Abstract
We report a rare case of acute human immunodeficiency virus (HIV) type 1 group O infection in a French Caucasian woman. Her sexual partner was secondarily diagnosed with HIV infection, and transmission was confirmed by phylogenetic analysis. The unequal performance of many of the serologic and molecular assays commercially available leads to delays in diagnosis and affects patient management.
CASE REPORT
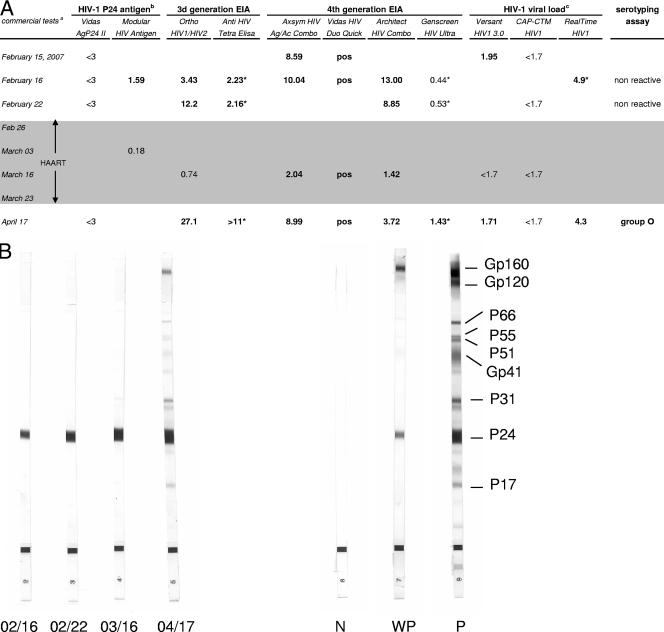
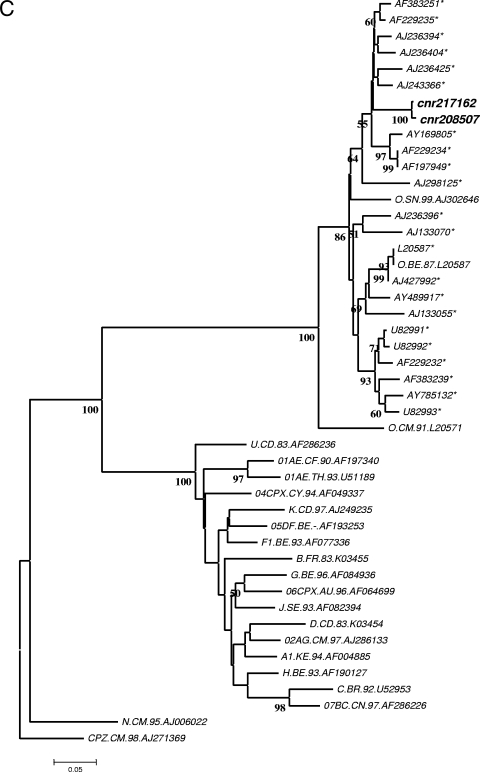
A 58-year-old Caucasian woman was admitted on 9 February 2007 to the internal medicine ward of the hospital of a medium-size city in central France for a hyperalgesic syndrome and a fever of 38.5°C that evolved over a 10-day period. Severe myalgia of the lower limbs required analgesic treatment and was associated with nonpainful jugular and axillary lymphadenopathy. She had not traveled abroad in the previous few months. The first biological investigation showed the presence of activated lymphocytes and biochemical signs of hepatic cytolysis (alanine aminotransferase level, 6 times normal; aspartate aminotransferase level, 3.5 times normal), cholestasis, and pancreatitis. Serological investigations for bacterial infections (syphilis, yersinia, Q fever, and mycoplasma) were negative. Tests for serological markers of viral hepatitis A (hepatitis A virus immunoglobulin M [IgM]), B (hepatitis B surface antigen and antibody to hepatitis B core antigen), and C (antibody to hepatitis C virus) were negative. Serological assays for cytomegalovirus, Epstein-Barr virus, and toxoplasmosis revealed the presence of IgG but not IgM antibodies, which is suggestive of past infections. A screening test for human immunodeficiency virus type 1 (HIV-1) or HIV-2 infection was performed 1 week later (15 February), when the patient reported having had sexual relations with an occasional partner at the beginning of January. Two routine fourth-generation assays, Axsym HIV Ag/Ac Combo (Abbott) and Vidas HIV Duo Quick (bioMérieux), were performed, as required by French law. They provided positive results. The HIV-1 p24 antigen assay (Vidas HIV Ag; bioMérieux) was negative. HIV-1 RNA was detected (90 copies/ml) with the Versant HIV-1 RNA 3.0 assay (Bayer) but was undetectable with the Cobas AmpliPrep-Cobas TaqMan HIV-1 test (Roche Diagnostics). Western blot analysis showed the presence of antibody to p24 only (Fig. 1B). An HIV-2 Western blot assay was negative. Although p24 antigenemia was undetectable, an acute primary HIV infection was suspected and the patient was started on highly active antiretroviral therapy (HAART) that combined Kivexa and Kaletra on 27 February and was switched to Truvada and Kaletra on 3 March because of a hypersensitivity reaction to abacavir. HAART was stopped after 3 weeks, on 23 March, because of severe adverse intestinal effects leading to an alteration in clinical status and because of confusing virological data that did not provide clear evidence of acute HIV infection. Indeed, at this date, the available virological results showed that enzyme-linked immunosorbent assays (ELISAs) were still positive but there was a decline in the positivity index (Axsym HIV Ag/Ac Combo and Architect HIV Combo; Abbott) (Fig. 1A) and no change in the Western blot assay reactivity profile, except for a slight increase in anti-p24 intensity (Fig. 1B). Results of additional analyses with other assays performed with sequential serum samples by (i) the local laboratory of the hospital where the patient was admitted, (ii) the laboratory of the regional teaching hospital, and (iii) the National Reference Center for HIV are summarized in Fig. 1A. On the basis of these data, a reverse transcription-PCR assay with group-specific primers (HIV-1 group M and group O) specific to a conserved region of the gp41 env gene was performed on a sample taken on 16 February as previously described (1). Unexpectedly, a positive reaction was obtained with group O primers only. The amplification product (415 bp) was sequenced and submitted to phylogenetic analysis, which confirmed HIV-1 group O infection (Fig. 1C).
FIG. 1.
Serological and molecular findings on sequential samples from the patient reported. (A) Results of the different immunoassays and viral load assays. Bold indicates reactivity. Results of serological tests are expressed as signal-to-cutoff ratios. Signal-to-cutoff ratios of ≥1.00 were considered reactive, except for Vidas HIV Duo Quick (positive at >0.25). The shaded area indicates the period under HAART. (Superscript a) The commercial tests used were Vidas AgP24 II (bioMérieux), Modular HIV Antigen (Roche Diagnostics), Ortho HIV1/HIV2 Antibody Capture (Ortho Diagnostics), Anti-HIV Tetra ELISA (Biotest), Axsym HIV Ag/Ac Combo (Abbott), Vidas HIV Duo Quick (bioMérieux), Architect HIV Combo (Abbott), Genscreen HIV Ultra (Bio-Rad), Versant HIV-1 TNA 3.0 (Bayer), Cobas AmpliPrep-CobasTaqMan HIV-1 (CAP-CTM; Roche Diagnostics), and RealTime HIV-1 (Abbott Molecular). (Superscript b) HIV-1 p24 antigen levels are expressed in picograms per milliliter. (Superscript c) HIV-1 viral loads are expressed as log10 numbers of copies per milliliter. Asterisks indicate values obtained with samples that were analyzed retrospectively. (B) Western blot analysis of sequential serum samples from February to April 2007 (HIV-1/2 Blot; Genelabs). From left to right, the strips are shown in chronological order (month/day). N, negative control; WP, weakly positive control; P, positive control. (C) Phylogenetic tree. The env sequences (415 bp) from the index patient (cnr208507) and her partner (cnr2171262) were analyzed by the neighbor-joining method. The amplified sequences from the two patients were amplified and sequenced separately and compared with the 22 closest group O sequences (indicated by asterisks) selected from the Los Alamos database (http://hiv-web.lanl.gov) and 3 group O reference sequences and 17 group M reference sequences representative of various clades (one strain per pure subtype and major circulating recombinant form). Distances were calculated by the Kimura two-parameter method (T/t ratio = 2.0) and the MEGA analysis program (http://www.megasoftware.net). Bootstrap analysis was used to test the reliability of the branching order. Bootstrap values above 50 are indicated. The tree was rooted with a SIVcpz sequence (CPZ.CM.98.AJ271369) as the outgroup.
A close clinical and virological follow-up was established. Fifteen days after discontinuation of treatment, the patient had persistent nausea and asthenia and her lymphadenopathy reappeared. On 17 April, immunoassays showed an increased ELISA index and an evolving Western blot assay profile, suggestive of seroconversion (Fig. 1B). The viral load was quantified at 4.3 log10 copies/ml with the RealTime HIV-1 test (Abbott Molecular). We therefore concluded that she had an acute HIV-1 group O infection. Her clinical status improved, with regressive but persistent lymphadenopathy, and antiretroviral treatment was not resumed. Her CD4+ T-cell count was 635/mm3 in May and 500/mm3 in July. Subsequent HIV-1 viral load measurements by the RealTime system in June and August were 5.4 log10 copies/ml.
One month after the seroconversion period, the sexual partner of this patient was identified and diagnosed with an HIV-1 group O infection. He was a Caucasian French resident and had made regular trips to Cameroon for several years. The phylogenetic analysis of the gp41 env amplification product from his plasma viral RNA confirmed the common origin of the strains. The two sequenced fragments showed 99.2% homology and cluster with a 100% bootstrap value (Fig. 1C).
HIV-1 group O infections are rare outside the endemicity region in west central Africa including Cameroon and neighboring countries (6). The French national surveillance system reported a 0.1% prevalence of HIV-1 group O infections among new HIV diagnoses between 2003 and 2006 (7). In spite of this low prevalence, systematic identification of group O infections, which is made easier by epidemiological data, would be necessary for an accurate assessment of viral replication and adapted HAART, given that most available viral load assays (3) are unable to quantify HIV RNA and that group O HIV-1 strains are naturally resistant to nonnucleoside reverse transcriptase inhibitors.
For physicians regularly involved in the management of HIV-infected patients, the clinical presentation of this patient was retrospectively strongly suggestive of acute HIV infection. However, the discordant results of various virological assays confused the interpretation. Discordant results of HIV antibody assays, p24 antigen assays, and nucleic acid-based assays might be suggestive of either false-positive or false-negative reactivities. They strongly complicated the early diagnosis of this acute HIV-1 group O infection and were largely instrumental in the decision to stop antiretroviral treatment soon after. One fourth-generation ELISA, Genscreen HIV Ultra (Bio-Rad), was negative during the 2 months following sexual exposure and was only weakly reactive more than 3 months later. Failure to detect HIV antibodies because of HIV genetic diversity has been regularly reported, even when using these highly sensitive assays. Single variant sequences were generally responsible for failure to detect HIV-1 group M or O infections (2, 8). In the present case, the sequence of the gp41 immunodominant epitope was similar to that of the group O consensus immunodominant epitope and it therefore could not explain the failure to detect antibodies. The sensitivity of p24 antigen detection of newly developed fourth-generation assays varies, and failures have been described, particularly for group O strains at p24 concentrations below 25 pg/ml (5). The Western blot assay profile showed only a reactive p24 band on early sequential samples, although with a slight increase in intensity. A significant drop in the positivity index of ELISAs was observed under therapy. This could be explained by early initiation of HAART. Of note, one third-generation assay, Ortho HIV1/HIV2 Antibody Capture (Ortho Diagnostics), although positive with earlier samples, became negative (Fig. 1A). Such a seroreversion was previously described in acutely infected individuals treated with early HAART (4).
Direct detection by p24 antigen assays or viral load assays is particularly useful in cases of indeterminate antibody response. However, the assays all provided discordant results in our case. p24 antigenemia was negative or slightly positive and became rapidly undetectable. HIV-1 RNA quantification was negative with the Cobas AmpliPrep-Cobas TaqMan HIV-1 assay and was particularly underestimated with the branched-DNA assay, since it was retrospectively determined at 4.9 log10 copies/ml with the RealTime HIV-1 assay (Fig. 1A). At present, only the Abbott RealTime system, among commercially available assays, can quantify HIV-1 group O RNA because it uses primers and probes designed to detect the highly conserved integrase region in the pol gene (3). Finally, we had no epidemiological information concerning our patient's sexual partner during the first weeks of diagnosis; it was the slight increase in reactivity to p24 shown by Western blot assay, associated with the various discordant results of the different assays, that prompted us to explore a possible group O infection with specific tools available at the National Reference Center.
This case of acute primary infection with an HIV-1 group O variant underlines the impact of the genetic diversity of HIV on virological diagnosis and its consequences for patient management. Divergent HIV strains must no longer be considered to be limited to developing countries, and the development of diagnostic tools better suited to the genetic diversity of HIV must be encouraged to improve the diagnosis and management of HIV-infected patients.
Acknowledgments
We thank Thomas Bourlet for his help in performing the HIV-1 viral load assay with the RealTime HIV-1 system, Catherine Gaudy-Graffin for her help in the phylogenetic analysis, and Jeffrey Watts for revision of the English manuscript.
Footnotes
Published ahead of print on 14 May 2008.
REFERENCES
- 1.Brand, D., A. Beby-Defaux, M. Mace, S. Brunet, A. Moreau, C. Godet, X. Jais, F. Cazein, C. Semaille, and F. Barin. 2004. First identification of HIV-1 groups M and O dual infections in Europe. AIDS 182425-2430. [PubMed] [Google Scholar]
- 2.Gaudy, C., A. Moreau, S. Brunet, J. M. Descamps, P. Deleplanque, D. Brand, and F. Barin. 2004. Subtype B human immunodeficiency virus (HIV) type 1 mutant that escapes detection in a fourth-generation immunoassay for HIV infection. J. Clin. Microbiol. 422847-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gueudin, M., J. C. Plantier, V. Lemée, M. P. Schmitt, L. Chartier, T. Bourlet, et al. 2007. Evaluation of the Roche Cobas TaqMan and Abbott real time extraction-quantification systems for HIV-1 subtypes. J. Acquir. Immune Defic. Syndr. 44500-505. [DOI] [PubMed] [Google Scholar]
- 4.Kassutto, S., M. N. Johnston, and E. S. Rosenberg. 2005. Incomplete HIV type 1 evolution and seroreversion in acutely infected individuals treated with early antiretroviral therapy. Clin. Infect. Dis. 40868-873. [DOI] [PubMed] [Google Scholar]
- 5.Ly, T. H., S. Laperche, C. Brennan, A. Vallari, A. Ebel, J. Hunt, L. Martin, D. Daghfal, G. Schochetman, and S. Devare. 2004. Evaluation of the sensitivity and specificity of six HIV combined p24 antigen and antibody assays. J. Virol. Methods 122185-194. [DOI] [PubMed] [Google Scholar]
- 6.Peeters, M., A. Gueye, F. Mboup, F. Bibollet-Ruche, E. Ekaza, R. Mulanga, R. Ouedraogo, R. Gandjii, P. Mpele, G. Dibanga, B. Koumare, M. Saidou, E. Esu-Williams, J. P. Lombart, W. Badombena, N. Luo, M. Vanden Haesevelde, and E. Delaporte. 1997. Geographical distribution of HIV-1 group O viruses in Africa. AIDS 11493-498. [DOI] [PubMed] [Google Scholar]
- 7.Semaille, C., F. Barin, F. Cazein, J. Pillonel, D. Brand, J. C. Plantier, P. Bernillon, S. Le Vu, R. Pinget, and J. C. Desenclos. 2007. Monitoring the dynamics of the HIV epidemic at a national level using assays for recent infection and serotyping among new HIV diagnoses: experience after 2 years in France. J. Infect. Dis. 196377-383. [DOI] [PubMed] [Google Scholar]
- 8.Zouhair, S., S. Roussin-Bretagne, A. Moreau, S. Brunet, S. Laperche, M. Maniez, F. Barin, and M. Harzic. 2006. Group O human immunodeficiency virus type 1 infection that escaped detection in two immunoassays. J. Clin. Microbiol. 44662-665. [DOI] [PMC free article] [PubMed] [Google Scholar]