Abstract
In the present study, we analyzed genetic variation in Cryptosporidium species from humans (n = 62) with clinical cryptosporidiosis in South Australia. Sequence variation was assessed in regions within the small subunit of nuclear rRNA (p-SSU), the 70-kDa heat shock protein (p-hsp70), and the 60-kDa glycoprotein (p-gp60) genes by employing single-strand conformation polymorphism analysis and sequencing. Based on the analyses of p-SSU and p-hsp70, Cryptosporidium hominis (n = 38) and Cryptosporidium parvum (n = 24) were identified. The analysis of p-gp60 revealed eight distinct subgenotypes, classified as C. hominis IaA17R1 (n = 3), IbA9G3R2 (n = 14), IbA10G2R2 (n = 20), and IfA12G1R1 (n = 1), as well as C. parvum IIaA18G3R1 (n = 15), IIaA20G3R1 (n = 6), IIaA22G4R1 (n = 2), and IIcA5G3R2 (n = 1). Subgenotypes IaA17R1 and IIaA22G4R1 are new. Of the six other subgenotypes, IbA10G2R2, IIaA18G3R1, IIaA20G3R1, and IIcA5G3R2 were reported previously from the state of Victoria. This is the fourth record in Australia of C. parvum subgenotype IIaA18G3R1 from humans, which, to date, has been isolated only from cattle in other countries. This subgenotype might be a significant contributor to sporadic human cryptosporidiosis and may indicate a greater zoonotic contribution to the infection of humans in the area of study. Comparative analyses revealed, for the first time, the differences in the genetic makeup of Cryptosporidium populations between two relatively close, major metropolitan cities.
Cryptosporidium species are parasitic protists (of the Apicomplexa) that are commonly found in surface water globally (45) and parasitize many vertebrate classes, including birds, fish, mammals, and reptiles (21, 81). These species are transmitted via a highly resistant oocyst and usually infect the gastrointestinal tract, causing cryptosporidiosis. Cryptosporidiosis is commonly associated with enteritis and is usually characterized by acute, watery, or steatorrheic diarrhea and colic (16, 39), although asymptomatic infection can occur (15, 32, 55, 59, 68, 69). In humans, the transmission of Cryptosporidium infection is hypothesized to be most commonly anthroponotic (human to human) and, to a lesser extent, zoonotic (animal to human) (12, 62, 82). Cryptosporidiosis is usually short lived (days or weeks) and, in immunocompetent persons, eventually eliminated by a combined cellular and humoral immune response (63). In high-risk patient groups, such as infants and the elderly or people suffering from severe malnutrition, advanced human immunodeficiency virus/AIDS, or having undergone immunosuppressive therapy, cryptosporidiosis can be chronic, leading to severe dehydration and/or malabsorption (34). Although there has been progress with the development of drugs, such as paromomycin (10, 44, 56) and nitazoxanide (8, 65), currently there is limited accessibility to efficacious treatment in many countries (9, 13, 29, 43, 86). As a result, chronic cryptosporidiosis can be fatal and substantially affects the health of human communities, particularly in developing countries (39, 45). Therefore, the prevention and control of cryptosporidiosis are heavily reliant on diagnosis and an understanding of the epidemiology and genetics of Cryptosporidium species.
In recent years, Cryptosporidium species have received significant attention as the cause of epidemic outbreaks of disease (21, 37), often transmitted via drinking and/or recreational waters in developed cities in Australia (11, 61), Canada (54, 66, 75), Europe (67), and the United States (19, 20, 41, 85). Although such outbreaks can be substantial in scale and impact (41), sporadic cases are likely to be more common than widespread outbreaks, particularly in the developed nations. Elucidating the specific sources of sporadic cryptosporidiosis is often challenging and can be impractical. However, investigating the genetic makeup of Cryptosporidium species that are linked to sporadic cryptosporidiosis in particular geographical areas and understanding the risk factors associated with the disease provide a means of establishing the potential contributors to epidemics, the risk of zoonotic versus anthroponotic transmission to municipal populations, and the persistence of endemicity versus the emergence of “exotic” genetic variants of Cryptosporidium in the local environment.
Human cryptosporidiosis, whether sporadic or epidemic, predominantly is caused by Cryptosporidium hominis (for which only anthroponotic transmission has been reported) or Cryptosporidium parvum (for which both anthroponotic and zoonotic transmission have been reported) (12, 50). The population genetic analysis of C. parvum and C. hominis has revealed intraspecific variability in host specificity and geographical distribution (6, 24, 33, 51, 82, 84). For C. parvum, some of these intraspecific variants have been associated with substantial variation in minimum infective dosage and/or virulence (52, 53). Such variability in specificity, distribution, or virulence likely confers an effect on the risk that different strains can pose to human populations, which in turn might vary depending on the environment (e.g., urban versus rural or developed versus developing country).
Recently, the transmission dynamics of sporadic cryptosporidiosis in municipal regions of Australia was examined through a case control study of human populations in two major cities: Adelaide, South Australia (population of ∼1 million), and Melbourne, Victoria (population of ∼3.5 million) (64). The major risk factors associated with sporadic cases were inferred on the basis of positive statistical correlation (i.e., potential infection source versus incidence) to the use of public swimming pools (particularly those catering to young children); direct human-to-human contact; exposure to farm animals (particularly cattle and sheep); exposure to some food groups, such as cheeses produced from unpasteurized milk; and the consumption of untreated river, lake, or dam water in rural areas. Although Robertson et al. (64) examined the risk factors associated with sporadic cryptosporidiosis in humans, this study did not investigate the species/genetic variants of Cryptosporidium causing these cases. It is not possible, based on statistical data alone, to determine the contribution of anthroponotic or zoonotic transmission to sporadic infection in these cities, particularly for sources with greater potential for transmission via either pathway, such as risks related to untreated rural watersheds or foods. Such determinations require extensive and reliable epidemiological data sets that can be supported through the use of advanced molecular tools (23, 35, 70).
Although extensive molecular epidemiological studies have been carried out in a range of countries, surprisingly, there has been very limited study of the genetics of Cryptosporidium species isolated from humans in Australia (14, 36, 51). In addition, to date no study has examined the variation in the genetic makeup of Cryptosporidium populations that contribute to human infection between or among neighboring cities within the same country across a relatively small geographical range. A recent study (36) classified, for the first time, genotypes and subgenotypes of Cryptosporidium that cause sporadic human cryptosporidiosis in the city of Melbourne and surrounding areas (in the state of Victoria, Australia). In the present study, we extended recent work to investigate the genetic variability in multiple loci within and among Cryptosporidium species from humans with sporadic cryptosporidiosis from Adelaide and surrounding areas in the state of South Australia (∼1,000 km from Melbourne). The resultant data were compared to those presently available for sporadic cryptosporidiosis cases in Australia (14, 36, 51) and provided a first glimpse of the difference in the genetic makeup of Cryptosporidium populations between these two metropolitan cities and the extent to which the (epidemiological) risk factors identified by Robertson et al. (64) may have contributed to this difference.
MATERIALS AND METHODS
Isolation and purification of genomic DNA from fecal samples.
Fecal samples (n = 62) from human patients with clinical cryptosporidiosis were collected (from 2001 to 2005) in Adelaide and surrounding areas in the state of South Australia; 26 of these samples originated from a previous epidemiological study but had not been genetically characterized (64). The parasitological diagnosis was made based on the detection of oocysts consistent with Cryptosporidium by using the modified Ziehl-Neelsen staining procedure (31). Genomic DNA was extracted from fecal samples as described previously (49). In brief, samples were suspended in a 1:4 ratio of phosphate-buffered saline (PBS). The suspension was homogenized, and 20 μl of suspension was aspirated and added to 80 μl of 10% polyvinylpyrrolidone (PVPP) (Sigma, St. Louis, MO) in H2O. After the addition of PVPP, samples were heated to 100°C for 10 min and then centrifuged at 10,000 × g for 30 s. Following centrifugation, each supernatant was transferred to a fresh tube and DNA purified using a minicolumn (Copro kit; Qiagen, Germany) according to the manufacturer's protocol.
PCR-based amplification.
All PCRs were set up in a PCR series laminar-flow cabinet (Clyde-APAC, Australia) using filter tips. The PCR was carried out in a 50-μl volume of 10 mM Tris-HCl (pH 8.4), 50 mM KCl, 2 mM MgCl2, 200 μM of each deoxynucleoside triphosphate, 25 pmol of each primer, and 1 U of GoTaq polymerase (Promega, Madison, WI) in a GeneAmp PCR 2400 (Perkin Elmer) thermal cycler. Approximately 10 to 20 ng of template was included in the (primary) PCR amplification from genomic DNA; samples without templates were included in each run (as negative controls).
To identify the species of Cryptosporidium present in each DNA sample, an ∼300-bp region of the small subunit of the nuclear rRNA (p-SSU) gene was amplified by PCR from genomic DNA (∼10 to 20 ng) using the oligonucleotide primers 18SiF (forward; AGTGACAAGAAATAACAATACAGG-3′) and 18SiR (reverse; 5′-CCTGCTTTAAGCACTCTAATTTTC-3′) (24, 49) and subjected to sequencing (described below). The relative specificity of these primers for the amplification of Cryptosporidium species by PCR has been demonstrated previously (24, 49). The cycling conditions were one cycle of 94°C for 5 min (denaturation), followed by 35 cycles of 94°C for 30 s (denaturation), 58°C for 20 s (annealing), and 72°C for 30 s (extension), and then a final extension of 72°C for 7 min. The sequences derived from p-SSU amplicons were compared with selected, publicly available sequences representing C. hominis (accession no. AF093492 [80]) and C. parvum (accession no. AF115377 [83]).
To independently identify the Cryptosporidium species in each DNA sample, a part of the heat shock protein 70 gene (p-hsp70; ∼450 bp) also was PCR amplified from genomic DNA using oligonucleotide primers HSPR4-F (forward; 5′-GGTGGTGGTACTTTTGATGTATC-3′) and HSPR4-R (reverse; 5′-GCCTGAACCTTTGGAATACG-3′). These primers were originally designed to target relatively conserved regions of the heat shock protein 70 (hsp70) gene (38, 48) and have been used in previous studies of Cryptosporidium (3, 26). Sequences derived from p-hsp70 amplicons were compared with selected publicly available sequences representing C. hominis (accession no. DQ886255 [27]) and C. parvum (accession no. XM_001388291 [2]).
In order to assess genetic variation within each sample, a so-called variable region (∼300 to 450 bp) within the gp60 gene (p-gp60) was amplified from DNA samples using a nested PCR approach. In the first round, the gp60 locus (∼950 to 1,000 bp) was amplified from genomic DNA using primers gp15-ATG (forward; 5′-ATGAGATTGTCGCTCATTATC-3′) and gp15-STOP (reverse; 5′-TTACAACACGAATAAGGCTGC-3′) (71); the specificity of these primer pairs in PCR for the amplification of C. hominis and C. parvum DNA has been demonstrated previously (36, 71). In the second round, 1 μl of the primary amplicon was transferred to a fresh tube (containing reagents) and subjected to PCR using primers gp15-15A (forward; 5′-GCCGTTCCACTCAGAGGAAC-3′) and gp15-15E (reverse; 5′-CCACATTACAAATGAAGTGCCGC-3′) (42) to amplify p-gp60 using the same cycling protocol, except that the annealing time was modified to 45 s. Known C. parvum (positive) controls and samples without DNA template (negative controls) as well as appropriate carryover controls were included in each PCR run.
Agarose gel electrophoretic analysis, sequencing of amplicons, and phylogenetic analysis of sequence data sets.
The quality and intensity of individual amplicons were examined on ethidium bromide-stained 1.5% agarose gels, using TBE (65 mM Tris-HCl, 27 mM boric acid, 1 mM EDTA, pH 9; Bio-Rad) as the buffer and ΦX174-HaeIII (Promega) as a size marker. Amplicons were purified over minicolumns (Wizard PCR Preps; Promega), eluted in 30 μl H2O, and then subjected to direct, automated sequencing (BigDye chemistry; Applied Biosystems, Foster City, CA) from both strands using the same primers (individually) as those used in the PCR (gp15-15A/gp15-15E for p-gp60). The sequences determined were compared with those available in current gene databases and published in quality, peer-reviewed scientific journals. For each locus (p-SSU, p-hsp70, or p-gp60), sequences were trimmed and then aligned using the program Clustal X (77), and the alignments were adjusted by employing the program BioEdit (30). Pairwise comparisons of the sequence differences (D) were made using the formula D = 1 − (M/L) (17), where M is the number of alignment positions at which the two sequences have a base in common and L is the total number of alignment positions over which the two sequences are compared; the differences were compared with automated calculations using PAUP*4.0b10 (74).
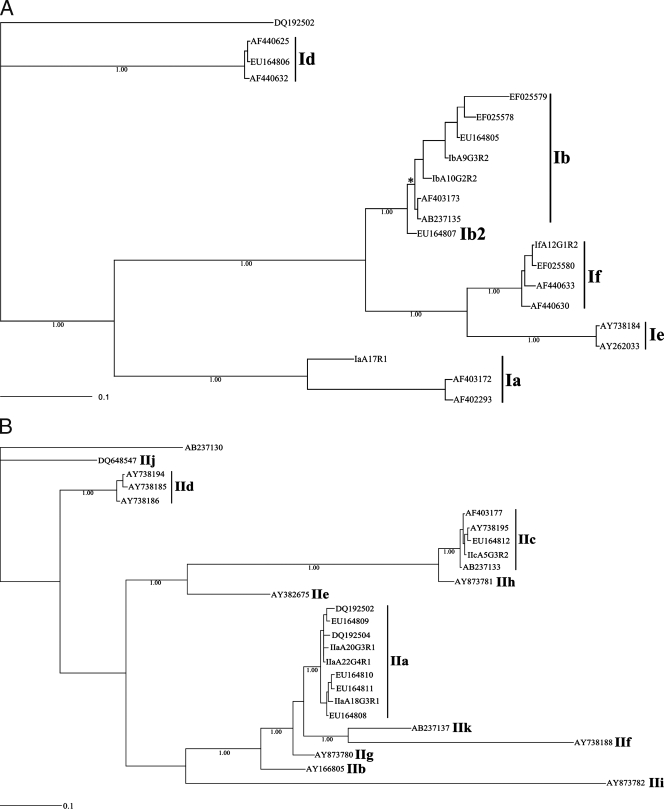
In the present study, we adopted the genotypic nomenclature proposed by Strong et al. (71), wherein each gp60 genotype is given the species-specific prefix I (C. hominis) or II (C. parvum), followed by an alphabetical letter (e.g., Ia) that is assigned in reference to the sequence of the locus. Also, the present study recognizes and adopts the subgenotypic nomenclature for the gp60 locus proposed by Sulaiman et al. (72), wherein the subgenotypic assignment is based on the trinucleotide repeats within the gp60 microsatellite region, which encodes a polyserine tract within the protein GP45 (71). These repeats are categorized according to the number of TCA (A), TCG (G), TCT (T), and/or additional rare (R) repeats. Rare repeats were considered to relate to codons within the microsatellite region with a single point mutation at the first nucleotide position of the TCA trinucleotide in C. hominis or C. parvum (36). For example, a sample with a sequence consisting of 10 TCA repeats, 3 TCG repeats, and 2 ACA repeats would be identified to the subgenotype as A10G3R2. As there currently is no consensus opinion on the precise definition of a genotype (or the threshold of intragenotypic versus intergenotypic sequence variation), a sample was assigned to a particular genotype based on its relationship with strongly supported clades after the phylogenetic analysis of the p-gp60 sequence data for all samples studied, including published sequences representing all currently recognized gp60 genotypes of C. hominis (Ia to If and Ib2 [7, 36, 71]) and C. parvum (IIa to IIk [1, 4, 7, 47, 58, 72, 76]) (for accession numbers, see Fig. 3).
FIG. 3.
Phylogenetic analysis of p-gp60 sequence data representing Cryptosporidium hominis (A) and C. parvum (B) from 62 human patients from Adelaide and surrounding areas of South Australia using key published sequence data for comparative purposes (1, 18, 40, 58, 71-73). From the present study, sequences representing C. hominis subgenotypes IaA17R1, IbA9G3R2, IbA10G2R2, and IfA12G1R2 have been deposited under accession numbers EU379544, EU379545, EU379546, and EU379547, respectively; those representing C. parvum subgenotypes IIaA18G3R1, IIaA20G3R1, IIaA22G4R1, and IIcA5G3R2 are available under accession numbers EU379548, EU379549, EU379550, and EU379551, respectively. The accession numbers of other (reference) sequences are indicated in the trees. Numbers beneath the node of each clade represent bootstrap support obtained using Bayesian inference; the bar indicates distance (0.1 substitutions per site). An asterisk indicates a pp of 1.00.
The phylogenetic analysis of the p-gp60 nucleotide sequence data was conducted (separately for each species) using Bayesian inference and by employing the software package MrBayes 3.1.2 (http://mrbayes.csit.fsu.edu/index.php). Posterior probabilities (pp) were calculated via 2,000,000 generations (ngen = 2,000,000; burnin = 20,000) using the Monte Carlo Markov chain method, which utilizes four simultaneous tree-building chains (nchains = 4) with every 100th tree saved (samplefreq = 100). The evolutionary distance was calculated using the general time reversible evolutionary model (nset = 6), which allows for a gamma-shaped variation in mutation rates between codons (rates = gamma). Cryptosporidium parvum IIa (GenBank accession number DQ192502 [78]) was used as the outgroup in the analysis of p-gp60 sequence data for C. hominis. Cryptosporidium hominis Ia (GenBank accession number AB273130 [1]) was used as the outgroup in the analysis of p-gp60 data for C. parvum. Upon the completion of the Bayesian analysis, a 50% majority-rule consensus tree for each species was constructed in Treeview X v.0.5.0 (http://darwin.zoology.gla.ac.uk/∼rpage/treeviewx/) and enhanced using InkScape 0.45 (www.inkscape.org).
Nonisotopic SSCP analysis.
The method of single-strand conformation polymorphism (SSCP) was carried out according to protocol B (25). In brief, 10 μl of each amplicon was mixed with 5 μl of DNA sequencing stop solution (Promega), heat denatured at 94°C for 30 min, snap-cooled on a freeze block (−20°C), and screened using precast GMA S-2x25 gels (Elchrom Scientific AG, Cham, Switzerland). Samples were subjected to electrophoresis in a SEA 2000 apparatus (Elchrom Scientific AG) containing TAE buffer (0.04 M Tris base, 0.04 M acetate, 0.001 M EDTA) and were run at 110 V and 7.4°C (constant) for 6 h. Samples representing all of the profile variations that were detectable were selected from an initial analysis and subjected, under the same conditions, to electrophoresis in a GMA S-2x13 gel (Elchrom Scientific AG), using 3 μl of product in 10 μl of DNA sequencing stop solution at 74 V and 7.4°C for 18 h. A control sample was included on each gel to ensure the reproducibility of profiles, thus allowing the direct comparison of gels.
Nucleotide sequence accession numbers.
The sequences determined in the course of this work have been deposited in GenBank under accession numbers EU379540 to EU379551.
RESULTS
Sixty-two samples from sporadic cases of cryptosporidiosis in humans from Adelaide and surrounding areas in South Australia were subjected to molecular analyses. Genetic variation was assessed in multiple loci (p-SSU, p-hsp70, and p-gp60) by SSCP analysis and targeted sequencing. Prior to SSCP and sequence-based analysis, all amplicons were subjected to agarose gel electrophoresis (not shown) and demonstrated to represent single, discrete bands. Amplicon sizes were ∼250 bp (p-SSU), ∼400 bp (p-hsp70), and ∼300 to 350 bp (p-gp60). For both p-SSU and p-hsp70, the SSCP analysis of all amplicons divided samples into two distinct groups; within each group, there was no evidence of variation in profiles among amplicons. The sequencing of the p-SSU and p-hsp70 amplicons identified C. hominis (n = 38) and C. parvum (n = 24) based on comparisons with selected reference sequences (see Materials and Methods for accession numbers) (Table 1). The p-SSU sequences determined for C. hominis and C. parvum all were 250 and 247 bp, respectively. The hsp70 sequences obtained for C. hominis and C. parvum all were 403 bp. For both p-SSU and hsp70, no intraspecific sequence variation or polymorphism was detected within either C. hominis or C. parvum. Between C. hominis and C. parvum, sequence differences were 2% (two substitutions and three indels) in p-SSU and 1.2% (five substitutions) in p-hsp70. Representative sequences have been deposited under accession numbers EU379540 (C. hominis) and EU379541 (C. parvum) for p-SSU and EU379542 (C. hominis) and EU379543 (C. parvum) for p-hsp70.
TABLE 1.
Initial categorization of C. hominis and C. parvum from human patients in Adelaide and surrounding areas (South Australia) based on SSCP analysis
| Species | p-SSU amplicon | No. detected | % of total | p-hsp70 amplicon | No. detected | % of total | p-gp60 genotype and subgenotypea | No. detected | % of total |
|---|---|---|---|---|---|---|---|---|---|
| C. hominis | Ch1-SSU | 38 | 61.3 | Ch1-hsp70 | 38 | 61.3 | IaA17R1 | 3 | 4.8 |
| IbA10G2R2 | 20 | 32.3 | |||||||
| IbA9G3R2 | 14 | 22.6 | |||||||
| IfA12G1R2 | 1 | 1.6 | |||||||
| C. parvum | Cp1-SSU | 24 | 38.7 | Cp1-hsp70 | 24 | 38.7 | IIaA18G3R1 | 15 | 24.2 |
| IIaA20G3R1 | 6 | 9.7 | |||||||
| IIaA22G4R1 | 2 | 3.2 | |||||||
| IIcA5G3R2 | 1 | 1.6 |
Genotypes and subgenotypes within each species were defined using p-gp60 sequence data.
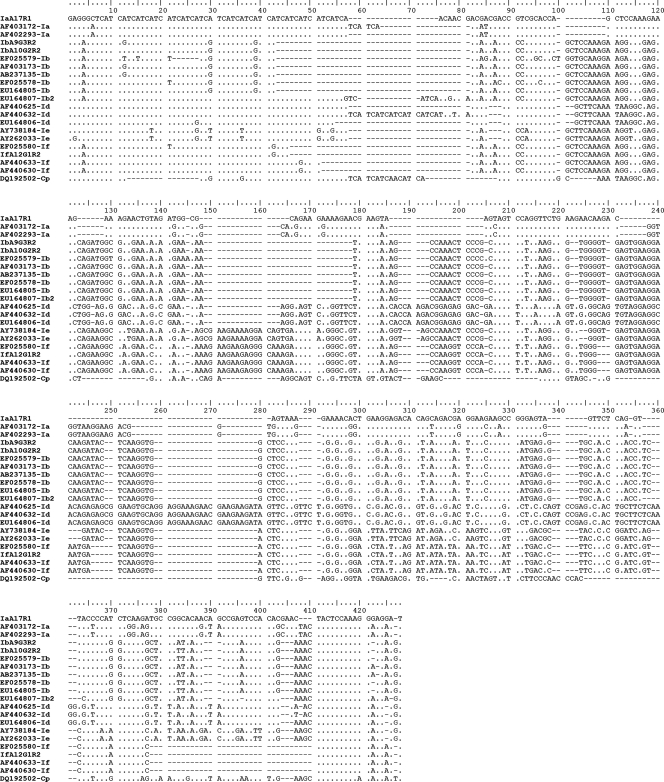
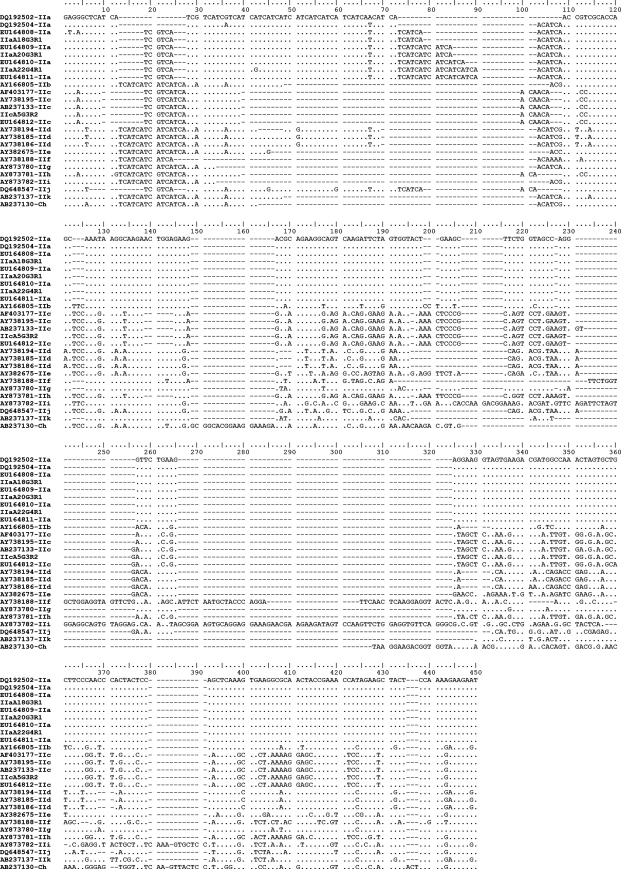
Having identified C. hominis and C. parvum within the set of 62 samples, genetic heterogeneity was investigated further using the gp60 locus. Amplicons representing all 62 samples were sequenced, and distinct p-gp60 sequence types were defined for C. hominis (n = 4) and C. parvum (n = 4) (Table 1). The p-gp60 sequences (accession numbers EU379544 to EU379551) varied in length from 302 to 326 bp (C. hominis) and from 263 to 314 bp (C. parvum) and had a G+C content of 45.1 to 49.2% (Fig. 1 and 2). According to the nomenclature proposed previously for Cryptosporidium genotypes/subgenotypes (36, 71, 72) and based on comparisons with reference sequences (Fig. 1 [C. hominis] and 2 [C. parvum] list the accession numbers), the 38 C. hominis samples were first classified genotypically/subgenotypically as Ia (IaA17R1), Ib (IbA10G2R2 and IbA9G3R2), or If (IfA12G1R2), and the C. parvum samples were classified as IIa (IIaA18G3R1, IIaA20G3R1, and IIaA22G4R1) or IIc (IIcA5G3R2) (Table 1). Sequence variation in p-gp60 was 0.3 to 52.4% (1 to 200 of 428 positions) within C. hominis and 2.4 to 51.3% (6 to 157 of 330 positions) within C. parvum (Tables 2 and 3). All sequence variation among all samples could be readily displayed visually by SSCP analysis (not shown).
FIG. 1.
Alignment of the four p-gp60 sequences representing Cryptosporidium hominis from 38 human patients from Adelaide and surrounding areas (South Australia). A dot indicates a nucleotide identical to that of the top sequence; a dash indicates an insertion/deletion event. Sequences representing subgenotypes IaA17R1, IbA9G3R2, IbA10G2R2, and IfA12G1R2 are from the present study and have been deposited in GenBank under accession numbers EU379544, EU379545, EU379546, and EU379547, respectively. All other sequences represent reference sequences (accession numbers are given in the alignment at the left). Cryptosporidium parvum (Cp) was used as the outgroup for the phylogenetic analysis, which is depicted in Fig. 3.
FIG. 2.
Alignment of the four p-gp60 sequences representing Cryptosporidium parvum from 24 human patients from Adelaide and surrounding areas (South Australia). A dot indicates a nucleotide identical to that of the top sequence; a dash indicates an insertion/deletion event. Sequences representing subgenotypes IIaA18G3R1, IIaA20G3R1, IIaA22G4R1, and IIcA5G3R2 are from the present study and have been deposited in GenBank under accession numbers EU379548, EU379549, EU379550, and EU379551, respectively. All other sequences represent reference sequences (accession numbers are given in the alignment at the left). Cryptosporidium hominis (Ch) was used as the outgroup for the phylogenetic analysis, which is depicted in Fig. 3.
TABLE 2.
Pairwise comparison of sequence variation in p-gp60 among 38 samples containing C. hominisa
| Subgenotype | IaA17R1 | IbA9G3R2 | IbA10G2R2 | IfA12G1R2 |
|---|---|---|---|---|
| IaA17R1 | - | 124 | 123 | 200 |
| IbA9G3R2 | 35.1 | - | 1 | 117 |
| IbA10G2R2 | 34.8 | 0.3 | - | 116 |
| IfA12G1R2 | 52.4 | 32.3 | 31.9 | - |
The samples were taken from humans from Adelaide and surrounding areas in South Australia. Values above the dashes are absolute numbers of nucleotide alterations (including insertion/deletion events); values below the dashes are percentages.
TABLE 3.
Pairwise comparison of sequence variation in p-gp60 among 24 samples containing C. parvuma
| Subgenotype | IIaA18G3R1 | IIaA20G3R1 | IIaA22G4R1 | IIcA5G3R3 |
|---|---|---|---|---|
| IIaA18G3R1 | - | 6 | 13 | 144 |
| IIaA20G3R1 | 2.1 | - | 7 | 150 |
| IIaA22G4R1 | 4.4 | 2.4 | - | 157 |
| IIcA5G3R3 | 48.9 | 50.0 | 51.3 | - |
The samples were taken from humans from Adelaide and surrounding areas in South Australia. Values above the dashes are absolute numbers of nucleotide alterations (including insertion/deletion events); values below the dashes are percentages.
For each species, all distinct p-gp60 sequence types that were determined were aligned (Fig. 1 and 2) with reference sequences representing all currently recognized gp60 genotypes, including some reported recently (36) from Melbourne, Australia (GenBank accession numbers EU164805 to EU164812). The nature and extent of sequence variation were evaluated. Among the four p-gp60 sequences determined for C. hominis (Fig. 1), relatively high levels of nucleotide variability, external to the microsatellite region (positions 7 to 81), were detected in regions between alignment positions 121 to 167, 171 to 229, 241 to 301, and 302 to 428. In regions ′121 to 167′ and ′171 to 229′, variation related predominantly to substitutions (42 and 90 sites, respectively) and some insertion/deletion events (16 and 45 sites, respectively). In regions ′241 to 301′ and ′302 to 428′, substitutions also were common (51 and 53 sites, respectively), but the number of insertion/deletion events was greater (47 and 54 sites, respectively) than that for the former two regions. Of the four p-gp60 sequences determined for C. parvum (Fig. 2), most of the nucleotide variation external to the microsatellite region (positions 7 to 108) was between alignment positions 121 to 230, 231 to 325, and 326 to 450. The nucleotide variability in regions ′121 to 230′ and ′326 to 450′ related predominantly to substitutions (56 and 87 sites, respectively), although insertion/deletion events were recorded (49 and 12 sites, respectively). In region ′231 to 325′, insertion/deletion events were more common (85 sites) than substitutions (54 sites) and related mainly to the insertions in the sequences representing genotypes IIb and IIi (Fig. 2).
Having characterized the nature and extent of nucleotide variation and aligned all gp60 sequences for each species to achieve maximal positional homology, phylogenetic analyses were conducted using Bayesian inference (Fig. 3). Given the substantial sequence variability in p-gp60 between C. hominis and C. parvum and the inability to reliably align all sequences to achieve positional homology, the sequence data set for each species was subjected to separate phylogenetic analyses. The analysis of gp60 data showed that each recognized genotype of C. hominis (Ia to If) (7, 36, 71) and of C. parvum (IIa to IIk (1, 4, 7, 47, 58, 72, 76) resolved as a separate, strongly supported clade (pp of 1.00 for individual genotypic clades). The detailed appraisal of the C. hominis tree (Fig. 1) revealed the following clades (all with maximum nodal support [pp = 1.00]): genotype If grouped with Ie, If/Ie with Ib, and If/Ie/Ib with Ia to the exclusion of Id. A similar analysis of the C. parvum tree revealed the following clades (all with maximum nodal support [pp = 1.00]): genotype IIc grouped with IIh and IIc/IIh with IIe; genotype IIf grouped with IIk, IIf/IIk with IIa, IIa/IIf/IIk with IIg, IIa/IIf/IIg/IIk with IIb, IIa/IIb/IIf/IIg/IIk with IIi, and IIa/IIb/IIf/IIg/IIi/IIk with IIc/IIe/IIh to the exclusion of IId and IIj. Although genotype IId was shown to form a clade with all other C. parvum genotypes to the exclusion of IIj, the posterior probabilities calculated for this large clade (IIa to IIk to the exclusion of IIj) was low (pp = 0.50) and is not strongly supported by the present analysis. Importantly, for both species of Cryptosporidium, all of the gp60 genotypes determined herein grouped according to their preliminary classification based on genetic similarity alone (e.g., IaA17R1 with Ia and IbA10G2R2 with Ib).
DISCUSSION
Using genotypic and subgenotypic definitions adopted from previous studies (36, 71, 72), the present C. hominis samples were initially classified as IaA17R1, IbA9G3R2, IbA10G2R2, and IfA12G1R2, and C. parvum samples were classified as IIaA18G3R1, IIaA20G3R1, IIaA22G4R1, and IIcA5G3R2. As there currently is no consensus view on the genetic boundaries or thresholds (based on nucleotide divergence) for the definition of gp60 genotypes or subgenotypes, we have suggested a cautious approach based on the phylogenetic analysis of sequence data (36). Thus, the classification of any new genetic variant within a species (or operational taxonomic unit) has been based on its relationship with known representatives (i.e., sequences from published studies and deposited in publicly available databases) and strong support at crucial nodes. In the present study, all previously identified gp60 genotypes of C. hominis and C. parvum were shown, based on Bayesian analysis, to form strongly supported clades consistent with their specific identification, as has been shown previously using other phylogenetic algorithms (1, 36, 60). In addition, the C. hominis and C. parvum gp60 variants from humans in Adelaide and surrounding areas in South Australia were positioned within these clades, supporting the initial classification based on sequence similarity.
Of the gp60 subgenotypes identified in the present study, four (C. hominis IbA9G3R2, C. hominis IbA10G2R2, C. parvum IIaA18G3R1, and C. parvum IIaA20G3R1) have been reported previously in Adelaide (14) from a distinct subset of samples from the study by Robertson et al. (64). In addition to the C. hominis and C. parvum gp60 subgenotypes reported in the present study, Chalmers et al. (14) also identified subgenotypes C. hominis IeA12G3T3R1, C. parvum IIaA15G2R1, and C. parvum IIaA19G3R1. Of the C. hominis and C. parvum gp60 subgenotypes detected in humans in Adelaide/South Australia, all but two (C. hominis IaA17R1 and C. parvum IIaA22G4R1) have been detected elsewhere. Within the C. hominis gp60 subgenotypes detected in Adelaide/South Australia, C. hominis IbA9G3R2 and IbA10G2R2 appear to be exceptionally common in humans and broadly distributed geographically, having been reported previously from numerous countries (6, 7, 18, 22, 40, 57, 72, 73, 87), including Australia (14, 36, 51). Cryptosporidium hominis IfA12G1R2 has been reported previously from humans in Australia (51) but appears to be much less prevalent than either IbA9G3R2 or IbA10G2R2 and has not been reported previously elsewhere in the world. In addition, C. hominis IeA12G3T3R1, reported previously in Adelaide (14) but not detected in the present study, has not been reported elsewhere in Australia, but it has been reported previously in the United States (87).
Within the C. parvum sample set examined in the present study, none of the subgenotypes detected were as prevalent in humans as C. hominis IbA9G3R2 or IbA10G2R2. However, some of the subgenotypes do appear to be distributed globally and may be significant contributors to human cryptosporidiosis. Three IIa subgenotypes of C. parvum (IIaA18G3R1, IIaA20G3R1, and IIaA22G4R1) were detected in the present study. Of these, the most common of the three, based on presently available data, was IIaA18G3R1, which has been reported previously from cattle in Australia (51), Canada (78), and Northern Ireland (76). Interestingly, the only other reports of this subgenotype in humans are from Australia (14, 36, 51). Of the remaining C. parvum IIa subgenotypes detected herein, one (IIaA20G3R1) has been reported previously in Australia as IIaA20G3R1 (36) and as IIaA19G3R1 (14, 51) (see the description of the subgenotypic nomenclature in Materials and Methods), whereas the other (IIaA22G4R1) appears to be a new subgenotype. One C. parvum IIa subgenotype reported previously from Adelaide (14) has not been detected herein. This subgenotype, previously identified as IIaA15G2R1, appears to be the most widely distributed of the C. parvum gp60 subgenotypes reported to date and appears to have the broadest host distribution, having been reported from humans (1, 6, 14, 18, 22, 57, 72, 87), cattle (1, 6, 7, 76, 78, 79, 84), deer (6), rodents (46), and sheep (6) from every continent except Antarctica. Lastly, C. parvum IIcA5G3R2, which is linked to one sample in the present study, has been reported previously in Australia (36, 51) and is recognized as a common subgenotype in humans globally (4, 6, 7, 40, 57, 58, 72).
Recently, we studied gp60 genotypes and subgenotypes among samples that represented sporadic cases of human cryptosporidiosis in Melbourne and surrounding areas (Victoria, Australia) (36). Four of the subgenotypes detected in humans in Adelaide (South Australia) in the present study, namely, C. hominis IbA10G2R2, C. parvum IIaA18G3R1, C. parvum IIaA20G3R1, and C. parvum IIcA5G3R2, were identical in sequence to those reported previously in Melbourne (36). Given the relatively close proximity of the cities Adelaide and Melbourne (∼1,000 km apart) and the states South Australia and Victoria and the high level of human transit that occurs between them, it was not surprising to encounter the same subgenotypes in both studies. Also interesting were the C. hominis and C. parvum gp60 subgenotypes that have been recorded either in Adelaide/South Australia or Melbourne/Victoria (36) but not in both cities or states. Subgenotypes (based on gp60 data) found in Adelaide/South Australia and not reported previously from Melbourne/Victoria were C. hominis IaA17R1, C. hominis IbA9G3R2, C. hominis IfA12G1R2, and C. parvum IIaA22G4R1. Subgenotypes found in the previous study in Melbourne/Victoria (36) but not yet reported from Adelaide/South Australia were C. hominis Ib2A18G1R4, C. hominis IdA15G1R2, C. parvum IIaA22G3R1, and C. parvum IIaA23G3R1. Interestingly, only 4 (28.6%) of the 14 subgenotypes presently described from either of these regions (14, 36, and the present study) were found in both geographical regions. However, it should be noted that all of the subgenotypes found either in Adelaide/South Australia or Melbourne/Victoria but not both, e.g., Ia17R1 (n = 3), IeA12G3T3R1 (n = 2) (14), and IIaA15G2R1 (n = 1) (14) in Adelaide/South Australia or IdA15G1R2 (n = 1), IIaA22G3R1 (n = 1), and IIaA23G3R1 (n = 1) (36) in Melbourne/Victoria, are relatively rare in the region from which they have been reported. The relatively small sample sizes in the present and a previous study (n = 62 to 97) may have contributed to some of the differences observed.
In the present study, a relatively high proportion (38.7%) of the sporadic cases of human cryptosporidiosis in Adelaide/South Australia were identified as C. parvum. Furthermore, all but one sample from this subset was identified as C. parvum genotype IIa. The only other published data available for gp60 genotypes and subgenotypes from cases of sporadic human cryptosporidiosis in Adelaide or South Australia also showed that C. parvum genotype IIa represented approximately one-third of the isolates investigated (14). Previous studies have reported that C. parvum genotype IIa is a major contributor to human cryptosporidiosis in Portugal, the United Kingdom, Slovenia, and parts of North America (6, 14, 28, 78), and it has been proposed in these studies that the predominance of this genotype is indicative of a high level of zoonotic transmission. In contrast, previous studies (12, 82) found that ∼80% of cryptosporidiosis infections in urban populations in developed countries were associated with C. hominis infection. This 4:1 ratio of C. hominis/C. parvum infections appears to be the case for samples from humans with sporadic cryptosporidiosis in Melbourne, as a recent study (36) revealed that 75.5% of 98 samples contained C. hominis. The high prevalence of C. parvum subgenotypes within genotype IIa in sporadic human cryptosporidiosis in Adelaide/South Australia also might indicate a significant zoonotic contribution. Indeed, a previous study (64) showed a positive correlation between samples from this region and likely zoonotic risk factors, such as human contact with calves, the consumption of some cheeses and unpasteurized milk, or the consumption of untreated lake, river, and dam water in rural areas. However, it is important that the available data (51, 84) do indicate that C. parvum can be transmitted both by anthroponotic and zoonotic routes, so an increased zoonotic risk to humans in this region cannot be concluded solely based upon the data presented here.
Interestingly, some subgenotypes, such as C. hominis IbA10G2R2 (6, 7, 18, 22, 40, 57, 72, 73, 87) and C. parvum IIaA15G2R1 (1, 6, 18, 57, 72, 76, 84, 87), have been reported from numerous countries and appear to be commonly associated with cryptosporidiosis in humans and/or animals globally. However, rare genetic variants, such as C. hominis IaA17R1, C. hominis IfA12G1R2, and C. parvum IIaA22G4R1 (present study), indicate that localized, potentially geographically unique, subgenotypes are present even in large, urban populations in cosmopolitan cities, emphasizing the need for the increased study of underrepresented countries (such as those in Africa, Asia, and South America, as well as most island nations). There have been some suggestions (24, 57) that strain variation within Cryptosporidium species is correlated with geographical origin. Also, various authors (1, 5, 7, 51, 84, 87) have reported gp60 genotypes or subgenotypes that infect both humans and animals and have formulated hypotheses regarding the importance of zoonotic transmission in causing sporadic cases or outbreaks of cryptosporidiosis in humans. However, to date neither the global distribution of gp60 genotypes and subgenotypes nor the range in host specificities of these types has been assessed comprehensively.
The present study classified Cryptosporidium species genotypes/subgenotypes based on phylogenetic analysis and the formation of strongly supported monophyletic clades (36). This approach was used because, although many gp60 genotypes and subgenotypes have been reported, threshold values for sequence variation (within species) and sequence difference (between species) have not yet been defined for the classification of genotypes/subgenotypes. As such, the decision as to whether a new sequence type constitutes a new genotype or subgenotype is largely subjective; while phylogenetic analyses conducted in the present and previous studies (1, 36, 60, 72) indicate that the currently recognized genotypes usually form strongly supported monophyletic clades and thus appear to be valid, the current lack of defined genetic thresholds does not provide a solid framework for the classification of new genetic variants of Cryptosporidium. A comprehensive assessment of the nature and extent of sequence variation within and among gp60 and other informative genetic loci is required to establish a more definitive classification system for genotypes and subgenotypes globally.
Acknowledgments
This study was supported by the Melbourne Water Corporation (R.B.G.) and the Australian Research Council (LP0561862).
We thank Ivan Bastian for providing some of the samples.
Footnotes
Published ahead of print on 30 April 2008.
REFERENCES
- 1.Abe, N., M. Matsubayashi, I. Kimata, and M. Iseki. 2006. Subgenotype analysis of Cryptosporidium parvum isolates from humans and animals in Japan using the 60-kDa glycoprotein gene sequences. Parasitol. Res. 99303-305. [DOI] [PubMed] [Google Scholar]
- 2.Abrahamsen, M. S., T. J. Templeton, S. Enomoto, J. E. Abrahante, G. Zhu, C. A. Lancto, M. Deng, C. Liu, G. Widmer, S. Tzipori, G. A. Buck, P. Xu, A. T. Bankier, P. H. Dear, B. A. Konfortov, H. F. Spriggs, L. Iyer, V. Anantharaman, L. Aravind, and V. Kapur. 2004. Complete genome sequence of the apicomplexan Cryptosporidium parvum. Science 304441-445. [DOI] [PubMed] [Google Scholar]
- 3.Abs EL-Osta, Y. G., R. M. Chalmers, and R. B. Gasser. 2003. Survey of Cryptosporidium parvum genotypes in humans from the UK by mutation scanning analysis of a heat shock protein gene region. Mol. Cell. Probes 17127-134. [DOI] [PubMed] [Google Scholar]
- 4.Akiyoshi, D. E., J. K. Tumwine, S. Bakeera-Kitaka, and S. Tzipori. 2006. Subtype analysis of Cryptosporidium isolates from children in Uganda. J. Parasitol. 921097-1100. [DOI] [PubMed] [Google Scholar]
- 5.Alves, M., A. M. Ribeiro, C. Neto, E. Ferreira, M. J. Benoliel, F. Antunes, and O. Matos. 2006. Distribution of Cryptosporidium species and subtypes in water samples in Portugal: a preliminary study. J. Eukaryot. Microbiol. 53(Suppl. 1)S24-S5. [DOI] [PubMed] [Google Scholar]
- 6.Alves, M., L. Xiao, F. Antunes, and O. Matos. 2006. Distribution of Cryptosporidium subtypes in humans and domestic and wild ruminants in Portugal. Parasitol. Res. 99287-292. [DOI] [PubMed] [Google Scholar]
- 7.Alves, M., L. Xiao, I. M. Sulaiman, A. A. Lal, O. Matos, and F. Antunes. 2003. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 412744-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson, V. R., and M. P. Curran. 2007. Nitazoxanide: a review of its use in the treatment of gastrointestinal infections. Drugs 671947-1967. [DOI] [PubMed] [Google Scholar]
- 9.Armson, A., R. C. A. Thompson, and J. A. Reynoldson. 2003. A review of chemotherapeutic approaches to the treatment of cryptosporidiosis. Expert Rev. Anti. Infect. Ther. 1297-305. [DOI] [PubMed] [Google Scholar]
- 10.Baishanbo, A., G. Gargala, C. Duclos, A. Francois, J. F. Rossignol, J. J. Ballet, and L. Favennec. 2006. Efficacy of nitazoxanide and paromomycin in biliary tract cryptosporidiosis in an immunosuppressed gerbil model. J. Antimicrob. Chemother. 57353-355. [DOI] [PubMed] [Google Scholar]
- 11.Black, M., and J. McAnulty. 2006. The investigation of an outbreak of cryptosporidiosis in New South Wales in 2005. N. S. W. Public Health Bull. 1776-79. [DOI] [PubMed] [Google Scholar]
- 12.Cacciò, S. M. 2005. Molecular epidemiology of human cryptosporidiosis. Parassitologia 47185-192. [PubMed] [Google Scholar]
- 13.Cacciò, S. M., and E. Pozio. 2006. Advances in the epidemiology, diagnosis and treatment of cryptosporidiosis. Expert Rev. Anti. Infect. Ther. 4429-443. [DOI] [PubMed] [Google Scholar]
- 14.Chalmers, R. M., C. Ferguson, S. M. Cacciò, R. B. Gasser, Y. G. Abs EL-Osta, L. Heijnen, L. Xiao, K. Elwin, S. Hadfield, M. I. Sinclair, and M. Stevens. 2005. Direct comparison of selected methods for genetic categorisation of Cryptosporidium parvum and Cryptosporidium hominis species. Int. J. Parasitol. 35397-410. [DOI] [PubMed] [Google Scholar]
- 15.Checkley, W., R. H. Gilman, L. D. Epstein, M. Suarez, J. F. Diaz, L. Cabrera, R. E. Black, and C. R. Sterling. 1997. Asymptomatic and symptomatic cryptosporidiosis: their acute effect on weight gain in Peruvian children. Am. J. Epidemiol. 145156-163. [DOI] [PubMed] [Google Scholar]
- 16.Chen, X. M., J. S. Keithly, C. V. Paya, and N. F. LaRusso. 2002. Cryptosporidiosis. N. Engl. J. Med. 3461723-1731. [DOI] [PubMed] [Google Scholar]
- 17.Chilton, N. B., R. B. Gasser, and I. Beveridge. 1995. Differences in a ribosomal DNA sequence of morphologically indistinguishable species within the Hypodontus macropi complex (Nematoda: Strongyloidea). Int. J. Parasitol. 25647-651. [DOI] [PubMed] [Google Scholar]
- 18.Cohen, S., F. Dalle, A. Gallay, M. Di Palma, A. Bonnin, and H. D. Ward. 2006. Identification of Cpgp40/15 type Ib as the predominant allele in isolates of Cryptosporidium spp. from a waterborne outbreak of gastroenteritis in South Burgundy, France. J. Clin. Microbiol. 44589-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craun, G. F., R. L. Calderon, and M. F. Craun. 2005. Outbreaks associated with recreational water in the United States. Int. J. Environ. Health Res. 15243-262. [DOI] [PubMed] [Google Scholar]
- 20.Dziuban, E. J., J. L. Liang, G. F. Craun, V. Hill, P. A. Yu, J. Painter, M. R. Moore, R. L. Calderon, S. L. Roy, and M. J. Beach. 2006. Surveillance for waterborne disease and outbreaks associated with recreational water-United States, 2003-2004. MMWR Surveill. Summ. 551-30. [PubMed] [Google Scholar]
- 21.Fayer, R. 2004. Cryptosporidium: a water-borne zoonotic parasite. Vet. Parasitol. 12637-56. [DOI] [PubMed] [Google Scholar]
- 22.Feltus, D. C., C. W. Giddings, B. L. Schneck, T. Monson, D. Warshauer, and J. M. McEvoy. 2006. Evidence supporting zoonotic transmission of Cryptosporidium spp. in Wisconsin. J. Clin. Microbiol. 444303-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasser, R. B. 2006. Molecular tools—advances, opportunities and prospects. Vet. Parasitol. 13669-89. [DOI] [PubMed] [Google Scholar]
- 24.Gasser, R. B., Y. G. Abs EL-Osta, and R. M. Chalmers. 2003. Electrophoretic analysis of genetic variability within Cryptosporidium parvum from imported and autochthonous cases of human cryptosporidiosis in the United Kingdom. Appl. Environ. Microbiol. 692719-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasser, R. B., M. Hu, N. B. Chilton, B. E. Campbell, A. R. Jex, D. Otranto, C. Cafarchia, I. Beveridge, and X. Q. Zhu. 2006. Single-strand conformation polymorphism (SSCP) for the analysis of genetic variation. Nat. Protoc. 13121-3128. [DOI] [PubMed] [Google Scholar]
- 26.Gasser, R. B., X. Q. Zhu, S. M. Cacciò, R. M. Chalmers, G. Widmer, U. M. Morgan, R. C. A. Thompson, E. Pozio, and G. F. Browning. 2001. Genotyping Cryptosporidium parvum by single-strand conformation polymorphism analysis of ribosomal and heat shock gene regions. Electrophoresis 22433-437. [DOI] [PubMed] [Google Scholar]
- 27.Gatei, W., C. A. Hart, R. H. Gilman, P. Das, V. Cama, and L. Xiao. 2006. Development of a multilocus sequence typing tool for Cryptosporidium hominis. J. Eukaryot. Microbiol. 53(Suppl. 1)S43-S48. [DOI] [PubMed] [Google Scholar]
- 28.Glaberman, S., J. E. Moore, C. J. Lowery, R. M. Chalmers, I. M. Sulaiman, K. Elwin, P. J. Rooney, B. C. Millar, J. S. Dooley, A. A. Lal, and L. Xiao. 2002. Three drinking-water-associated cryptosporidiosis outbreaks, Northern Ireland. Emerg. Infect. Dis. 8631-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greif, G., A. Harder, and A. Haberkorn. 2001. Chemotherapeutic approaches to protozoa: Coccidiae-current level of knowledge and outlook. Parasitol. Res. 87973-975. [DOI] [PubMed] [Google Scholar]
- 30.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 4195-98. [Google Scholar]
- 31.Henriksen, S. A., and J. F. L. Pohlenz. 1981. Staining of cryptosporidia by a modified Ziehl-Neelsen technique. Acta Vet. Scand. 22594-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houpt, E. R., O. Y. Bushen, N. E. Sam, A. Kohli, A. Asgharpour, C. T. Ng, D. P. Calfee, R. L. Guerrant, V. Maro, S. Ole-Nguyaine, and J. F. Shao. 2005. Short report: asymptomatic Cryptosporidium hominis infection among human immunodeficiency virus-infected patients in Tanzania. Am. J. Trop. Med. Hyg. 73520-522. [PubMed] [Google Scholar]
- 33.Hunter, P. R., S. J. Hadfield, D. Wilkinson, I. R. Lake, F. C. Harrison, and R. M. Chalmers. 2007. Subtypes of Cryptosporidium parvum in humans and disease risk. Emerg. Infect. Dis. 1382-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter, P. R., and G. Nichols. 2002. Epidemiology and clinical features of Cryptosporidium infection in immunocompromised patients. Clin. Microbiol. Rev. 15145-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jex, A. R., H. V. Smith, P. Monis, B. E. Campbell, and R. B. Gasser. Cryptosporidium—biotechnological advances in detection, diagnosis and analysis of genetic variation. Biotechnol. Adv., in press. [DOI] [PubMed]
- 36.Jex, A. R., M. Whipp, B. E. Campbell, S. M. Cacciò, M. Stevens, G. Hogg, and R. B. Gasser. 2007. A practical and cost-effective mutation scanning-based approach for investigating genetic variation in Cryptosporidium. Electrophoresis 283875-3883. [DOI] [PubMed] [Google Scholar]
- 37.Karanis, P., C. Kourenti, and H. V. Smith. 2007. Waterborne transmission of protozoan parasites: a worldwide review of outbreaks and lessons learnt. J. Water Health 51-38. [DOI] [PubMed] [Google Scholar]
- 38.Khramtsov, N. V., M. Tilley, D. S. Blunt, B. A. Montelone, and S. J. Upton. 1995. Cloning and analysis of a Cryptosporidium parvum gene encoding a protein with homology to cytoplasmic form Hsp70. J. Eukaryot. Microbiol. 42416-422. [DOI] [PubMed] [Google Scholar]
- 39.Kosek, M., C. Alcantara, A. A. Lima, and R. L. Guerrant. 2001. Cryptosporidiosis: an update. Lancet Infect. Dis. 1262-269. [DOI] [PubMed] [Google Scholar]
- 40.Leav, B. A., M. R. Mackay, A. Anyanwu, R. M. O'Connor, A. M. Cevallos, G. Kindra, N. C. Rollins, M. L. Bennish, R. G. Nelson, and H. D. Ward. 2002. Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infect. Immun. 703881-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacKenzie, W. R., W. L. Schell, K. A. Blair, D. G. Addiss, D. E. Peterson, N. J. Hoxie, J. J. Kazmierczak, and J. P. Davis. 1995. Massive outbreak of waterborne Cryptosporidium infection in Milwaukee, Wisconsin: recurrence of illness and risk of secondary transmission. Clin. Infect. Dis. 2157-62. [DOI] [PubMed] [Google Scholar]
- 42.Mallon, M., A. MacLeod, J. Wastling, H. V. Smith, B. Reilly, and A. Tait. 2003. Population structures and the role of genetic exchange in the zoonotic pathogen Cryptosporidium parvum. J. Mol. Evol. 56407-417. [DOI] [PubMed] [Google Scholar]
- 43.Mead, J. R. 2002. Cryptosporidiosis and the challenges of chemotherapy. Drug Resist. Updat. 547-57. [DOI] [PubMed] [Google Scholar]
- 44.Meamar, A. R., M. Rezaian, S. Rezaie, M. Mohraz, E. B. Kia, E. R. Houpt, and S. Solaymani-Mohammadi. 2006. Cryptosporidium parvum bovine genotype oocysts in the respiratory samples of an AIDS patient: efficacy of treatment with a combination of azithromycin and paromomycin. Parasitol. Res. 98593-595. [DOI] [PubMed] [Google Scholar]
- 45.Medema, G., P. Teunis, M. Blokker, D. Deere, A. Davison, P. Charles, and J.-F. Loret. 2006. WHO guidelines for drinking water quality: Cryptosporidium. WHO, New York, NY.
- 46.Meireles, M. V., R. M. Soares, F. Bonello, and S. M. Gennari. 2007. Natural infection with zoonotic subtype of Cryptosporidium parvum in Capybara (Hydrochoerus hydrochaeris) from Brazil. Vet. Parasitol. 147166-170. [DOI] [PubMed] [Google Scholar]
- 47.Misic, Z., and N. Abe. 2007. Subtype analysis of Cryptosporidium parvum isolates from calves on farms around Belgrade, Serbia and Montenegro, using the 60 kDa glycoprotein gene sequences. Parasitology 134351-358. [DOI] [PubMed] [Google Scholar]
- 48.Morgan, U. M., C. C. Constantine, D. A. Forbes, and R. C. A. Thompson. 1997. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J. Parasitol. 83825-830. [PubMed] [Google Scholar]
- 49.Morgan, U. M., K. D. Sargent, P. Deplazes, D. A. Forbes, F. Spano, H. Hertzberg, A. Elliot, and R. C. A. Thompson. 1998. Molecular characterization of Cryptosporidium from various hosts. Parasitology 11731-37. [DOI] [PubMed] [Google Scholar]
- 50.Morgan, U. M., L. Xiao, I. M. Sulaiman, R. Weber, A. A. Lal, R. C. A. Thompson, and P. Deplazes. 1999. Which genotypes/species of Cryptosporidium are humans susceptable to? J. Eukaryot. Microbiol. 4642S-43S. [PubMed] [Google Scholar]
- 51.O'Brien, E., L. McInnes, and U. M. Ryan. 2008. Cryptosporidium GP60 genotypes from humans and domesticated animals in Australia, North America and Europe. Exp. Parasitol. 118118-121. [DOI] [PubMed] [Google Scholar]
- 52.Okhuysen, P. C., and C. L. Chappell. 2002. Cryptosporidium virulence determinants-are we there yet? Int. J. Parasitol. 32517-525. [DOI] [PubMed] [Google Scholar]
- 53.Okhuysen, P. C., C. L. Chappell, J. H. Crabb, C. R. Sterling, and H. L. DuPont. 1999. Virulence of three distinct Cryptosporidium parvum isolates for healthy adults. J. Infect. Dis. 1801275-1281. [DOI] [PubMed] [Google Scholar]
- 54.Ong, C. S., D. L. Eisler, S. H. Goh, J. Tomblin, F. M. Awad-El-Kariem, C. B. Beard, L. Xiao, I. M. Sulaiman, A. Lal, M. Fyfe, A. King, W. R. Bowie, and J. L. Isaac-Renton. 1999. Molecular epidemiology of cryptosporidiosis outbreaks and transmission in British Columbia, Canada. Am. J. Trop. Med. Hyg. 6163-69. [DOI] [PubMed] [Google Scholar]
- 55.Palit, A., D. Sur, K. MitraDhar, and M. R. Saha. 2005. Asymptomatic cryptosporiosis in a periurban slum setting in Kolkata, India-a pilot study. Jpn. J. Infect. Dis. 58110-111. [PubMed] [Google Scholar]
- 56.Palmieri, F., S. Cicalini, N. Froio, E. B. Rizzi, D. Goletti, A. Festa, G. Macri, and N. Petrosillo. 2005. Pulmonary cryptosporidiosis in an AIDS patient: successful treatment with paromomycin plus azithromycin. Int. J. STD AIDS 16515-517. [DOI] [PubMed] [Google Scholar]
- 57.Peng, M. M., O. Matos, W. Gatei, P. Das, M. Stantic-Pavlinic, C. Bern, I. M. Sulaiman, S. Glaberman, A. A. Lal, and L. Xiao. 2001. A comparison of Cryptosporidium subgenotypes from several geographic regions. J. Eukaryot. Microbiol. Suppl.:28S-31S. [DOI] [PubMed]
- 58.Peng, M. M., S. R. Meshnick, N. A. Cunliffe, B. D. Thindwa, C. A. Hart, R. L. Broadhead, and L. Xiao. 2003. Molecular epidemiology of cryptosporidiosis in children in Malawi. J. Eukaryot. Microbiol. 50(Suppl.)557-559. [DOI] [PubMed] [Google Scholar]
- 59.Pettoello-Mantovani, M., L. Di Martino, G. Dettori, P. Vajro, S. Scotti, M. T. Ditullio, and S. Guandalini. 1995. Asymptomatic carriage of intestinal Cryptosporidium in immunocompetent and immunodeficient children: a prospective study. Pediatr. Infect. Dis. J. 141042-1047. [DOI] [PubMed] [Google Scholar]
- 60.Plutzer, J., and P. Karanis. 2007. Genotype and subtype analyses of Cryptosporidium isolates from cattle in Hungary. Vet. Parasitol. 146357-362. [DOI] [PubMed] [Google Scholar]
- 61.Puech, M. C., J. M. McAnulty, M. Lesjak, N. Shaw, L. Heron, and J. M. Watson. 2001. A statewide outbreak of cryptosporidiosis in New South Wales associated with swimming at public pools. Epidemiol. Infect. 126389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramirez, N. E., L. A. Ward, and S. Sreevatsan. 2004. A review of the biology and epidemiology of cryptosporidiosis in humans and animals. Microbes. Infect. 6773-785. [DOI] [PubMed] [Google Scholar]
- 63.Riggs, M. W. 2002. Recent advances in cryptosporidiosis: the immune response. Microbes Infect. 41067-1080. [DOI] [PubMed] [Google Scholar]
- 64.Robertson, B., M. I. Sinclair, A. B. Forbes, M. Veitch, M. Kirk, D. Cunliffe, J. Willis, and C. K. Fairley. 2002. Case-control studies of sporadic cryptosporidiosis in Melbourne and Adelaide, Australia. Epidemiol. Infect. 128419-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rossignol, J. F., S. M. Kabil, Y. El-Gohary, and A. M. Younis. 2006. Effect of nitazoxanide in diarrhea and enteritis caused by Cryptosporidium species. Clin. Gastroenterol. Hepatol. 4320-324. [DOI] [PubMed] [Google Scholar]
- 66.Schuster, C. J., A. G. Ellis, W. J. Robertson, D. F. Charron, J. J. Aramini, B. J. Marshall, and D. T. Medeiros. 2005. Infectious disease outbreaks related to drinking water in Canada, 1974-2001. Can. J. Public Health 96254-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Semenza, J. C., and G. Nichols. 2007. Cryptosporidiosis surveillance and water-borne outbreaks in Europe. Eur. Surveill. 12E13-E14. [DOI] [PubMed] [Google Scholar]
- 68.Siwila, J., I. G. Phiri, J. Vercruysse, F. Goma, S. Gabriel, E. Claerebout, and T. Geurden. 2007. Asymptomatic cryptosporidiosis in Zambian dairy farm workers and their household members. Trans. R. Soc. Trop. Med. Hyg. 101733-734. [DOI] [PubMed] [Google Scholar]
- 69.Skerrett, H. E., and C. V. Holland. 2001. Asymptomatic shedding of Cryptosporidium oocysts by red deer hinds and calves. Vet. Parasitol. 94239-246. [DOI] [PubMed] [Google Scholar]
- 70.Smith, H. V., S. M. Cacciò, A. Tait, J. McLauchlin, and R. C. A. Thompson. 2006. Tools for investigating the environmental transmission of Cryptosporidium and Giardia infections in humans. Trends Parasitol. 22160-167. [DOI] [PubMed] [Google Scholar]
- 71.Strong, W. B., J. Gut, and R. G. Nelson. 2000. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 684117-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sulaiman, I. M., P. R. Hira, L. Zhou, F. M. Al-Ali, F. A. Al-Shelahi, H. M. Shweiki, J. Iqbal, N. Khalid, and L. Xiao. 2005. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 432805-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sulaiman, I. M., A. A. Lal, and L. Xiao. 2001. A population genetic study of the Cryptosporidium parvum human genotype parasites. J. Eukaryot. Microbiol. Suppl:24S-27S. [DOI] [PubMed]
- 74.Swofford, D. L. 1999. PAUP*, 4.0b10 ed. Sinauer Associates, Sunderland, MA.
- 75.Thomas, K. M., D. F. Charron, D. Waltner-Toews, C. Schuster, A. R. Maarouf, and J. D. Holt. 2006. A role of high impact weather events in waterborne disease outbreaks in Canada, 1975-2001. Int. J. Environ. Health Res. 16167-180. [DOI] [PubMed] [Google Scholar]
- 76.Thompson, H. P., J. S. Dooley, J. Kenny, M. McCoy, C. J. Lowery, J. E. Moore, and L. Xiao. 2007. Genotypes and subtypes of Cryptosporidium spp. in neonatal calves in Northern Ireland. Parasitol. Res. 100619-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 244876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trotz-Williams, L. A., D. S. Martin, W. Gatei, V. Cama, A. S. Peregrine, S. W. Martin, D. V. Nydam, F. Jamieson, and L. Xiao. 2006. Genotype and subtype analyses of Cryptosporidium isolates from dairy calves and humans in Ontario. Parasitol. Res. 99346-352. [DOI] [PubMed] [Google Scholar]
- 79.Wu, Z., I. Nagano, T. Boonmars, T. Nakada, and Y. Takahashi. 2003. Intraspecies polymorphism of Cryptosporidium parvum revealed by PCR-restriction fragment length polymorphism (RFLP) and RFLP-single-strand conformational polymorphism analyses. Appl. Environ. Microbiol. 694720-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiao, L., L. Escalante, C. Yang, I. M. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 651578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiao, L., R. Fayer, U. M. Ryan, and S. J. Upton. 2004. Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 1772-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiao, L., and U. M. Ryan. 2004. Cryptosporidiosis: an update in molecular epidemiology. Curr. Opin. Infect. Dis. 17483-490. [DOI] [PubMed] [Google Scholar]
- 83.Xiao, L., U. M. Ryan, T. K. Graczyk, J. Limor, L. Li, M. Kombert, R. Junge, I. M. Sulaiman, L. Zhou, M. J. Arrowood, B. Koudela, D. Modry, and A. A. Lal. 2004. Genetic diversity of Cryptosporidium spp. in captive reptiles. Appl. Environ. Microbiol. 70891-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiao, L., L. Zhou, M. Santin, W. Yang, and R. Fayer. 2007. Distribution of Cryptosporidium parvum subtypes in calves in eastern United States. Parasitol. Res. 100701-706. [DOI] [PubMed] [Google Scholar]
- 85.Yoder, J. S., and M. J. Beach. 2007. Cryptosporidiosis surveillance-United States, 2003-2005. MMWR Surveill. Summ. 561-10. [PubMed] [Google Scholar]
- 86.Zardi, E. M., A. Picardi, and A. Afeltra. 2005. Treatment of cryptosporidiosis in immunocompromised hosts. Chemotherapy 51193-196. [DOI] [PubMed] [Google Scholar]
- 87.Zhou, L., A. Singh, J. Jiang, and L. Xiao. 2003. Molecular surveillance of Cryptosporidium spp. in raw wastewater in Milwaukee: implications for understanding outbreak occurrence and transmission dynamics. J. Clin. Microbiol. 415254-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]