Abstract
The molecular characterization of partial- length genomic segment 2 of porcine picobirnavirus (PBV) strains and the development of a specific reverse transcription-PCR (RT-PCR) assay for detection of virus in feces are reported. Phylogenetic analysis indicated that the studied porcine isolates were more closely related (>85% identity) to human PBV belonging to genogroup I than to the other porcine PBV described so far. Analysis by RT-PCR and polyacrylamide gel electrophoresis of fecal samples collected in Venezuela and Argentina showed that PBV circulate at high frequencies in piglets.
Picobirnaviruses (PBV) are a group of small viruses whose genome is composed of two segments of double-stranded RNA (dsRNA), ranging in size from 2.2 to 2.6 kbp and from 1.5 to 1.9 kbp for the larger and smaller segment, respectively. The virion is nonenveloped, with a diameter of 33 to 35 nm (21). The larger RNA segment, or segment 1, contains two open reading frames encoding 224 and 552 amino acids, while the small RNA segment, or segment 2, contains a single open reading frame encoding 534 amino acids (29). The small segment encodes amino acid motifs typical of those encoded by the RNA-dependent RNA polymerase genes (23, 27, 29). Because of its genomic and morphological characteristics, this novel group of viruses is a likely candidate for a new taxon.
PBV have been identified in fecal samples of humans practically worldwide (2, 4, 5, 8, 10, 20, 23, 25, 27), in feces of a wide variety of farm mammals and birds, including pigs (3, 9, 12, 21, 26), calves (7), rabbits (11, 14, 22), and chickens (19), and even in feces of wild animals keep in captivity (16, 24, 30). Despite the numerous reports of the presence of PBV in fecal samples from vertebrates, the pathogenicity of these viruses has not been established. However, studies conducted with human immunodeficiency virus-infected persons (13, 15) suggest that PBV may be an opportunistic pathogen that may cause diarrhea in immunosuppressed individuals.
Molecular characterization of PBV is limited. Epidemiological studies carried out in several countries have made it clear that human PBV strains are highly diverse and can be grouped into at least two genogroups (2, 5, 23, 27). PBV sequences from pigs have recently been described (3), being genetically related to the human genogroup I. In this work, we report the partial molecular characterization of porcine isolates of PBV and the development of a specific reverse transcription-PCR (RT-PCR) assay for the direct detection of PBV in porcine feces. Our results suggest that porcine PBV strains are genetically diverse, are related to human strains, and cause frequent infections among young pigs.
For cloning purposes, stool samples were collected from piglets 7 to 56 days of age and kept at −70°C until processed. RNA was extracted from clarified fecal suspensions using Trizol LS (Invitrogen). PBV-positive samples were identified by 7% polyacrylamide gel electrophoresis (PAGE) and silver nitrate staining. The clarified supernatant from a sample that tested strongly positive by PAGE was centrifuged through a 20% (wt/vol) sucrose cushion, and the pelleted virions were purified by centrifugation in a CsCl gradient (21). dsRNA extracted from purified virions was subjected to DNase I and RNase T1 digestion prior to cloning assays.
Based on human and rabbit PBV sequences available in GenBank, degenerated primers were designed for PBV genomic segments 2 and 1, respectively. The sequences, locations, and specificities of these primers are shown in the supplemental material. The cDNA synthesis was performed in a one-tube RT-PCR assay (18) and amplicons cloned into the plasmid vector pCR 2.1-TOPO. The obtained clones were sequenced in both directions at least twice. The sequences were edited and aligned using the program DNAMAN, version 5.2.2, and compared to PBV reference strains deposited in GenBank using the BLAST program. Phylogenetic trees were constructed with the program MEGA 4 (28) using the minimum-evolution method, with evolutionary distances computed using the maximum-composite-likelihood method. Five clones, derived from amplicons generated with primers directed to segment 2, could be established. The clones were named PBV1-POR (442 bp), PBV2-POR (307 bp), PBV3-POR (363 bp), PBV4-POR (300 bp), and PBV1a-POR (116 bp). BLAST analysis and alignments mapped the clones between nucleotides 443 and 1059 of the human reference strain 1-CHN-97 and between nucleotides 466 and 1091 of the human reference strain Hy005102, thus indicating that all five clones contained sequences corresponding to internal portions of PBV segment 2. The nucleotide identity of the clones with the human PBV reference strains 1-CHI-97 and Hy005102 ranged between 71 and 77%. Sequence analysis revealed that each had a unique sequence with the exception of clone PBV1a-POR, which was identical (100% nucleotide identity) to clone PBV-1. Clones PBV3-POR and PBV4-POR showed nucleotide identities with clone PBV1-POR of 61% and 71.3%, respectively. Finally, clone PBV2-POR showed an overlapping region of only 82 nucleotides with the 3′ end of clone PBV1-POR, and within this region the nucleotide identity was 72%. The presence of significantly different sequences within the same virus sample was unexpected, since the original sample showed a single electrophoretic pattern. One possible explanation is that the degenerated primers were able to recognize several strains with the identical electrophoretic pattern present in the sample. An alternative explanation is that these strains were present in the sample in viral loads below the detection limit of the PAGE technique, which is an insensitive technique.
The deduced amino acid sequences showed that all the clones encoded the conserved residues D-S-D and clone PBV2-POR encoded additionally the conserved residues SG-T—E, corresponding, respectively, to motifs 1 and 2 of the RNA-dependent RNA polymerases of dsRNA viruses (6), thus confirming previous results obtained with human PBV which postulated that genomic segment 2 encodes the polymerase of these viruses (23, 27, 29).
A single clone of 199 bp was established from amplicons derived from segment 1. However, BLAST analyses failed to identify any identity between this clone and sequences deposited in GenBank.
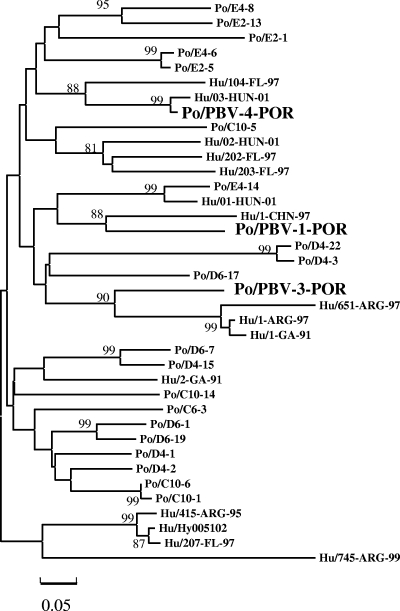
Phylogenetic trees were constructed to study the relationship between these and previously reported porcine and human PBV isolates. Clone PBV2-POR was left out of the analysis, since mostly PBV sequences containing RNA polymerase motif 1 were available in GenBank. The phylogenetic analysis included sequences from 19 human strains, 18 representative of genogroup I (2, 23, 27, 29), and 43 recently reported sequences from porcine picobirnaviruses isolated in Hungary, also classified as genogroup I (3). Figure 1 shows a phylogenetic tree based on a 168-nucleotides fragment which included clones PBV1-POR, PVB3-POR, and PBV4-POR. For clarity, only 19 of the 43 porcine sequences available in GenBank are included in the figure, excluding in each clade redundant sequences (>97.5% nucleotide sequence identity). The branching order of the basal nodes could not be resolved, since they were not supported by significant bootstrap values, not allowing a further subdivision scheme within genogroup I based on the sequence of this region. Nevertheless, several clades were identified with high bootstrap values (>80%). The three porcine PBV isolates from Venezuela were not clustered within a single clade, nor with the porcine strains isolated in Hungary, but each one was found to be closely related to a different clade of human strains. Clone PBV1-POR showed maximum nucleotide identity with the reference strain 1-CHN-97 (77%) and clone PBV3-POR with strain 651-ARG-97 (76%), while clone PBV4-POR was found to be very similar to strain 03-HUN-01 (96%). Thus, these porcine isolates from Venezuela were distributed quite differently from the porcine isolates from Hungary, which all clustered, with one exception (Po/E4-14), on branches distinct from those of human isolates. These results strongly suggest that PBV strains may circulate between humans and pigs. Of note, a similar distribution between porcine and human strains was observed when the phylogenetic analyses were based on deduced amino acid sequences (data not shown). The zoonotic potential of PBV is an interesting question that deserves further attention.
FIG. 1.
Phylogenetic analysis of genogroup I human and porcine picobirnavirus strains based on a 168-nucleotide fragment of the RNA polymerase gene, corresponding to positions 714 to 881 of the RNA segment 2 sequence of human strain Hy005102. The tree was constructed using the minimum-evolution method as implemented in the package Mega 4, with evolutionary distances computed using the maximum-composite-likelihood method. Bars are in units of base substitutions per site. Bootstrap values of >80% are indicated for the corresponding nodes, based on a resampling analysis of 1,000 replicates. The “Po” or “Hu” prefix to strain names indicates porcine or human origin, respectively. Venezuelan isolates are indicated in bold.
The high genetic variability for porcine PBV has been previously suggested by epidemiological studies based on PAGE which found high electrophoretic mobility among PBV isolates (12, 21) and more recently by phylogenetic analysis (3). In agreement, high genetic variability has also been observed among human PBV isolates, even among isolates from single locations and homogeneous populations (2, 4, 5, 23, 27). The significance of extended genetic variability among PBV is currently unknown.
To develop an RT-PCR assay for the detection of porcine PBV in fecal samples, several primer pairs were tested with PBV genomes extracted from CsCl-purified virions. However, the most consistent results were obtained with primer pair PBV2-19/PBV2-281, which generated clean amplicons of 262 bp. The amplicon was mapped between nucleotides 824 and 1086 of the human reference strain Hy005102. Thus, the primer pair PBV2-19/PBV2-281 was further tested against a panel of 144 fecal samples collected from piglets in farms in Venezuela (n = 80) and Argentina (n = 64). Nucleic acids were directly extracted from clarified fecal samples (10% [wt/vol]) without further processing. Reverse transcription was carried out using random primers (17). PCR amplification was carried out in a 50-μl reaction mixture which contained 1× PCR buffer, 2 mM MgCl2, 0.4 mM deoxynucleoside triphosphates, 0.5 μM of each primer, 0.25 U of Taq polymerase, and 5 μl of cDNA. The thermal cycling conditions were as follows: an initial denaturation at 95°C for 5 min, followed by 35 cycles of amplification (denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension for 72°C for 1 min) and a final extension of 72°C for 7 min. Samples had been previously analyzed for the presence of PBV by PAGE. The results are shown in Table 1. Overall, the RT-PCR assay was capable of identifying as positive for PBV 26 (67%) of the 39 samples that were positive by PAGE. In addition, the RT-PCR assay identified as positive for PBV 58% (61/105) of the samples that tested negative by PAGE. Thus, a total 60% (87/144) of the samples tested were found to be positive for PBV by RT-PCR (versus 27% by PAGE). To corroborate the RT-PCR results, six amplicons obtained from the PAGE-negative samples were sequenced in both senses and found to correspond to PBV. Noteworthily, all the 13 samples that were positive by PAGE but failed to be amplified with primers PBV2-19/PBV2-281 were from the Argentinean collection. Since all primers were initially derived from the same clones, no other primer set was tested with those samples. Finally, none of the samples that were found to be positive for rotavirus by PAGE (n = 6) or lysates from cells infected with rotavirus included as controls generated amplicons.
TABLE 1.
Detection by PAGE and RT-PCR of porcine PBV in porcine fecal samples collected in Venezuela and Argentina
| PAGE result | No. (%) of samples with RT-PCR result
|
Total no. (%) of samples | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 26 | 13a | 39 (27) |
| Negative | 61 | 44 | 105 |
| Total | 87 (60) | 57 | 144 |
Samples positive by PAGE but negative by RT-PCR were all of Argentinean origin.
The reactivities observed with the primers developed in this study for porcine PBV resemble the limited reactivity pattern observed for the two primer pairs developed by Rosen et al. (27) for the detection of human PBV (2, 4, 23). The broad genetic variability observed for PBV most likely is the cause of the limited reactivity shown by the primers developed so far. On the other hand, these results suggest the existence of more than one genogroup for porcine PBV circulating in Argentina. Thus, as is currently the situation for human noroviruses, where no universal primers capable of detecting all the genogroups and genotypes are available and the analysis of a single sample with several primer pairs is customary (1), it is likely that reliable detection of PBV by molecular techniques will call for the combined used of several primers covering different specificities.
Finally, based on PAGE detection, the prevalence of PBV among piglets has been estimated to be around 10 to 12% (12, 21). Although this study was not designed to evaluate prevalence or disease association, the results obtained by RT-PCR suggest that the actual prevalence of PBV in piglets may be at least four or five times higher and that PBV is frequently excreted by piglets without any sign of disease.
Nucleotide sequence accession number.
The nucleotide sequences determined in this study were deposited in GenBank under the following accession numbers: for PBV1-POR, EU104358; for PBV1a-POR, EU104359; for PBV2-POR, EU104360; for PBV3-POR, EU104361; and for PBV4-POR, EU104362.
Acknowledgments
We are in debt to Morella de Rolo (INIA, Maracay) for her help in collection of the samples.
This work was partially supported by BID-FONACIT II (Subproyecto de Biotecnología no. 2004000386), Venezuela.
Footnotes
Published ahead of print on 28 May 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Atmar, R. L., and M. K. Estes. 2001. Diagnosis of noncultivable gastrointesinal viruses. The human calicivirus. Clin. Microbiol. Rev. 1415-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bányai, K., F. Jakab, G. Reuter, J. Bene, M. Új, B. Melegh, and G. Szücs. 2003. Sequence heterogeneity among human picobirnaviruses detected in a gastroenteritis outbreak. Arch. Virol. 1482281-2291. [DOI] [PubMed] [Google Scholar]
- 3.Bányai, K., V. Martella, A. Bogdán, P. Forgách, F. Jakab, E. Meleg, H. Bíró, B. Melegh, and G. Szücs. 2008. Genogroup I picobirnaviruses in pigs: evidence for genetic diversity and relatedness to human strains. J. Gen. Virol. 89534-539. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya, R., G. C. Sahoo, M. K. Nayak, D. R. Saha, D. Sur, T. N. Naik, S. K. Bhattacharya, and T. Krishnan. 2006. Molecular epidemiology of human picobirnaviruses among children of slum community in Kolkata, India. Infect. Genet. Evol. 6453-458. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharya, R., G. C. Sahoo, M. K. Nayak, K. Rajendran, P. Dutta, U. Mitra, M. K. Bhattacharya, T. N. Naik, S. K. Bhattacharya, and T. Krishnan. 2007. Detection of genogroup I and II human picobirnaviruses showing small genomic RNA profile causing watery diarrhoea among children in Kolkata, India. Infect. Genet. Evol. 7229-238. [DOI] [PubMed] [Google Scholar]
- 6.Bruenn, J. A. 1991. Relationships among the positive strand and double-stranded RNA viruses as viewed through their RNA-dependent-RNA polymerases. Nucleic Acids Res. 19217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buzinaro, M. G., P. P. Freitas, J. J. Kisiellus, M. Ueda, and J. A. Jerez. 2003. Identification of a bisegmented double-stranded RNA virus (picobirnavirus) in calf faeces. Vet. J. 166185-187. [DOI] [PubMed] [Google Scholar]
- 8.Cascio, A., M. Bosco, E. Vizzi, A. Giammanco, D. Ferraro, and S. Arista. 1996. Identification of picobirnavirus from faeces of Italian children suffering from acute diarrhea. Eur. J. Epidemiol. 12545-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chasey, D. 1990. Porcine picobirnavirus in UK. Vet. Record. 126465. [PubMed] [Google Scholar]
- 10.Gallimore, C., H. Appleton, D. Lewis, J. Green, and D. W. Brown. 1995. Detection and characterisation of bisegmented double-stranded RNA viruses (Picobirnavirus) in human faecal specimens. J. Med. Virol. 45135-140. [DOI] [PubMed] [Google Scholar]
- 11.Gallimore, C., D. Lewis, and D. W. Brown. 1993. Detection and characterization of a novel bisegmented double-stranded RNA virus (picobirnavirus) from rabbit faeces. Arch. Virol. 13363-73. [DOI] [PubMed] [Google Scholar]
- 12.Gatti, M. S., A. Pestana de Castro, and M. Ferraz. 1989. Viruses with bisegmented double-stranded RNA in pig faeces. Res. Vet. Sci. 47397-398. [PubMed] [Google Scholar]
- 13.Giordano, M., L. Martínez, D. Rinaldi, C. Espul, N. Martínez, M. Isa, A. Depetris, S. Medeot, and S. Nates. 1999. Diarrhea and enteric emerging viruses in HIV-infected patients. AIDS Res. Hum. Retrovir. 151427-1432. [DOI] [PubMed] [Google Scholar]
- 14.Green, J., C. I. Gallimore, J. P. Clewley, and D. W. Brown. 1999. Genomic characterisation of the large segment of a rabbit picobirnavirus and comparison with the atypical picobirnavirus of Cryptosporidium parvum. Arch. Virol. 1442457-2465. [DOI] [PubMed] [Google Scholar]
- 15.Grohmann, G., R. I. Glass, H. G. Pereira, S. S. Monroe, A. W. Hightower, R. Weber, and R. T. Bryan. 1993. Enteric viruses and diarrhea in HIV-infected patients. N. Engl. J. Med. 32914-20. [DOI] [PubMed] [Google Scholar]
- 16.Haga, I. R., S. S. Martins, S. T. Hosomi, F. Vicentini, H. Tanaka, and M. S. Gatti. 1999. Identification of bisegmented double-stranded RNA virus (Picobirnavirus) in faeces of giant anteaters (Myrmecophaga tridactyla). Vet. J. 158234-236. [DOI] [PubMed] [Google Scholar]
- 17.Iturriza-Gomara, M., J. Green, D. W. Brown, U. Desselberger, and J. J. Gray. 1999. Comparison of specific and random priming in the reverse transcriptase polymerase chain reaction for genotyping group A rotaviruses. J. Virol. Methods 7893-103. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, X., P. W. Huang, W. M. Zhong, T. Farkas, D. W. Cubitt, and D. O. Matson. 1999. Design and evaluation of a primer pair that detects both Norwalk-and Sapporo-like caliciviruses by RT-PCR. J. Virol. Methods 83145-154. [DOI] [PubMed] [Google Scholar]
- 19.Leite, J. P. G., S. P. Monteiro, A. M. Fialho, and H. G. Pereira. 1990. A novel avian virus with trisegmented double-stranded RNA and further observations on previously described similar viruses with bisegmented genome. Virus Res. 16119-126. [DOI] [PubMed] [Google Scholar]
- 20.Ludert, J. E., and F. Liprandi. 1993. Identification of viruses with bi- and trisegmented double stranded RNA genome in faeces of children with gastroenteritis. Res. Virol. 144219-224. [DOI] [PubMed] [Google Scholar]
- 21.Ludert, J. E., M. Hidalgo, F. Gil, and F. Liprandi. 1991. Identification in porcine faeces of a novel virus with a bisegmented double stranded RNA genome. Arch. Virol. 11797-107. [DOI] [PubMed] [Google Scholar]
- 22.Ludert, J. E., L. Abdul-Latif, and F. Liprandi. 1995. Identification of picobirnavirus, viruses with double-stranded RNA in rabbits faeces. Res. Vet. Sci. 59222-225. [DOI] [PubMed] [Google Scholar]
- 23.Martínez, L. C., M. O. Giordano, M. B. Isa, L. Alvarado, J. V. Pavan, D. Rinaldi, and S. V. Nates. 2003. Molecular diversity of partial-length genomic segment 2 of human picobirnavirus. Intervirology 46207-213. [DOI] [PubMed] [Google Scholar]
- 24.Masachessi, G., L. C. Martínez, M. O. Giordano, P. A. Barril, B. M. Isa, L. Ferreyra, D. Villareal, M. Carello, C. Asis, and S. V. Nates. 2007. Picobirnavirus (PBV) natural hosts in captivity and virus excretion pattern in infected animals. Arch. Virol. 152989-998. [DOI] [PubMed] [Google Scholar]
- 25.Novikova, N. A., N. V. Epifanova, O. F. Fedorova, L. N. Golitsyna, and N. V. Kupriyanova. 2003. Detection of picobirnavirus by electrophoresis of RNA in polyacrylamide gel. Vopr. Virusol. 4841-43. [PubMed] [Google Scholar]
- 26.Pongsuwanna, Y., K. Taniguchi, M. Chiwakul, T. Urasawa, F. Wakasugi, C. Jayavasu, and S. Urasawa. 1996. Serological and genomic characterization of porcine rotaviruses in Thailand: detection of a G10 porcine rotavirus. J. Clin. Microbiol. 341050-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosen, B. I., Z. Y. Fang, R. I. Glass, and S. S. Monroe. 2000. Cloning of human picobirnavirus genomic segments and development of an RT-PCR detection assay. Virology 277316-329. [DOI] [PubMed] [Google Scholar]
- 28.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 29.Wakuda, M., Y. Pongsuwanna, and K. Taniguchi. 2005. Complete nucleotide sequences of two RNA segments of human picobirnavirus. J. Virol. Methods 126165-169. [DOI] [PubMed] [Google Scholar]
- 30.Wang, Y., X. Tu, C. Humphrey, H. McClure, X. Jiang, C. Qin, R. I. Glass, and B. Jiang. 2007. Detection of viral agents in fecal specimens of monkeys with diarrhea. J. Med. Primatol. 36101-107. [DOI] [PMC free article] [PubMed] [Google Scholar]