helpful discussions.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 2 April 2008.
REFERENCES
- 1.Anderson, K. F., D. R. Lonsway, J. K. Rasheed, J. Biddle, B. Jensen, L. K. McDougal, R. B. Carey, A. Thompson, S. Stocker, B. Limbago, and J. B. Patel. 2007. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J. Clin. Microbiol. 452723-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, P. A., S. Bratu, C. Urban, M. Visalli, N. Mariano, D. Landman, J. J. Rahal, S. Brooks, S. Cebular, and J. Quale. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin. Infect. Dis. 3955-60. [DOI] [PubMed] [Google Scholar]
- 3.Bratu, S., D. Landman, M. Alam, E. Tolentino, and J. Quale. 2005. Detection of KPC carbapenem-hydrolyzing enzymes in Enterobacter spp. from Brooklyn, New York. Antimicrob. Agents Chemother. 49776-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bratu, S., D. Landman, R. Haag, R. Recco, A. Eramo, M. Alam, and J. Quale. 2005. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City. Arch. Intern. Med. 1651430-1435. [DOI] [PubMed] [Google Scholar]
- 5.Bratu, S., M. Mooty, S. Nichani, D. Landman, C. Gullans, B. Pettinato, U. Karumudi, P. Tolaney, and J. Quale. 2005. Emergence of KPC-possessing Klebsiella pneumoniae in Brooklyn, New York: epidemiology and recommendations for detection. Antimicrob. Agents Chemother. 493018-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bratu, S., P. Tolaney, U. Karumudi, J. Quale, M. Mooty, S. Nichani, and D. Landman. 2005. Carbapenemase-producing Klebsiella pneumoniae in Brooklyn, N.Y.: molecular epidemiology and in vitro activity of polymyxin B and other agents. J. Antimicrob. Chemother. 56128-132. [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 7th ed. CLSI document M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial disk susceptibility tests; approved standard, 9th ed. CLSI document M2-A9. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement. CLSI document M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Cuzon, G., T. Naas, M. C. Demachy, and P. Nordmann. 2008. Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC-2 in Klebsiella pneumoniae isolate from Greece. Antimicrob. Agents Chemother. 52796-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deshpande, L. M., P. R. Rhomberg, H. S. Sader, and R. N. Jones. 2006. Emergence of serine carbapenemases (KPC and SME) among clinical strains of Enterobacteriaceae isolated in the United States medical centers: report from the MYSTIC Program (1999-2005). Diagn. Microbiol. Infect. Dis. 56367-372. [DOI] [PubMed] [Google Scholar]
- 12.Hong, T., E. S. Moland, B. Abdalhamid, N. D. Hanson, J. Wang, C. Sloan, D. Fabian, A. Farajallah, J. Levine, and K. S. Thomson. 2005. Escherichia coli: development of carbapenem resistance during therapy. Clin. Infect. Dis. 40e84-e86. [DOI] [PubMed] [Google Scholar]
- 13.Hossain, A., M. J. Ferraro, R. M. Pino, R. B. Dew III, E. S. Moland, T. J. Lockhart, K. S. Thomson, R. V. Goering, and N. D. Hanson. 2004. Plasmid-mediated carbapenem-hydrolyzing enzyme KPC-2 in an Enterobacter sp. Antimicrob. Agents Chemother. 484438-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miriagou, V., L. S. Tzouvelekis, S. Rossiter, E. Tzelepi, F. J. Angulo, and J. M. Whichard. 2003. Imipenem resistance in a Salmonella clinical strain due to plasmid-mediated class A carbapenemase KPC-2. Antimicrob. Agents Chemother. 471297-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moland, E. S., N. D. Hanson, V. L. Herrera, J. A. Black, T. J. Lockhart, A. Hossain, J. A. Johnson, R. V. Goering, and K. S. Thomson. 2003. Plasmid-mediated, carbapenem-hydrolysing β-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J. Antimicrob. Chemother. 51711-714. [DOI] [PubMed] [Google Scholar]
- 16.Naas, T., P. Nordmann, G. Vedel, and C. Poyart. 2005. Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob. Agents Chemother. 494423-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navon-Venezia, S., I. Chmelnitsky, A. Leavitt, M. J. Schwaber, D. Schwartz, and Y. Carmeli. 2006. Plasmid-mediated imipenem-hydrolyzing enzyme KPC-2 among multiple carbapenem-resistant Escherichia coli clones in Israel. Antimicrob. Agents Chemother. 503098-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pope, J., J. Adams, Y. Doi, D. Szabo, and D. L. Paterson. 2006. KPC type β-lactamase, rural Pennsylvania. Emerg. Infect. Dis. 121613-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasheed, J. K., G. J. Anderson, H. Yigit, A. M. Queenan, A. Doménech-Sanchez, J. M. Swenson, J. W. Biddle, M. J. Ferraro, G. A. Jacoby, and F. C. Tenover. 2000. Characterization of the extended-spectrum β-lactamase reference strain Klebsiella pneumoniae K6 (ATCC 700603), which produces the novel enzyme SHV-18. Antimicrob. Agents Chemother. 442382-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villegas, M. V., K. Lolans, A. Correa, C. J. Suarez, J. A. Lopez, M. Vallejo, J. P. Quinn, and the Colombian Nosocomial Resistance Study Group. 2006. First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob. Agents Chemother. 502880-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villegas, M. V., K. Lolans, A. Corrrea, J. N. Kattan, J. A. Lopez, J. P. Quinn, and the Colombian Nosocomial Resistance Study Group. 2007. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing β-lactamase. Antimicrob. Agents Chemother. 511553-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei, Z.-Q., X.-X. Du, Y.-S. Yu, P. Shen, Y.-G. Chen, and L.-J. Li. 2007. Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob. Agents Chemother. 51763-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 451151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yigit, H., A. M. Queenan, J. K. Rasheed, J. W. Biddle, A. Domenech-Sanchez, S. Alberti, K. Bush, and F. C. Tenover. 2003. Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing β-lactamase KPC-2. Antimicrob. Agents Chemother. 473881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]