Abstract
In search of novel genes expressed in metastatic prostate cancer, we subtracted cDNA isolated from benign prostatic hypertrophic tissue from cDNA isolated from a prostate cancer xenograft model that mimics advanced disease. One novel gene that is highly expressed in advanced prostate cancer encodes a 339-amino acid protein with six potential membrane-spanning regions flanked by hydrophilic amino- and carboxyl-terminal domains. This structure suggests a potential function as a channel or transporter protein. This gene, named STEAP for six-transmembrane epithelial antigen of the prostate, is expressed predominantly in human prostate tissue and is up-regulated in multiple cancer cell lines, including prostate, bladder, colon, ovarian, and Ewing sarcoma. Immunohistochemical analysis of clinical specimens demonstrates significant STEAP expression at the cell–cell junctions of the secretory epithelium of prostate and prostate cancer cells. Little to no staining was detected at the plasma membranes of normal, nonprostate human tissues, except for bladder tissue, which expressed low levels of STEAP at the cell membrane. Protein analysis located STEAP at the cell surface of prostate-cancer cell lines. Our results support STEAP as a cell-surface tumor-antigen target for prostate cancer therapy and diagnostic imaging.
Keywords: cancer, six-transmembrane-domain protein, epithelial marker
Prostate cancer is the most frequently diagnosed cancer and the second leading cause of cancer death in men in North America. The efficiency of early detection of prostate cancer has increased dramatically with a serum test for the prostate-specific antigen (PSA) (1). However, PSA may not distinguish prostate cancer from benign diseases such as benign prostatic hyperplasia (BPH) and prostatitis (2, 3). Although locally confined disease is treatable, recurrent and metastasized prostate cancer is essentially incurable. Androgen ablation therapy may palliate advanced disease, as prostate cancer cells are androgen-responsive. However, the majority of patients inevitably progress to incurable, androgen-independent disease (4).
The identification of novel markers and therapeutic targets in advanced prostate cancer and androgen-independent disease is critical for improving diagnosis and therapy. Ideal targets for prostate cancer therapy would include proteins that are exclusively expressed in dispensable normal tissues such as prostate, that are highly expressed in metastatic disease, and that are accessible to therapeutic modalities at the cell surface. Progress in identifying such markers for prostate cancer has been improved by the generation of prostate cancer cell lines (5) and xenografts that can recapitulate different stages of the disease in mice (6–9). Prostate-specific membrane antigen (PSM) (10) and prostate carcinoma tumor antigen (PCTA-1) (11) are two cell-surface antigens that are up-regulated in prostate cancer and were identified from the androgen-responsive cell line LNCaP (5). Prostate stem cell antigen (PSCA), a glycosylphosphatidylinositol-linked marker that is up-regulated in prostate cancer (12), was identified by using the LAPC (Los Angeles prostate cancer) xenografts, which have the capacity to mimic the transition from androgen dependence to androgen independence and metastasis to distal sites (9).
The LAPC xenografts represent advanced prostate cancer specimens that were derived from bone and lymph node metastases (9, 13). To isolate novel genes that are up-regulated in metastatic prostate cancer, we performed suppression subtractive hybridizations (SSHs) (14) with the LAPC xenografts as a source of cDNA. With this strategy, a prostate-specific gene encoding a serpentine transmembrane protein, named STEAP (six-transmembrane epithelial antigen of the prostate), was identified. STEAP is unique among the currently known prostate cancer markers because of its putative secondary structure, from which one may predict that it functions as a potential channel or transporter protein. STEAP is also distinct in that it is expressed in multiple cancers, suggesting that it is a general tumor antigen. Its strong expression in advanced prostate cancer, its cell surface localization, and its predicted secondary structure suggest that STEAP may be an ideal target for tumor therapy and diagnosis.
Materials and Methods
Cell Lines and Human Tissues.
All human cancer cell lines used in this study were obtained from the American Type Culture Collection. All cell lines were maintained in DMEM with 10% fetal calf serum. Primary prostate epithelial cells were obtained from Clonetics (San Diego) and were grown in PrEBM (prostate epithelial cell basal medium) supplemented with growth factors (Clonetics).
All human prostate cancer xenografts were originally provided by Charles Sawyers (University of California, Los Angeles) (9). LAPC-4 androgen-dependent (AD) and LAPC-9 AD xenografts were routinely passaged as small tissue chunks in recipient severe combined immunodeficient male mice. LAPC-4 androgen-independent (AI) and LAPC-9 AI xenografts were derived as described previously (9) and were passaged in castrated males or in severe combined immunodeficient female mice.
Human tissues for RNA and protein analyses were obtained from the Human Tissue Resource Center at the University of California, Los Angeles; from the National Disease Research Interchange (Philadelphia); and from QualTek Molecular Labs (Santa Barbara, CA).
SSH.
Tumor tissue and cell lines were homogenized in Trizol reagent (GIBCO/BRL) by using 10 ml/g of tissue or 10 ml/108 cells to isolate total RNA. Poly(A)-RNA was purified from total RNA by using Qiagen’s Oligotex mRNA Mini and Midi kits (Chatsworth, CA).
SSH was performed as described by Diatchenko et al. (14) with the PCR-Select cDNA Subtraction Kit (CLONTECH). SSH-derived gene fragments were inserted into pCR2.1 by using the T/A vector cloning kit (Invitrogen). Transformed Escherichia coli were subjected to blue/white and ampicillin selection. White colonies were picked and arrayed into 96-well plates and stored in 20% glycerol. Plasmid DNA was prepared, sequenced, and searched for homology against public databases.
Expression Analysis.
The STEAP cDNA was isolated by screening a human prostate phage library (CLONTECH) by using a STEAP SSH-derived fragment as a probe. Northern blotting was performed on 10 μg of total RNA prepared from cell lines and LAPC xenografts with random hexamer-labeled (Roche Molecular Biochemicals) STEAP cDNA. Human multitissue Northern blots were purchased from CLONTECH and probed with STEAP cDNA.
Protein Analysis.
Secondary protein structure prediction for STEAP was performed by using the web tools SOSUI at http://www.tuat.ac.jp/∼mitaku/adv_sosui/submit.html and PSORT at http://psort.nibb.ac.jp:8800/form.html. Sheep polyclonal antiserum (anti-STEAP) (Capralogics, Hardwick, MA) was generated toward the amino-terminal peptide WKMKPRRNLEEDDYL, coupled to keyhole limpet hemocyanin, and affinity-purified by using the peptide coupled to Affi-Gel 10 (Bio-Rad). To express STEAP in NIH 3T3 cells, STEAP cDNA was cloned into the retroviral vector pSRαtkneo (15). The retrovirus was generated in human 293T cells (16) and was used to infect NIH 3T3 cells, which were selected in G418 for 2 weeks to generate stable lines. For protein expression in 293T cells, STEAP was cloned into pcDNA 3.1 Myc-His (Invitrogen). Transfected cells were cell-surface-labeled with biotin-7-NHS as described in the Cellular Labeling and Immunoprecipitation Kit (Roche Molecular Biochemicals). Immunoprecipitation of STEAP was performed with 10 μg of anti-STEAP antibodies, anti-His antibodies (Santa Cruz Biotechnology), or anti-human transferrin receptor antibodies. Biotinylated proteins were affinity-purified by using streptavidin gel (Roche Molecular Biochemicals). Western blotting of xenograft and cancer cell line lysates was performed on 20 μg of cell lysate protein. Normalization of extracts was achieved by probing cell extracts with antibodies to the Grb-2 protein (Transduction Laboratories, Lexington, KY).
Immunohistochemical analysis of formalin-fixed, paraffin-embedded tissues with anti-STEAP polyclonal antibody was performed on 4-μm tissue sections. After steam treatment in sodium citrate (10 mM, pH 6.0), slides were incubated with 3 μg/ml anti-STEAP antibodies followed by incubation with biotinylated rabbit anti-sheep IgG. The reactions were visualized by using avidin-conjugated horseradish peroxidase (Vector Laboratories, Burlingame, CA).
Chromosomal Mapping.
Chromosomal localization of STEAP was determined by using the GeneBridge 4 Human/Hamster radiation hybrid panel (17). STEAP gene product was amplified by PCR with the following primers: 5′-ACTTTGTTGATGACCAGGATTGGA-3′ and 5′-CAGAACTTCAGCACACACAGGAAC-3′. The resulting mapping vector for the 93-radiation hybrid panel DNAs was: 210000020110101000100000010111010122100011100111011010100010001000101001021000001111001010000. This vector and the mapping program at http://carbon.wi.mit.edu:8000/cgi-bin/contig/rhmapper.plplaced STEAP on chromosome 7p22.3 telomeric to D7S531.
Results
Cloning of STEAP, a Prostate-Specific Gene Highly Expressed in Advanced Prostate Cancer.
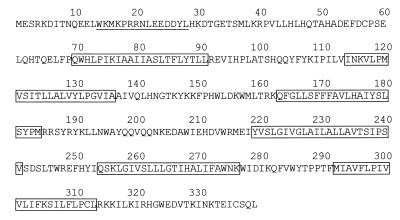
The LAPC-4 xenograft was derived from a lymph node metastasis of stage D prostate cancer and exhibits AD and AI sublines (9). In pursuit of genes that are up-regulated in advanced metastatic prostate cancer, cDNA derived from BPH tissue was subtracted from cDNA generated from LAPC-4 AD xenograft. BPH is often characterized by a hyperplasia of the smooth muscle and fibroblast component of prostate tissue, suggesting that our subtraction strategy may also enrich for epithelial markers. Extensive expression analysis was performed on a selected set of gene fragments to look for genes that are differentially regulated between tester and driver cDNA. One of the novel gene fragments that was highly expressed in the LAPC xenografts exhibited an ORF of 145 amino acids containing two potential transmembrane domains. A full-length cDNA of 1195 bp was isolated from a normal prostate library revealing an ORF of 339 amino acids with a predicted molecular mass of 40 kDa and with no significant homology to any known genes (Fig. 1). Analysis of the ORF predicts that the protein contains six potential transmembrane domains.
Figure 1.
Amino acid sequence of human STEAP. Boxed regions correspond to the putative membrane-spanning domains. The amino and carboxyl termini are both predicted to be intracellular. The amino-terminal peptide used for generating the anti-STEAP antibody is underlined.
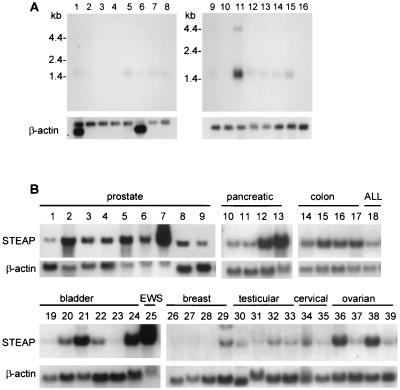
Northern blotting of normal human tissue RNA showed expression of two transcripts of 1.4 kb and 4.0 kb primarily in prostate (Fig. 2A, lane 11), with 5- to 10-fold lower levels detected in colon and liver (Fig. 2A, lanes 15 and 5, respectively). The major 1.4-kb transcript was found to encode the mature protein, whereas the larger transcript was shown to be unprocessed RNA containing an intron of 2399 bp. The prostate-specific expression combined with secondary-structure prediction of six transmembrane domains led us to name this gene STEAP.
Figure 2.
Gene expression of STEAP in tissues and cell lines. (A) Human normal tissue filters contain 2 μg of mRNA per lane. Lanes are: 1, heart; 2, brain; 3, placenta; 4, lung; 5, liver; 6, skeletal muscle; 7, kidney; 8, pancreas; 9, spleen; 10, thymus; 11, prostate; 12, testis; 13, ovary; 14, small intestine; 15, colon; and 16, peripheral blood leukocytes. (B) Xenograft and cell-line filters were prepared with 10 μg of total RNA per lane. Lanes are: 1, primary prostate epithelial cells; 2, prostate; 3, LAPC-4 AD; 4, LAPC-4 AI; 5, LAPC-9 AD; 6, LAPC-9 AI; 7, LNCaP; 8, PC-3; 9, DU145; 10, PANC-1; 11, BxPC-3; 12, HPAC; 13, Capan-1; 14, CACO-2; 15, LOVO; 16, T84; 17, COLO-205; 18, KCL-22 (acute lymphocytic leukemia, ALL); 19, HT1197; 20, SCABER; 21, UM-UC-3; 22, TCCSUP; 23, J82; 24, 5637; 25, RD-ES (Ewing sarcoma, EWS); 26, CAMA-1; 27, DU4475; 28, MCF-7; 29, MDA-MB-435s; 30, NTERA-2; 31, NCCIT; 32, TERA-1; 33, TERA-2; 34, A431; 35, HeLa; 36, OV-1063; 37, PA-1; 38, SW 626; and 39, CAOV-3. The blots were probed by using an SSH-derived gene fragment. All RNA samples were normalized by ethidium bromide staining and subsequent analysis with a β-actin probe.
The initial STEAP gene fragment was isolated from LAPC-4 AD. Analysis of the LAPC xenografts and prostate cancer cell lines demonstrated strong expression in all xenografts and cell lines analyzed (Fig. 2B). The highest levels were detected in LNCaP, a cell line derived from a lymph-node metastasis of prostate cancer (5), and LAPC-9 (Fig. 2B, lane 7), a xenograft derived from a bone metastasis of prostate cancer (13). No significant difference in expression was observed between AD and AI xenograft sublines (Fig. 2B, lanes 3–6). Expression of STEAP was equally high in LAPC-4 and LAPC-9 xenografts grown locally in the bone of mice to mimic bone metastasis (data not shown). To examine whether STEAP expression is androgen-regulated, AD LNCaP cells were androgen-deprived for 1 week and were stimulated with mibolerone (synthetic androgen) for various time periods. Analysis indicated that STEAP expression did not vary with androgen deprivation and/or androgen stimulation (data not shown). In contrast, expression of PSA, an androgen-regulated gene, correlated with the presence of androgen. These data suggest that STEAP is a hormone-independent, prostate-specific gene that is highly expressed in advanced prostate cancer.
STEAP Gene Is Expressed in Multiple Cancer Cell Lines.
To determine the expression level of STEAP in other cancers, Northern blot analysis was performed on an extensive panel of cancer cell lines. In contrast to the prostate-specific expression detected in normal tissues, STEAP expression was also seen in several colon, bladder, ovarian, and pancreatic cancer cell lines, suggesting that this gene may be generally up-regulated in cancers (Fig. 2). RD-ES, a Ewing sarcoma-derived cell line, expressed the highest level of STEAP mRNA in the non-prostate-derived cell lines (Fig. 2B, lane 25).
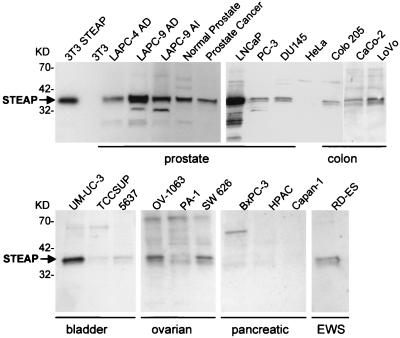
To analyze STEAP protein expression, polyclonal antibodies were raised toward a 15-mer peptide designed from the STEAP amino terminus (anti-STEAP). Antibody specificity was tested by using NIH 3T3 cells and NIH 3T3 cells infected with retroviruses encoding STEAP. Western blot analysis of protein extracts of infected and uninfected NIH 3T3 cells showed expression of a protein with an apparent molecular mass of 36 kDa only in STEAP retrovirus-infected cells (Fig. 3). All cell extracts were normalized by probing the Western blots with anti-Grb-2-specific antibodies (data not shown).
Figure 3.
Analysis of STEAP protein expression in cell lines and tissues. Western blots of cell lysates were prepared from xenografts, and cell lines were probed with anti-STEAP antibodies. The samples were normalized with anti-Grb-2 probing of the Western blots (data not shown).
STEAP protein expression was detected in all the LAPC xenografts, prostate cancer cell lines, a primary prostate cancer specimen, and its matched normal prostate control (Fig. 3). The highest protein expression was found in the LAPC-9 xenograft and in LNCaP, in good correlation with Northern blot analysis. Among the non-prostate-derived cells, expression was highest in the bladder carcinoma cell line UM-UC-3. Lower but significant expression of STEAP was detected in RD-ES cells, despite the high levels of STEAP message in these cells. Protein expression was also observed in colon and ovarian cancer cell lines, with no protein expression detected in pancreatic cancer cells. Interestingly, the prostate cancer cells PC-3 and DU145, which exhibit significantly lower RNA expression (Fig. 2), display equal or greater protein expression compared with the colon and pancreatic cancer cell lines (Fig. 3). It seems that protein expression of STEAP is differentially regulated in nonprostate cell lines as compared with prostate-derived cell lines and tissues.
STEAP Is Expressed in the Epithelia of Prostate Cancer Specimens.
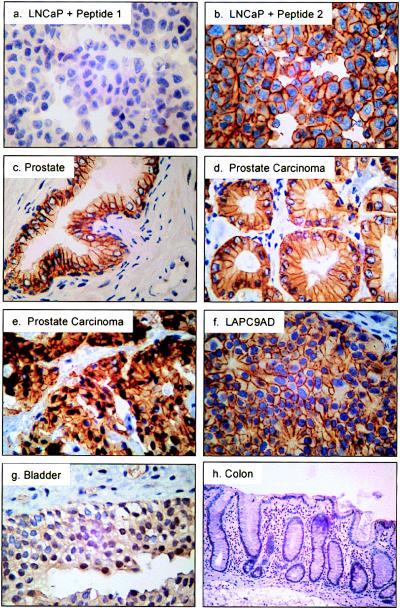
Western blotting analysis of a matched cancer-normal tissue pair showed expression of STEAP in both samples (Fig. 3). To examine the expression of STEAP at the cellular level in prostate cancer biopsies and surgical samples, tissue sections were prepared for immunohistochemical analysis. As a positive control for staining, LNCaP cells were fixed, embedded in paraffin, and stained with anti-STEAP antibodies. Anti-STEAP staining of LNCaP cells showed strong pericellular staining in all cells (Fig. 4b). The addition of excess STEAP amino-terminal peptide (peptide 1) was able to competitively inhibit the staining (Fig. 4a), whereas a STEAP peptide (YQQVQQNKEDAWIEH, peptide 2) designed from a different region of the molecule had no effect on the staining (Fig. 4b). Similarly strong pericellular staining is seen in the LAPC-9 (Fig. 4f) and LAPC-4 xenograft cells (data not shown). This demonstrates that the staining is specific and localizes STEAP to the plasma membrane, suggesting that STEAP is expressed at the surface of these cells.
Figure 4.
Immunohistochemical analysis of patient samples with anti-STEAP antibodies. Samples include: LNCaP cells probed in the presence of amino-terminal STEAP peptide 1 (a), LNCaP plus nonspecific peptide 2 (b), normal prostate tissue (c), grade 3 prostate carcinoma (d), grade 4 prostate carcinoma (e), LAPC-9 AD xenograft (f), normal bladder (g), and normal colon (h). (×400.)
Analysis of 26 clinical specimens of prostate tissue showed moderate to strong staining in glandular epithelia of all prostate cancer samples tested and in all samples derived from normal prostate or benign disease (Table 1). The signal appears to be strongest at the cell membrane of the epithelial cells, especially at the cell–cell junctions (Fig. 4 c–e), and is also inhibited with excess STEAP amino-terminal peptide (data not shown). STEAP seems to be expressed at all stages of prostate cancer, because low grades, high grades, and metastatic prostate cancer (represented by LAPC-9) specimens all exhibit strong staining (Fig. 4 d–f, respectively).
Table 1.
Immunohistochemical staining of human tissues with anti-STEAP antibodies
| Staining intensity | Tissues* |
|---|---|
| None | Cerebellum, cerebral cortex, spinal cord, heart, skeletal muscle (2), artery, thymus, spleen (4), bone marrow, lymph node (3), lung (3), liver (4), ileum, kidney (2), testis (2), ovary, placenta, breast, adrenal gland (2), thyroid gland (2), skin (2) |
| Light cytoplasmic | Ureter, bladder cancer, colon (4/7), colon cancer (4/7), fallopian tubes (2/2), pituitary gland, pancreas, stomach (1/2), uterus (1/2) |
| Moderate to strong membrane | Prostate (6/6), BPH (5/5), bladder (2/5), prostate cancer† (15/15) |
In cases where more than one sample was analyzed per tissue, the number in parentheses indicate how many samples showed positive staining/total analyzed.
†Prostate cancer grades ranged from Gleason grades 3 to 5 (40).
Immunohistochemical staining of normal nonprostate tissues showed no detectable STEAP expression in 21 of 30 tissues (Table 1). Low levels of cell-surface staining were detected in the transitional epithelium of two of five normal bladder specimens (Table 1; Fig. 4g). Seven additional normal tissues, ureter, fallopian tubes, uterus, pituitary, pancreas, stomach, and colon, showed a diffuse type of light cytoplasmic staining (Table 1). The staining intensities did not correlate with RNA expression, because pituitary, pancreas, and stomach, for instance, show little to no RNA expression for STEAP (Fig. 2 and data not shown). The staining in these tissues also appeared sporadic, because not all samples tested exhibited specific staining. This result is in contrast to the prostate tissues, which consistently showed very clear membrane-associated staining. These findings suggest that some nonprostate tissues may express STEAP protein at low levels in the cytoplasm.
Light, diffuse cytoplasmic staining was also observed for primary colon and bladder cancer specimens (Table 1), indicating a low level of expression. This finding is in contrast to colon and bladder cancer cell lines, some of which showed high expression (see Fig. 3). The primary colon and bladder cancers came from localized disease and were generally well to moderately differentiated, although the cell lines were derived from metastatic sites. Therefore, it is possible that the difference in STEAP expression is related to the stage and grade of the disease.
STEAP Localizes to the Cell Surface of Cancer Cells.
Immunohistochemical analysis and secondary-structure prediction of STEAP suggest that it localizes to the plasma membrane. To analyze the subcellular localization of STEAP protein, the full-length cDNA was cloned into an expression vector that provides a 6-His tag at the carboxyl terminus. The construct was transfected into 293T cells, which were analyzed by flow cytometry using anti-His and anti-STEAP antibodies. Staining of cells was performed on intact cells as well as permeabilized cells. The results indicated that only permeabilized cells stained with both antibodies (data not shown). This result suggested that both termini are localized intracellularly, raising the possibility that the protein may be associated with intracellular organelles rather than the plasma membrane.
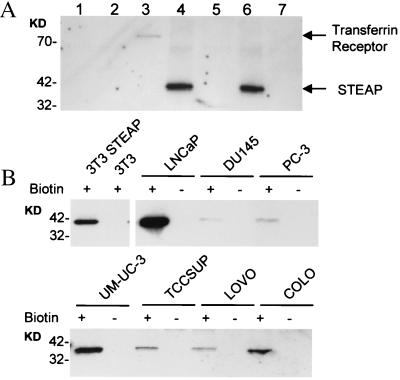
To determine whether STEAP protein is indeed expressed at the cell surface, intact cells were labeled with a water-soluble biotinylation reagent that is excluded from live cells. STEAP was immunoprecipitated from cell extracts by using anti-His and anti-STEAP antibodies. Simian virus 40 large T antigen, an intracellular protein that is expressed at high levels in 293T cells, and the endogenous cell-surface transferrin receptor were immunoprecipitated as controls. After immunoprecipitation, the proteins were transferred to a membrane and visualized with horseradish peroxidase-conjugated streptavidin. The results demonstrate that the transferrin receptor and STEAP were labeled with biotin, whereas the large T antigen was not measurably labeled (Fig. 5). Because only cell-surface proteins are labeled with this technique, we can conclude that STEAP is a cell-surface marker with intracellular amino and carboxyl termini.
Figure 5.
Cell-surface biotinylation of STEAP in transfected cells and in cancer cell lines. (A) Cell-surface biotinylation of 293T cells transfected with vector alone or with vector containing cDNA encoding 6-His-tagged STEAP. Cell lysates were immunoprecipitated with specific antibodies, transferred to a membrane, and probed with horseradish peroxidase-conjugated streptavidin. Lanes 1–4 and 6 correspond to immunoprecipitates from lysates prepared from STEAP-expressing 293T cells. Lanes 5 and 7 are immunoprecipitates from vector-transfected cells. The immunoprecipitations were performed using the following antibodies: 1, sheep nonimmune; 2, anti-Large T antigen; 3, anti-CD71 (transferrin receptor); 4, anti-His; 5, anti-His; 6, anti-STEAP; and 7, anti-STEAP. (B) Prostate cancer (LNCaP, DU145, PC-3), bladder cancer (UM-UC-3, TCCSUP), and colon cancer (LOVO, COLO) cell lines were either biotinylated (+) or not (−) before lysis. Western blots of streptavidin-purified proteins were probed with anti-STEAP antibodies. Molecular mass markers are indicated in kDa.
To examine cell-surface expression in prostate, bladder, and colon cancer cells, biotinylated cell-surface proteins were affinity-purified with streptavidin-Sepharose and probed with anti-STEAP antibodies. Western blotting of streptavidin-purified proteins clearly shows cell-surface biotinylation of endogenous STEAP in all prostate, bladder, and colon cancer cells tested, and in NIH 3T3 cells infected with a STEAP-encoding retrovirus, but not in nonexpressing NIH 3T3 cells (Fig. 5). In additional controls, STEAP protein was not detected in streptavidin precipitates from nonbiotinylated STEAP-expressing cells (Fig. 5). Our data combined with sequence analysis led us to predict STEAP to be a type IIIa membrane protein with a molecular topology of six potential transmembrane domains, three extracellular loops, two intracellular loops, and two intracellular termini.
Localization of STEAP Gene to the Telomeric Region of Chromosome 7.
To investigate possible mechanisms of STEAP up-regulation in cancer, chromosomal mapping was performed by using radiation hybrid analysis. STEAP localized to chromosome 7p22.3, a region close to the telomere. This region is within a large region of allelic gain reported for both primary and recurrent prostate cancer (18, 19). However, Southern blot analysis of genomic DNA derived from the xenografts, LNCaP, PC-3, and DU145, and normal human cells showed no evidence of amplification or rearrangement of STEAP (data not shown).
Discussion
Our objective was to identify novel markers and therapeutic targets for prostate cancer by using advanced metastatic cancer as a source of genetic material. The LAPC xenografts represent a unique source of tissue, because they were derived from metastatic disease and recapitulate different stages of prostate cancer. The LAPC-4 xenograft was derived from a lymph-node metastasis of prostate cancer (9), whereas the LAPC-9 xenograft was derived from a bone metastasis (13). Our strategy led to the identification of STEAP, which was chosen for further study because of its singularity, strong expression in prostate cancer specimens, restricted expression pattern in normal tissues, and cell-surface localization.
STEAP shows no homology to any known proteins, but biochemical analysis and secondary-structure prediction suggest that it is a cell-surface molecule with six transmembrane domains. Cell-surface molecules that contain six transmembrane domains are often ion channels (20) or water channels (aquaporins) (21). Structural studies show that both types of molecules assemble into tetrameric complexes to form functional channels (22–24). Immunohistochemical staining of STEAP in the prostate gland seems to be concentrated at the cell–cell boundaries, with less staining detected at the luminal side of the epithelia. It seems possible that STEAP may perform a function as a channel in tight junctions, in gap junctions, or in cell adhesion. Ion channels have been implicated in the proliferation and invasiveness of prostate cancer cells (25). Both rat and human prostate cancer cells have been shown to contain subpopulations of cells with higher expression levels of sodium channels and a more invasive phenotype in vitro (26). The specific blockade of sodium channels inhibited the invasiveness of PC-3 cells in vitro (27), whereas the inhibition of potassium channels in LNCaP cells decreased cell proliferation (28). Although the function of STEAP remains to be determined, its expression pattern in advanced metastatic disease and its structural prediction as a potential channel or transporter protein support STEAP as a potential drug target in cancer therapy.
STEAP is localized to the cell surface in prostate cancer, making it also a potential target for monoclonal antibody-mediated therapy and diagnosis. Antibody-mediated therapies toward cell-surface antigens such as CD20 and HER2/neu are being used as treatments for non-Hodgkin’s lymphoma (29, 30) and metastatic breast cancer (31), respectively. Antigens suitable for antibody-mediated therapy should be highly expressed in cancer tissue, and ideally expressed only in normal tissues that are dispensable for life. STEAP meets these criteria, as it is strongly expressed in prostate cancer, whereas cell-surface expression in normal tissues is restricted to the prostate, a dispensable organ, and the bladder, a tissue with high regenerative capacity.
STEAP belongs to a small “family” of cell-surface antigens that are expressed in prostate cancer and are thought to be potential targets for antibody-mediated therapy and diagnosis. These include PSM (10), PCTA-1 (11), and PSCA (12). PSM, a type II transmembrane protein with hydrolase activity and 85% identity to a rat neuropeptidase (32, 33), is also expressed in the small intestine and the brain (34) and may have a potential role in neuropeptide catabolism in the brain (32). Anti-PSM antibodies are currently used to detect metastatic disease as the Prostascint scan (35) and are being evaluated for prostate cancer treatment (36–38). However, in a study looking at bone metastasis of prostate cancer, only 8 of 18 specimens expressed PSM, indicating that other prostate-specific cell-surface markers may be needed to manage metastatic disease (39). PCTA-1, a galectin that is highly expressed in prostate cancer, is largely secreted into the media of expressing cells and may be more promising as a diagnostic serum marker for prostate cancer (11). PSCA, a member of the Thy-1/Ly-6 family of glycosylphosphatidylinositol-anchored cell-surface antigens, is unique in that it is expressed primarily in basal cells of normal prostate tissue, suggesting that it is a potential stem-cell marker (12). PSCA expression is up-regulated in cancer epithelia and is detected in 80% of prostate cancer cases analyzed (12).
STEAP is unique among this group of cell-surface antigens because of its secondary-structure prediction as a potential channel or transport protein. STEAP is highly expressed at all stages of prostate cancer and does not seem to be modulated by hormones, a property that is beneficial when managing hormone-refractory prostate cancer or during anti-androgen therapy for advanced metastatic disease. STEAP is also expressed in multiple cancers while showing restricted expression in normal human tissues. Combined, these features suggest that STEAP is a suitable target for a variety of clinical applications that include antibody therapy, cancer-vaccine therapy, small-molecule therapy, and diagnostic imaging.
Acknowledgments
We thank Drs. Robert Eisenman, Owen Witte, William Isaacs, Inder Verma, Charles Sawyers, and Robert Reiter for helpful suggestions and for critical readings of our manuscript; Frank Lynch and Page Erickson at QualTek Molecular Labs for their expertise in immunohistochemistry; and Lianna Doan for her help in preparation of the manuscript.
Abbreviations
- STEAP
six-transmembrane epithelial antigen of the prostate
- BPH
benign prostatic hyperplasia
- LAPC xenografts
Los Angeles prostate cancer xenografts
- SSH
suppression subtractive hybridization
- AD
androgen-dependent
- AI
androgen-independent
- PSA
prostate-specific antigen
- PSM
prostate-specific membrane antigen
- PSCA
prostate stem cell antigen
- PCTA
prostate carcinoma tumor antigen
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF186249).
References
- 1.Oesterling J E. J Am Med Assoc. 1992;267:2236–2238. doi: 10.1001/jama.267.16.2236. [DOI] [PubMed] [Google Scholar]
- 2.van Iersel M P, Witjes W P, de la Rosette J J, Oosterhof G O. Br J Urol. 1995;76:47–53. doi: 10.1111/j.1464-410x.1995.tb07830.x. [DOI] [PubMed] [Google Scholar]
- 3.Jung K, Meyer A, Lein M, Rudolph B, Schnorr D, Loening S A. J Urol. 1998;159:1595–1598. doi: 10.1097/00005392-199805000-00050. [DOI] [PubMed] [Google Scholar]
- 4.Afrin L B, Stuart R K. J S C Med Assoc. 1994;90:231–236. [PubMed] [Google Scholar]
- 5.Horoszewicz J S, Leong S S, Kawinski E, Karr J P, Rosenthal H, Chu T M, Mirand E A, Murphy G P. Cancer Res. 1983;43:1809–1818. [PubMed] [Google Scholar]
- 6.Pretlow T G, Wolman S R, Micale M A, Pelley R J, Kursh E D, Resnick M I, Bodner D R, Jacobberger J W, Delmoro C M, Giaconia J M, et al. J Natl Cancer Inst. 1993;85:394–398. doi: 10.1093/jnci/85.5.394. [DOI] [PubMed] [Google Scholar]
- 7.Nagabhushan M, Miller C M, Pretlow T P, Giaconia J M, Edgehouse N L, Schwartz S, Kung H J, de Vere White R W, Gumerlock P H, Resnick M I, et al. Cancer Res. 1996;56:3042–3046. [PubMed] [Google Scholar]
- 8.van Weerden W M, de Ridder C M, Verdaasdonk C L, Romijn J C, van der Kwast T H, Schroder F H, van Steenbrugge G J. Am J Pathol. 1996;149:1055–1062. [PMC free article] [PubMed] [Google Scholar]
- 9.Klein K A, Reiter R E, Redula J, Moradi H, Zhu X L, Brothman A R, Lamb D J, Marcelli M, Belldegrun A, Witte O N, et al. Nat Med. 1997;3:402–408. doi: 10.1038/nm0497-402. [DOI] [PubMed] [Google Scholar]
- 10.Israeli R S, Powell C T, Fair W R, Heston W D. Cancer Res. 1993;53:227–230. [PubMed] [Google Scholar]
- 11.Su Z Z, Lin J, Shen R, Fisher P E, Goldstein N I, Fisher P B. Proc Natl Acad Sci USA. 1996;93:7252–7257. doi: 10.1073/pnas.93.14.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reiter R E, Gu Z, Watabe T, Thomas G, Szigeti K, Davis E, Wahl M, Nisitani S, Yamashiro J, Le Beau M M, et al. Proc Natl Acad Sci USA. 1998;95:1735–1740. doi: 10.1073/pnas.95.4.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whang Y E, Wu X, Suzuki H, Reiter R E, Tran C, Vessella R L, Said J W, Isaacs W B, Sawyers C L. Proc Natl Acad Sci USA. 1998;95:5246–5250. doi: 10.1073/pnas.95.9.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diatchenko L, Lau Y F, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, et al. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller A J, Young J C, Pendergast A M, Pondel M, Landau N R, Littman D R, Witte O N. Mol Cell Biol. 1991;11:1785–1792. doi: 10.1128/mcb.11.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter M A, Spillett D J, Thomas P, Weissenbach J, Goodfellow P N. Nat Genet. 1994;7:22–28. doi: 10.1038/ng0594-22. [DOI] [PubMed] [Google Scholar]
- 18.Visakorpi T, Kallioniemi A H, Syvanen A-C, Hyytinen E R, Karhu R, Tammela T, Isola J J, Kallioniemi O-P. Cancer Res. 1995;55:342–347. [PubMed] [Google Scholar]
- 19.Nupponen N N, Kakkola L, Koivisto P, Visakorpi T. Am J Pathol. 1998;153:141–148. doi: 10.1016/S0002-9440(10)65554-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolly J O, Parcej D N. J Bioenerg Biomembr. 1996;28:231–253. doi: 10.1007/BF02110698. [DOI] [PubMed] [Google Scholar]
- 21.Reizer J, Reizer A, Saier M H., Jr Crit Rev Biochem Mol Biol. 1993;28:235–257. doi: 10.3109/10409239309086796. [DOI] [PubMed] [Google Scholar]
- 22.Christie M J. Clin Exp Pharmacol Physiol. 1995;22:944–951. doi: 10.1111/j.1440-1681.1995.tb02331.x. [DOI] [PubMed] [Google Scholar]
- 23.Walz T, Hirai T, Murata K, Heymann J B, Mitsuoka K, Fujiyoshi Y, Smith B L, Agre P, Engel A. Nature (London) 1997;387:624–627. doi: 10.1038/42512. [DOI] [PubMed] [Google Scholar]
- 24.Cheng A, van Hoek A N, Yeager M, Verkman A S, Mitra A K. Nature (London) 1997;387:627–630. doi: 10.1038/42517. [DOI] [PubMed] [Google Scholar]
- 25.Lalani E N, Laniado M E, Abel P D. Cancer Metastasis Rev. 1997;16:29–66. doi: 10.1023/a:1005792206377. [DOI] [PubMed] [Google Scholar]
- 26.Smith P, Rhodes N P, Shortland A P, Fraser S P, Djamgoz M B, Ke Y, Foster C S. FEBS Lett. 1998;423:19–24. doi: 10.1016/s0014-5793(98)00050-7. [DOI] [PubMed] [Google Scholar]
- 27.Laniado M E, Lalani E N, Fraser S P, Grimes J A, Bhangal G, Djamgoz M B, Abel P D. Am J Pathol. 1997;150:1213–1221. [PMC free article] [PubMed] [Google Scholar]
- 28.Skryma R N, Prevarskaya N B, Dufy-Barbe L, Odessa M F, Audin J, Dufy B. Prostate. 1997;33:112–122. doi: 10.1002/(sici)1097-0045(19971001)33:2<112::aid-pros5>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 29.Maloney D G, Grillo-Lopez A J, White C A, Bodkin D, Schilder R J, Neidhart J A, Janakiraman N, Foon K A, Liles T M, Dallaire B K, et al. Blood. 1997;90:2188–2195. [PubMed] [Google Scholar]
- 30.Leget G A, Czuczman M S. Curr Opin Oncol. 1998;10:548–551. doi: 10.1097/00001622-199811000-00012. [DOI] [PubMed] [Google Scholar]
- 31.Ross J S, Fletcher J A. Stem Cells. 1998;16:413–428. doi: 10.1002/stem.160413. [DOI] [PubMed] [Google Scholar]
- 32.Carter R E, Feldman A R, Coyle J T. Proc Natl Acad Sci USA. 1996;93:749–753. doi: 10.1073/pnas.93.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bzdega T, Turi T, Wroblewska B, She D, Chung H S, Kim H, Neale J H. J Neurochem. 1997;69:2270–2277. doi: 10.1046/j.1471-4159.1997.69062270.x. [DOI] [PubMed] [Google Scholar]
- 34.Israeli R S, Powell C T, Corr J G, Fair W R, Heston W D. Cancer Res. 1994;54:1807–1811. [PubMed] [Google Scholar]
- 35.Sodee D B, Conant R, Chalfant M, Miron S, Klein E, Bahnson R, Spirnak J P, Carlin B, Bellon E M, Rogers B. Clin Nucl Med. 1996;21:759–767. doi: 10.1097/00003072-199610000-00002. [DOI] [PubMed] [Google Scholar]
- 36.Gregorakis A K, Holmes E H, Murphy G P. Semin Urol Oncol. 1998;16:2–12. [PubMed] [Google Scholar]
- 37.Liu H, Rajasekaran A K, Moy P, Xia Y, Kim S, Navarro V, Rahmati R, Bander N H. Cancer Res. 1998;58:4055–4060. [PubMed] [Google Scholar]
- 38.Murphy G P, Greene T G, Tino W T, Boynton A L, Holmes E H. J Urol. 1998;160:2396–2401. doi: 10.1097/00005392-199812020-00006. [DOI] [PubMed] [Google Scholar]
- 39.Silver D A, Pellicer I, Fair W R, Heston W D, Cordon-Cardo C. Clin Cancer Res. 1997;3:81–85. [PubMed] [Google Scholar]
- 40.Brawn P N, Ayala A G, Von Eschenbach A C, Hussey D H, Johnson D E. Cancer. 1982;49:525–532. doi: 10.1002/1097-0142(19820201)49:3<525::aid-cncr2820490321>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]