Vibrio parahaemolyticus is a gram-negative, halophilic bacterium widely distributed in coastal waters worldwide that is associated with gastroenteritis due to the consumption of raw or improperly cooked seafood. The thermostable direct hemolysin (TDH) and TDH-related hemolysin are the major virulence factors. In the past decades, diverse serotypes, including O3:K6, have been related to human gastrointestinal diseases. However, since 1996 the emergence of a new clone of serotype O3:K6 with pandemic potential, which has accounted for an increased incidence of cases in Asia, Africa, and America, has been reported (5). Previous studies have demonstrated that V. parahaemolyticus clinical isolates belonging to both the old and new O3:K6 clones have distinct molecular patterns but do not differ in biological characteristics such as TDH production and susceptibilities to antibiotics and environmental stresses (4, 10). In Europe, pandemic V. parahaemolyticus O3:K6 strains have been detected in clinical samples in France (8), Russia (5), and Spain (9).
Here, we report the characterization of a pandemic V. parahaemolyticus O3:K6 strain isolated from a stool sample of a diarrheal patient hospitalized in central Italy in the summer of 2007. The reported epidemiological information indicated that the patient had eaten fresh shellfish, bought from a local seller, 24 h before hospitalization. The microbiology laboratory of the hospital isolated a gram-negative, oxidase-positive curved bacterium in the absence of other enteric pathogens. The isolate, presumptively identified as belonging to the Vibrio genus, was sent to the Italian Reference Laboratory for Bacteriological Contamination of Shellfish (CEREM) for definitive identification. Suspect colonies grown on thiosulfate citrate bile salt agar (Oxoid Ltd., Basingstoke, England) were subcultured on Trypticase soy agar (Oxoid) with 1% NaCl and biochemically identified at the genus and species levels (6). Moreover, all sucrose-negative colonies were tested by PCR for the presence of V. parahaemolyticus species-specific genes (toxR and tlh) and the tdh and trh genes (3, 7). A group-specific PCR (GS-PCR) method, based on the sequence variation in the toxRS gene, was also performed to detect the pandemic clone (4). O (lipopolysaccharide) and K (capsular) serotypes were determined by using specific commercial antisera (Denka; Seiken Corp., Tokyo, Japan). The susceptibilities of the isolate to 11 antimicrobial agents were determined by the disk diffusion method on Mueller-Hinton agar plates according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (2). The antibiotics tested were amoxicillin-clavulanic acid, ampicillin, cefoperazone, cephalexin, cephalothin, colistin, gentamicin, kanamycin, oxolinic acid, tetracycline, and trimethoprim-sulfamethoxazole. The interpretative criteria to determine the susceptibilities to the tested antimicrobials were as published by the CLSI (2) or followed the recommendations of the antimicrobial agent suppliers.
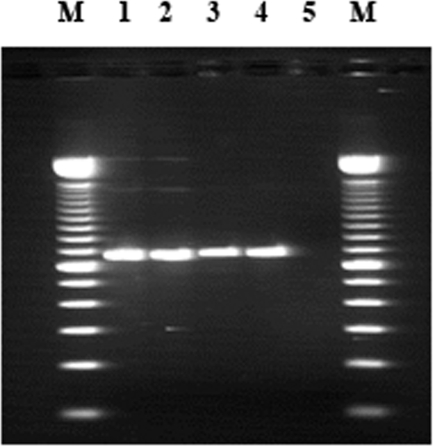
The strain, identified by the biochemical protocol as V. parahaemolyticus, had the toxR and tlh genes and was serotyped as O3:K6. This isolate was also characterized as tdh positive, trh negative, and GS-PCR positive (Fig. 1) and was resistant to amoxicillin-clavulanic acid, ampicillin, cephalexin, and cephalothin. The pattern of antimicrobial susceptibilities was similar to those of other O3:K6 clinical strains previously isolated (10). GS-PCR analysis indicated that the isolate was unequivocally linked to the pandemic O3:K6 clone of V. parahaemolyticus (4) (Fig. 1). In further investigations, this V. parahaemolyticus O3:K6 strain will be characterized by molecular typing methods in order to compare it with other pandemic strains of environmental and clinical origins circulating worldwide.
FIG. 1.
Results of the GS-PCR assay. Lanes: M, molecular size markers (100-bp DNA ladder; Invitrogen); 1 and 2, V. parahaemolyticus O3:K6 strain isolated in this study; 3 and 4, V. parahaemolyticus ATCC BAA-238 (positive control); 5, PCR water (negative control).
To our knowledge, this report documents the first clinical isolation of a pandemic V. parahaemolyticus O3:K6 strain in Italy, with local shellfish as the most probable source of the infection. This evidence, in association with the previous reports of the presence of the pandemic O3:K6 clone in other European countries, demonstrates the spread of this clone to Italian coastal areas. Recently, we isolated toxigenic V. parahaemolyticus strains, serotyped as O1:K untypeable (data not shown), from local shellfish in central Italy. Previous studies have shown the pandemic potential of this serotype and its close phylogenetic relationship to the new O3:K6 clone (4). Moreover, in May 2007, a V. parahaemolyticus strain with pandemic potential was detected in a seawater sample from the northern Adriatic Sea (Italy) (1). All this evidence should prompt official veterinary and health authorities to pay more attention to the epidemiological roles of environmental microorganisms in local food-borne diseases and to increase the microbial surveillance of pathogenic V. parahaemolyticus strains isolated from environmental, seafood, and clinical sources.
Footnotes
Published ahead of print on 30 April 2008.
REFERENCES
- 1.Caburlotto, G., V. Ghidini, M. Gennari, M. C. Tafi, and M. M. Lleo. 13 March 2008, posting date. Isolation of a Vibrio parahaemolyticus pandemic strain from a marine water sample obtained in the northern Adriatic. Eurosurveillance 13pii=8068. http://www.eurosurveillance.org/edition/v13n11/080313_3.asp. [DOI] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2006. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. Approved standard M45-A. Clinical and Laboratory Standards Institute, Wayne, PA. [DOI] [PubMed]
- 3.Kim, Y. B., J. Okuda, C. Matsumoto, N. Takahashi, S. Hashimoto, and M. Nishibuchi. 1999. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J. Clin. Microbiol. 371173-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsumoto, C., J. Okuda, M. Ishibashi, M. Iwanaga, P. Garg, T. Ramamurthy, H. C. Wong, A. DePaola, Y. B. Kim, M. J. Albert, and M. Nishibuchi. 2000. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nair, G. B., T. Ramamurthy, S. K. Bhattacharya, B. Dutta, Y. Takeda, and D. A. Sack. 2007. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 2039-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ottaviani, D., L. Masini, and S. Bacchiocchi. 2003. A biochemical protocol for the isolation and identification of current species of Vibrio in seafood. J. Appl. Microbiol. 951277-1284. [DOI] [PubMed] [Google Scholar]
- 7.Ottaviani, D., S. Santarelli, S. Bacchiocchi, L. Masini, C. Ghittino, and I. Bacchiocchi. 2005. Presence of pathogenic Vibrio parahaemolyticus strains in mussels from the Adriatic Sea, Italy. Food Microbiol. 22585-590. [DOI] [PubMed] [Google Scholar]
- 8.Quilici, M. L., A. Robert-Pillot, J. Picart, and J. M. Fournier. 2005. Pandemic Vibrio parahaemolyticus O3:K6 spread, France. Emerg. Infect. Dis. 111148-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urtaza, J. M., L. Simental, D. Velasco, A. DePaola, M. Ishibashi, Y. Nakaguchi, M. Nishibuchi, D. Carrera-Flores, C. Rey-Alvarez, and A. Pousa. 2005. Pandemic Vibrio parahaemolyticus O3:K6, Europe. Emerg. Infect. Dis. 81319-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong, H. C., S. H. Liu, T. K. Wang, C. L. Lee, C. S. Chiou, D. P. Liu, M. Nishibuchi, and B. K. Lee. 2000. Characteristics of Vibrio parahaemolyticus O3:K6 from Asia. Appl. Environ. Microbiol. 663981-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]