Abstract
Drug resistance in Plasmodium falciparum is a serious public health threat in the countries where this organism is endemic. Since resistance has been associated with specific single-nucleotide polymorphisms (SNPs) in parasite genes, molecular markers are becoming useful surrogates for monitoring the emergence and dispersion of drug resistance. In this study, a multiplex PCR (mPCR) and oligonucleotide microarray method was developed for the detection of these SNPs in genes encoding chloroquine resistance transporter (Pfcrt), multidrug resistance 1 (Pfmdr1), dihydrofolate reductase (Pfdhfr), dihydropteroate synthetase (Pfdhps), and ATPase 6 (PfATPase6) of P. falciparum. The results show that DNA microarray technology, combined with mPCR, is a promising and time-saving tool that supports conventional detection methods, allowing sensitive, accurate, simultaneous analysis of the SNPs associated with drug resistance in P. falciparum.
Malaria is still one of the most serious public health problems in the world; more than one million people worldwide die from it every year (35). This trouble is largely caused by the resistance of Plasmodium falciparum to most antimalarial drugs currently used, including chloroquine (CQ), sulfadoxine-pyrimethamine (SP), mefloquine (MQ), amodiaquine (AQ), and lumefantrine (LUM); furthermore, reduced susceptibility of P. falciparum to artemisinin and its derivatives has been also reported (15, 16, 34).
Antimalarial drug resistance has been associated with single-nucleotide polymorphisms (SNPs), in particular P. falciparum genes. The P. falciparum CQ resistance transporter (Pfcrt) gene K76T mutation has been linked to CQ and possibly AQ resistance (6, 17, 20); the P. falciparum multidrug resistance 1 (Pfmdr1) gene N86Y mutation has been associated with the response of the parasite to amino quinolines such as MQ (9, 25, 27) and LUM (26); mutations N51I, C59R, S108N/T, and I164L in the P. falciparum dihydrofolate reductase (Pfdhfr) gene have been associated with resistance to pyrimethamine (13, 14, 19, 21, 22); mutations S436A, A437G, K540E, A581G, and A613 S/T in the P. falciparum dihydropteroate synthetase (Pfdhps) gene have been shown to confer resistance to sulfadoxine (13, 14, 21, 22); and P. falciparum ATPase-6 (PfATPase6) S769N mutation has been associated with increased 50% inhibitory concentrations of artemether (11, 16, 18). These SNPs are believed to represent molecular epidemiology surveillance tools of antimalarial drug resistance, which can complement the more conventional and logistically complex in vitro or in vivo test (7, 24, 33, 34).
Existing molecular methods for analysis of these SNPs include allele-specific PCR, PCR-restriction fragment length polymorphism (RFLP) analysis, multiplex PCR (mPCR)-RFLP, DNA sequencing, dot blot hybridization techniques, the molecular beacon method, real-time PCR, PCR-enzyme-linked immunosorbent assay, and pyrosequencing (1, 2, 6, 8, 10, 28, 31, 37, 38). Each of these techniques offers its own advantages and limitations. Among the techniques, PCR-RFLP protocols constitute the most common methodological approach for the analyses of these SNPs, but this approach is laborious and time-consuming. A rapid and high-throughput genotyping method would be ideal for large-scale population-based studies.
The oligonucleotide microarray technology, also known as DNA chip, has provided an incredible technical development in rapid, large-scale, and automatic analysis of SNPs (5, 36). Briefly, the principle of oligonucleotide microarray is based on reverse Southern hybridization on a glass or silicon substrate. The fluorescently labeled products hybridized to the immobilized probes on the substrate can be easily detected by using a fluorescence-detecting scanner, and the readable fluorescent signals obtained are combined to determine the genotype of genes (12). In recent years, a microarray technique has been successfully applied to the detection of resistance genes or resistance-associated SNPs in various pathogens (3, 4, 23, 29).
Here, we introduce a one-step mPCR in combination with an oligonucleotide microarray method for rapid, high-throughput detection of the antimalarial resistance-associated SNPs in Pfcrt, Pfmdr1, Pfdhps, Pfdhfr, and PfATPase6.
MATERIALS AND METHODS
Samples.
Three P. falciparum laboratory strains (3D7, HB3, and DD2) (30) and 92 field samples (positive for P. falciparum by microscopy) collected on 3MM filter paper (Whatman, Maidstone, United Kingdom) from China's Yunnan province (40 samples), Hainan province (27 samples), and Myanmar's Shan state (25 samples) were used in the present study. The genomic DNA of P. berghei (CS strain), P. cynomolgi (B strain) and P. vivax (collected from Anhui province, China) were used as controls. The research protocol was approved by the Chinese National Institute of Parasitic Diseases ethics committee.
DNA extraction.
Genomic DNAs of the P. falciparum laboratory strains 3D7, HB3, and Dd2 were provided by the Malaria Research and Reference Reagent Resource Center (http://www.mr4.org/). Extraction of DNA from bloodspots on filter paper was carried out by the Chelex-100 (Bio-Rad Laboratories, Hercules, CA) method described by Wooden et al. (32) with some modifications described by Pearce et al. (22). The quality of the DNA samples was tested by determining the optical density at 260 and 280 nm.
SNPs studied.
The following 21 SNPs were analyzed: Pfcrt 391T/A, 392G/C, 399G/T, 400A/G, 402T/A, and 404A/C (corresponding to codons C72S, M74I, N75E, and K76T); Pfmdr1 256A/T and 257A/T (N86Y/F); Pfdhps 1482 T/G, 1483C/T/G, 1486 C/G, 1794 A/G, 1918 C/G, and 2013G/T/A (S436A/F/C, A437G, K540E, A581G, and A613S/T); Pfdhfr 148T/C, 152A/T, 153T/C, 175T/C, 323G/A/C, and 490A/T (C50R, N51I, C59R, S108N/T, and I164L); and PfATPase6 2306G/A (S769N). These SNPs are located at 11 nucleotide positions since some positions include two or more SNPs.
mPCR.
To reduce the time and cost of DNA amplification, an mPCR was developed to amplify the DNA fragments containing all of the SNPs studied in one reaction. Five pairs of primers were designed by using Primer 5.0 software and checked for specificity in a BLAST search available through the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/BLAST/). The primers used in the mPCR are shown in Table 1. mPCR was performed with the PCR amplification kit of Genscript (Piscataway, NJ) according to the manufacturer's instructions. Briefly, the mPCR was performed by using the PTC 200 cycler (MJ Research, Inc., Waltham, MA) in a total volume of 50 μl of reaction mix containing 2.5 μl of template DNA, 25 μl of 2× Genscript master mix, and 1.0 μl of primer mixture (consisting of 3.2 μl of 12.5 μM P1F, 3.2 μl of 12.5 μM P1R, 2.0 μl of 12.5 μM P2F, 2.0 μl of 12.5 μM P2R, 2.0 μl of 12.5 μM P3F, 2.0 μl of 12.5 μM P3R, 2.0 μl of 12.5 μM P4F, 2.0 μl of 12.5 μM P4R, 2.0 μl of 12.5 μM P5F, and 2.0 μl of 12.5 μM P5R). The mPCR cycling conditions were 94°C for 15 min for 1 cycle; followed by 30 cycles of denaturation at 94°C for 40 s, annealing at 50°C for 2 min, and extension (ramping from 50 to 72°C by 0.1°C/s) for about 3 min 40 s; followed in turn by 72°C for 3 min for 1 cycle and holding at 4°C. The resulting amplicon covers all of the 21 observed SNPs. A total of 3 μl of PCR product was identified by electrophoresis on a 20-g/liter agarose gel to confirm the successful amplification.
TABLE 1.
Primers used in the mPCR
| Gene | Primer | Sequence (5′-3′) | Amplicon size (bp) | Source or reference |
|---|---|---|---|---|
| Pfcrt | P1F | GGAGGTTCTTGTCTTGGTAAAT | 315 | This study |
| P1R | ATATTGGTAGGTGGAATAGATTCT | This study | ||
| Pfmdr1 | P2F | TGTTGAAAGATGGGTAAAGAGCAGA | 514 | This study |
| P2R | TCGTACCAATTCCTGAACTCACTT | This study | ||
| Pfdhps | P3F | GATTCTTTTTCAGATGGAGG | 770 | 22 |
| P3R | TTCCTCATGTAATTCATCTGA | 22 | ||
| Pfdhfr | P4F | TGATGGAACAAGTCTGCGACGTT | 594 | 22 |
| P4R | CTGGAAAAAATACATCACATTCATATG | 22 | ||
| PfATPase6 | P5F | AAAATAAATACCACATCAACACAT | 437 | This study |
| P5R | TCAATAATACCTAATCCACCTAAA | This study |
Fragmentation and labeling of mPCR product.
mPCR products were purified with a QIAquick PCR purification kit (Qiagen, Inc.) and treated with DNase I (Fermentas) at 0.001 U of DNA/μg to generate fragments of 50 to 200 bp. Reaction mixtures contained 1 mM Tris-HCl, 0.25 mM MgCl2, and 0.05 mM CaCl2. The fragmentation reaction was performed at 37°C for 18 min and then at 95°C for 10 min. Labeling of DNA was performed using terminal deoxynucleotidyltransferase, and reaction mixtures (25 μl) contained 200 mM potassium cacodylate, 25 mM Tris-HCl (pH 7.2), 1 mM CoCl2, 0.01% Triton X-100, 20 μM Cy3-dCTP (GE Healthcare), and 20 U of deoxynucleotidyltransferase (Fermentas). The mixtures were incubated for 1 h at 37°C, followed by denaturation at 95°C for 10 min.
Oligonucleotide probe design and microarray manufacture.
For each of the polymorphic sites associated with antimalarial drug resistance, a group of probes was designed, differentiated by the interrogated base(s) positioned in the central region of the probes. To reduce the number of potential ambiguous hybridization results, some oligonucleotide probes were designed to represent combinations of SNPs rather than one SNP. In all, 11 groups of probes were designed. The specificity of the oligonucleotide probes was verified through a BLAST search. Based on initial hybridization experiments, some probes were optimized by modifying their length and sequence to achieve more uniform fluorescence intensities across the microarray. Probes were commercially synthesized and modified (Table 2). Each probe contained a 5′-amino group for immobilization chemistry and a 15-mer poly(dT) spacer, followed by the nucleotide hybridization sequence.
TABLE 2.
Oligonucleotide probes used in the microarray
| Gene and oligonucleotide | Corresponding genotype | Sequence (5′-3′) |
|---|---|---|
| Pfdhfr | ||
| A1 | 50CN | NH2-(T)15-CCATGGAAATGTAATTCCCTAGATAT |
| A2 | 50CI | NH2-(T)15-CCATGGAAATGTATTTCCCTAGATAT |
| A3 | 50CN2 | NH2-(T)15-CATGGAAATGTAACTCCCTAGATATG |
| A4 | 50RN | NH2-(T)15-CCATGGAAACGTAATTCCCTAGATAT |
| A5 | 50RN2 | NH2-(T)15-CATGGAAACGTAACTCCCTAGATAT |
| A6 | 50RI | NH2-(T)15-CCATGGAAACGTATTTCCCTAGATAT |
| B1 | 59C | NH2-(T)15-ATATGAAATATTTTTGTGCAGTTACAACAT |
| B2 | 59R | NH2-(T)15-ATGAAATATTTTCGTGCAGTTACAAC |
| B3 | 59 control | NH2-(T)15-ATGAAATATTTTGGTGCAGTTACAAC |
| C1 | 108S | NH2-(T)15-GAAGAACAAGCTGGGAAAGC |
| C2 | 108N | NH2-(T)15-GGAAGAACAAACTGGGAAAGC |
| C3 | 108T | NH2-(T)15-GAAGAACAACCTGGGAAAGC |
| C4 | 108 control | NH2-(T)15-GGAAGAACAATCTGGGAAAGC |
| D1 | 164I | NH2-(T)15-AAATGTTTTATTATAGGAGGTTCCGT |
| D2 | 164L | NH2-(T)15-AAATGTTTTATTTTAGGAGGTTCCGT |
| D3 | 164 control | NH2-(T)15-ATGTTTTATTCTAGGAGGTTCCG |
| Pfmdr1 | ||
| E1 | 86N | NH2-(T)15-ATTAAAGAACATGAATTTAGGTGATGAT |
| E2 | 86Y | NH2-(T)15-ATTAAAGAACATGTATTTAGGTGATGAT |
| E3 | 86F | NH2-(T)15-TTAAAGAACATGTTTTTAGGTGATGATA |
| E4 | 86 control | NH2-(T)15-TTAAAGAACATGCATTTAGGTGATGA |
| PfATPase6 | ||
| F1 | 769S | NH2-(T)15-TTTGCTTATAAAAAATTAAGTAGTAAAGATTTAAATAT |
| F2 | 769N | NH2-(T)15-CTTTGCTTATAAAAAATTAAATAGTAAAGATTTAAATAT |
| F3 | 769 control | NH2-(T)15-TTTGCTTATAAAAAATTAACTAGTAAAGATTTAAATAT |
| Pfdhps | ||
| G1 | 436AA | NH2-(T)15-AGAATCCGCTGCTCCTTTTGT |
| G2 | 436AG | NH2-(T)15-AGAATCCGCTGGTCCTTTTGT |
| G3 | 436SA | NH2-(T)15-GAGAATCCTCTGCTCCTTTTG |
| G4 | 436SG | NH2-(T)15-GAGAATCCTCTGGTCCTTTTG |
| G5 | 436FA | NH2-(T)15-GAGAATCCTTTGCTCCTTTTGT |
| G6 | 436FG | NH2-(T)15-GAGAATCCTTTGGTCCTTTTGT |
| G7 | 436CA | NH2-(T)15-GAGAATCCTGTGCTCCTTTTG |
| H1 | 540K | NH2-(T)15-CACATACAATGGATAAACTAACAAATTA |
| H2 | 540E | NH2-(T)15-CACATACAATGGATGAACTAACAAATTA |
| H3 | 540 control | NH2-(T)15-CACATACAATGGATCAACTAACAAATT |
| I1 | 581A | NH2-(T)15-GATTAGGATTTGCGAAGAAACATG |
| I2 | 581G | NH2-(T)15-GATTAGGATTTGGGAAGAAACATGA |
| I3 | 581 control | NH2-(T)15-GATTAGGATTTGAGAAGAAACATG |
| J1 | 613A | NH2-(T)15-AAAAGATTTATTGCCCATTGCATGA |
| J2 | 613S | NH2-(T)15-AAAAAGATTTATTTCCCATTGCATGAAT |
| J3 | 613T | NH2-(T)15-AAAAAGATTTATTACCCATTGCATGAAT |
| J4 | 613 control | NH2-(T)15-AAAAGATTTATTCCCCATTGCATGA |
| Pfcrt | ||
| K1 | 72CVMNK | NH2-(T)15-GTGTATGTGTAATGAATAAAATTTTTGCTA |
| K2 | 72CVIET | NH2-(T)15-TGTATGTGTAATTGAAACAATTTTTGCTAA |
| K3 | 72SVMNT | NH2-(T)15-TATTTATTTAAGTGTAAGTGTAATGAATACAATT |
| K4 | 72CVIEK | NH2-(T)15-GTGTATGTGTAATTGAAAAAATTTTTGCTA |
| K5 | 72S2VMNT | NH2-(T)15-TATTTATTTAAGTGTATCTGTAATGAATACAATTTTTG |
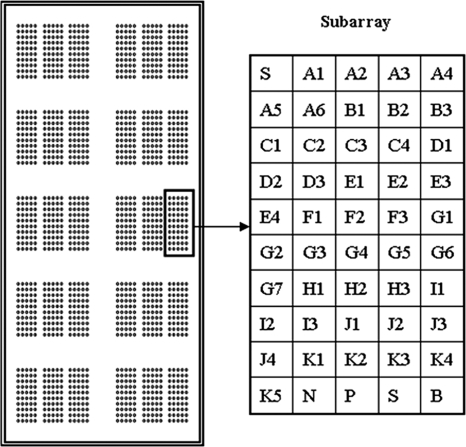
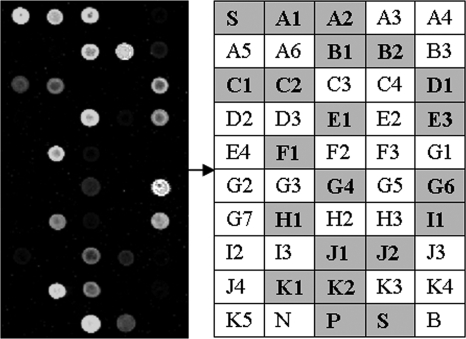
The probes were suspended in 100 mM phosphate-buffered solution containing 0.05% sodium dodecyl sulfate (SDS) at a concentration of 25 μM and printed on silylated slides (OPAldehydeSlide; CapitalBio Corp., Beijing, China) by an OmniGrid 100 Microarrayer (GeneMachine, San Carlos, CA). Each slide was spotted with 10 sets of three identical subarrays consisting of a 5 × 10 matrix; a matrix includes 45 specific probes and 2 Cy3-labeled spotting control probes (sequence, ATTGCTTGCGGCGGTAACG-Cy3), a positive hybridization control probe (TCTGCTTCTGCTTCTGCTT), a negative hybridization control probe (TCTGCTTCTCCTTCTGCTT), and a blank control probe (100 mM phosphate-buffered solution containing 0.05% SDS). A schematic diagram of the probe positions on the microarray is shown in Fig. 1.
FIG. 1.
Schematic diagram of the probe positions on the microarray. A1 to K5, specific probes for detection of the SNPs; S, spotting control probe; N, negative control probe; P, positive control probe; B, blank control probe. Each probe was spotted three times.
Hybridization and washing.
Hybridization of fluorescently labeled single-stranded DNA samples to the oligonucleotide microarray was performed in 1× hybridization buffer (6× sodium chloride-sodium phosphate-EDTA buffer, 5% dimethyl sulfoxide, 0.1% Triton X-100) at 52°C for 1 h. Just before hybridization, 10 μl of the Cy3-labeled single-stranded DNA sample was mixed with an equal volume of 2× hybridization buffer containing 1 nM Cy3-labeled control oligonucleotide (sequence, AAGCAGAAGCAGAAGCAGA), followed by denaturation at 95°C for 10 min and chilling on ice. After hybridization, the slides were washed at 42°C for 6 min with 2× saline sodium citrate (SSC) buffer containing 1% SDS, for 6 min with 1× SSC buffer containing 0.2% SDS, and for 3 min with 0.6× SSC buffer. Traces of buffer were removed by air stream.
Microarray scanning and analysis.
Slides were scanned by using a scanner GenePix 4000B (Axon Instruments), and data acquisition and processing were performed by using GenePix Pro 3.0 software. The median of local background-corrected feature intensities (henceforth referred to as net intensities) and their signal-to-noise ratios (net intensities/standard deviation of the local background) were used for further analyses. Each signal was ranked from highest to lowest in each probe group according to their net intensities. The highest ranked signal in each group was regarded as a positive signal, and a signal with an intensity more than one-third of the strongest signal in that group was also considered a positive signal, whereas signals with intensities less than one-third of the strongest signal were considered negative. When hybridized to single-infection samples, only one probe in each group produced a positive signal as the perfect match. In multiple infections, two or more probes with the highest signal intensities in a group might be selected as positive.
Microarray analyses of synthesized oligonucleotides.
Because genotype variations of the five genes from the samples studied here were limited, we also used a set of 37 synthesized 50-bp oligonucleotides (Table 3) harboring all known genotypes of the loci studied to evaluate the microarray. Twenty combinations (Table 4) of the synthesized oligonucleotides were detected by the microarray to test whether the method can correctly identify all possible genotypes in single or mixed infection. The labeling and hybridization conditions were as described above.
TABLE 3.
Thirty-seven synthesized oligonucleotides
| Gene and oligonucleotide | Harboring genotype | Sequence (5′-3′) |
|---|---|---|
| Pfdhfr | ||
| A1 | 50CN | TGTAACTGCACGAAAATATTTCATATCTAGGGAATTACATTTCCATGGTA |
| A2 | 50CI | TGTAACTGCACGAAAATATTTCATATCTAGGGAAATACATTTCCATGGTA |
| A3 | 50CN2 | TGTAACTGCACGAAAATATTTCATATCTAGGGAGTTACATTTCCATGGTA |
| A4 | 50RN | TGTAACTGCACGAAAATATTTCATATCTAGGGAATTACGTTTCCATGGTA |
| A5 | 50RN2 | TGTAACTGCACGAAAATATTTCATATCTAGGGAGTTACGTTTCCATGGTA |
| A6 | 50RI | TGTAACTGCACGAAAATATTTCATATCTAGGGAAATACGTTTCCATGGTA |
| B1 | 59C | TGATTCATTCACATATGTTGTAACTGCACAAAAATATTTCATATCTAGGG |
| B2 | 59R | TGATTCATTCACATATGTTGTAACTGCACGAAAATATTTCATATCTAGGG |
| C1 | 108S | AAATTTTTTTGGAATGCTTTCCCAGCTTGTTCTTCCCATAACTACAACAT |
| C2 | 108N | AAATTTTTTTGGAATGCTTTCCCAGTTTGTTCTTCCCATAACTACAACAT |
| C3 | 108T | AAATTTTTTTGGAATGCTTTCCCAGGTTGTTCTTCCCATAACTACAACAT |
| D1 | 164I | TCTTGATAAACAACGGAACCTCCTATAATAAAACATTTATAGTAATTTAA |
| D2 | 164L | TCTTGATAAACAACGGAACCTCCTAAAATAAAACATTTATAGTAATTTAA |
| Pfmdr1 | ||
| E1 | 86N | TAGGATTAATATCATCACCTAAATTCATGTTCTTTAATATTACACCAAAC |
| E2 | 86Y | TAGGATTAATATCATCACCTAAATACATGTTCTTTAATATTACACCAAAC |
| E3 | 86F | TAGGATTAATATCATCACCTAAAAACATGTTCTTTAATATTACACCAAAC |
| PfATPase6 | ||
| F1 | 769S | TTCTTAATATTTAAATCTTTACTACTTAATTTTTTATAAGCAAAGCTAAG |
| F2 | 769N | TTCTTAATATTTAAATCTTTACTATTTAATTTTTTATAAGCAAAGCTAAG |
| Pfdhps | ||
| G1 | 436AA | TTGGATTAGGTATAACAAAAGGAGCAGCGGATTCTCCACCTATATCTATA |
| G2 | 436AG | TTGGATTAGGTATAACAAAAGGACCAGCGGATTCTCCACCTATATCTATA |
| G3 | 436SA | TTGGATTAGGTATAACAAAAGGAGCAGAGGATTCTCCACCTATATCTATA |
| G4 | 436SG | TTGGATTAGGTATAACAAAAGGACCAGAGGATTCTCCACCTATATCTATA |
| G5 | 436FA | TTGGATTAGGTATAACAAAAGGAGCAAAGGATTCTCCACCTATATCTATA |
| G6 | 436FG | TTGGATTAGGTATAACAAAAGGACCAAAGGATTCTCCACCTATATCTATA |
| G7 | 436CA | TTGGATTAGGTATAACAAAAGGAGCACAGGATTCTCCACCTATATCTATA |
| H1 | 540K | ACTAGATTATCATAATTTGTTAGTTTATCCATTGTATGTGGATTTCCTCT |
| H2 | 540E | ACTAGATTATCATAATTTGTTAGTTCATCCATTGTATGTGGATTTCCTCT |
| I1 | 581A | TTTAATAGATTGATCATGTTTCTTCGCAAATCCTAATCCAATATCAAATA |
| I2 | 581G | TTTAATAGATTGATCATGTTTCTTCCCAAATCCTAATCCAATATCAAATA |
| J1 | 613A | ACATTTTGATCATTCATGCAATGGGCAATAAATCTTTTTCTTGAATATCC |
| J2 | 613S | ACATTTTGATCATTCATGCAATGGGAAATAAATCTTTTTCTTGAATATCC |
| J3 | 613T | ACATTTTGATCATTCATGCAATGGGTAATAAATCTTTTTCTTGAATATCC |
| Pfcrt | ||
| K1 | 72CVMNK | GTTCTTTTAGCAAAAATTTTATTCATTACACATACACTTAAATAAATAAT |
| K2 | 72CVIET | GTTCTTTTAGCAAAAATTGTTTCAATTACACATACACTTAAATAAATAAT |
| K3 | 72SVMNT | GTTCTTTTAGCAAAAATTGTATTCATTACACTTACACTTAAATAAATAAT |
| K4 | 72CVIEK | GTTCTTTTAGCAAAAATTTTTTCAATTACACATACACTTAAATAAATAAT |
| K5 | 72S2VMNT | GTTCTTTTAGCAAAAATTGTATTCATTACAGATACACTTAAATAAATAAT |
TABLE 4.
Twenty combinations of the synthesized oligonucleotides
| Combination no. | Oligonucleotide combinationa |
|---|---|
| 1 | A1, B1, C1, D1, E1, F1, G1, H1, I1, J1, K1 |
| 2 | A2, B2, C2, D2, E2, F2, G2, H2, I2, J2, K2 |
| 3 | A3, B1, C3, D1, E3, F1, G3, H1, I1, J3, K3 |
| 4 | A4, B2, C1, D2, E1, F2, G4, H2, I2, J1, K4 |
| 5 | A5, B1, C2, D1, E2, F1, G5, H1, I1, J2, K5 |
| 6 | A6, B2, C3, D2, E3, F2, G6, H2, I2, J3, K1 |
| 7 | A1, B1, C1, D1, E1, F1, G7, H1, I1, J1, K2 |
| 8 | A2, B2, C2, D2, E2, F2, G1, H2, I2, J2, K3 |
| 9 | A3, B1, C3, D1, E3, F1, G2, H1, I1, J3, K4 |
| 10 | A4, B2, C1, D2, E1, F2, G3, H2, I2, J1, K5 |
| 11 | A1, B1, C1, D1, E1, F1, G4, H1, I1, J1, K1, A2 |
| 12 | A1, B1, C1, D1, E1, F1, G4, H1, I1, J1, K1, B2 |
| 13 | A1, B1, C1, D1, E1, F1, G4, H1, I1, J1, K1, C2 |
| 14 | A1, B1, C1, D1, E1, F1, G4, H1, I1, J1, K1, D2 |
| 15 | A1, B1, C1, D1, E1, F1, G4, H1, I1, J1, K1, E2 |
| 16 | A1, B1, C1, D1, E1, F1, G4, H1, I1, J1, K1, F2 |
| 17 | A1, B1, C1, D1, E1, F1, G4, H1, I1, J1, K1, C2, G3 |
| 18 | A1, B1, C1, D1, E1, F1, G4, H1, I1, J1, K1, A2, B2, C2, E3, G6, J2, K2 |
| 19 | A1, B1, C1, D1, E1, F1, G4, H1, I1, J1, K1, B2, C2, E2, G5, K2 |
| 20 | A1, B1, C1, D1, E1, F1, G4, H1, I1, J1, K1, D2, K5 |
Combinations 1 to 10 simulate single infections with various genotypes; combinations 11 to 20 simulate mixed infections.
Traditional nested PCR-sequencing assay.
To validate the results of microarray analysis, nested PCR-sequencing assays were performed in parallel. Nested PCRs described by Djimde et al. (6) were used to amplify fragments of the Pfcrt and Pfmdr1 genes from genomic DNA. Nested PCRs as described by Pearce et al. (22) were used to amplify fragments of the Pfdhfr and Pfdhps genes. A nested PCR used to amplify fragments of the PfATPase6 gene will be described elsewhere (L. H. Tang et al., unpublished data). Sequencing reactions were carried out by using an ABI Prism BigDye terminator v3.1 cycle sequencing kit (Applied Biosystems) as specified by the manufacturer's protocol. The sequences of the amplicons were compared to the published data of the NCBI database by BLAST analysis.
RESULTS
mPCR.
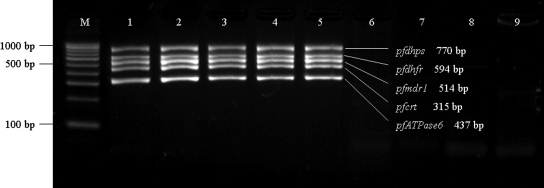
We began our analysis with mPCR amplification of the five antimalarial resistance related genes from laboratory strains and field samples. Gel electrophoresis of the resulting mPCR products indicated that they were of the expected sizes (Fig. 2). Under these amplification conditions, no nonspecific bands were observed for most samples. Moreover, the genomic DNAs of P. vivax, P. berghei, and P. cynomolgi did not yield any PCR products of the expected length.
FIG. 2.
Agarose gel electrophoresis of mPCR products. M, 100-bp DNA ladder. Lane 1, P. falciparum 3D7; lane 2, P. falciparum Dd2; lane 3, P. falciparum HB3; lanes 4 to 5, P. falciparum samples collected from field; lane 6, P. vivax; lane 7, P. berghei; lane 8, P. cynomolgi; lane 9, blank control (H2O).
Microarray analyses of laboratory strains and field samples.
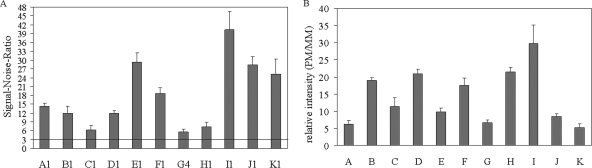
The net intensities achieved at the different oligonucleotide probes varied substantially, which is reflected by the wide range of signal-to-noise ratios from 5.6 ± 0.9 to 40.3 ± 6.4. However, upon hybridization to perfectly matching target DNA, the signal-to-noise ratios at all probes were larger than three (Fig. 3A). This threshold is considered indicative of sufficient signal in fluorescence-based microarray hybridization (3). Also, the net intensities at matching probes were always at least five times as stronger than those at probes with mismatch(es) to the respective targets (Fig. 3B), which will suffice for robust genotyping.
FIG. 3.
Signal-to-noise ratios and mismatch discrimination. The data are based on three independent hybridizations of P. falciparum 3D7. (A) Signal-to-noise ratios (means and standard errors) of perfectly matching probes; (B) relative intensities of mismatched probes (means and standard errors).
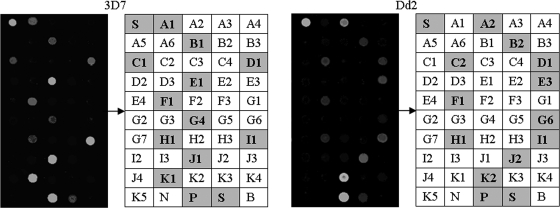
All of the SNPs in the three P. falciparum laboratory strains could be correctly analyzed by the mPCR-microarray system. In 8 of 92 field samples, results could not be obtained due to failure of DNA extraction or mPCR amplification. Among 957 genotyped positions of the 87 successful hybridizations, 943 (98.5%) yielded usable data, and 12 (1.3%) had inconsistent genotyping results between the nested PCR-sequencing assay and the microarray assay (see Table S1 in the supplemental material). Four discrepancies were due to errors in the sequencing assay, three discrepancies were due to insufficient probe-spotting quality, three discrepancies were due to a nonspecific microarray signal, and two discrepancies were due to false determination of mixed infection by microarray. Scanning images and genotyping results of P. falciparum 3D7 and Dd2 are shown in Fig. 4.
FIG. 4.
Scanning images of P. falciparum 3D7 and Dd2. For 3D7, the positive probes were A1, B1, C1, D1, E1, F1, G4, H1, I1, J1, K1, P, and S (genotypes: Pfdhfr, 50/51CN, 59C, 108S, and 164I; Pfmdr1, 86N; PfATPase6, 769S; Pfdhps, 436/437 SG, 540K, 581A, and 613A; Pfcrt, 72-76CVMNK). For Dd2, the positive probes were A2, B2, C2, D1, E3, F1, G6, H1, I1, J2, K2, P, and S (genotypes: Pfdhfr, 50/51CI, 59R, 108N, and 164I; Pfmdr1, 86F; PfATPase6, 769S; Pfdhps, 436/437 FG, 540K, 581A, and 613S; Pfcrt, 72-76CVIET).
Serial 10-fold dilutions of genomic DNA ranging from 60 ng to 0.6 pg were used to test the sensitivity of the mPCR-microarray system. The results demonstrated that when the DNA was not less than 0.06 ng, all of the SNPs in the five genes could be identified. When the DNA was diluted to 6 pg, SNPs at codons 50, 51, 59, 108, and 164 of gene Pfdhfr could not be identified, but other SNPs were identified successfully. When the DNA was diluted to 0.6 pg, only the SNPs at codons 436, 437, 540, and 613 of Pfdhps and the SNP at codon 86 of Pfmdr1 could be detected.
The specificity of microarray assay was tested by using mPCR products from P. berghei, P. cynomolgi, and P. vivax. For these three parasites no positive signal was obtained with any specific probe except the spotting control probe and the positive hybridization control probe.
To determine at what level minor SNPs in mixed infections can be detected, we mixed the genomic DNA of the 3D7 and Dd2 strains. These two strains have different genotypes at Pfdhfr codons 51, 59, and 108, Pfmdr1 codon 86, Pfdhps codons 436 and 613, and Pfcrt codons 74, 75, and 76. Although the DNA concentration of one parasite strain was fixed at 3 ng/μl, the DNA concentration of the other strain was serially 10-fold diluted from 30 ng/μl to 0.3 pg/μl. When the DNA concentration of 3D7 was fixed, Dd2 could be satisfactorily analyzed at 30 pg/μl. When the DNA concentration of Dd2 was fixed, 3D7 could also be satisfactorily analyzed at 30 pg/μl.
There was 98.2% (926/943) agreement among the genotypes obtained from three identical subarrays in the same slide for 87 samples. Thirty samples were investigated in two separate tests by the microarray method and interpreted blindly, and the reproducibility between the repeated typing was 96.1% (317/330).
Microarray analyses of the synthesized oligonucleotides.
All SNPs in the 20 combinations of the synthesized oligonucleotides were identified without ambiguity by the microarray except for two genotyped positions of Pfdhps 540 that could not give usable signals due to spotting failure. The net intensity ratios of matching probes to the mismatch ones were more than five times for 83.9% of the total and between 2.7 and 5.0 times for the others. Figure 5 shows a scanning image of the combination of 18 of the synthesized oligonucleotides (simulation of mixed infection of P. falciparum 3D7 and P. falciparum Dd2).
FIG. 5.
Scanning image of simulated mixed infection of P. falciparum 3D7 and P. falciparum Dd2 (combination of 18 synthesized oligonucleotides). The positive probes were A1, B1, C1, D1, E1, F1, G4, H1, I1, J1, K1, A2, B2, C2, E3, G6, J2, K2, P, and S.
DISCUSSION
In this study, we describe the development of an mPCR-based oligonucleotide microarray method that can simultaneously detect a set of SNPs, covering the genetic markers associated with the resistance of P. falciparum to the most common used antimalarial drugs, such as CQ, AQ, MQ, LUM, pyrimethamine, sulfadoxine, and artemether.
A high degree of concordance was observed when we compared the new microarray assay with the existing nested PCR-sequencing method, probably the most credible method currently used for detection of these SNPs. This indicates that the microarray technique for drug resistance-associated genes genotyping is reliable and accurate.
Traditional nested PCR method requires 10 PCRs for application of the five genes, which may take five workdays to accomplish. Here, the use of mPCR to amplify the five genes in one reaction leads to a significant decrease in time, cost, and number of manipulations. It takes a total of about 8 h to perform the entire detection procedure, from mPCR reaction (4.0 h), DNA labeling (2.0 h), hybridization (1.5 h), and microarray washing (0.25 h) to microarray scanning and analysis (0.25 h). There are 30 subarrays in a slide, allowing the genotyping of 10 different samples thrice by each assay, so approximately 100 samples could be analyzed for the 21 resistance-associated SNPs in one workday by two technicians. Therefore, this method is especially suitable for large-scale surveillance of the molecular markers. Moreover, the method requires less genomic DNA, which is propitious for analyzing samples with low parasite density.
The high throughput of the microarray method substantially reduces the cost. In estimating the cost, we included expenses associated with DNA isolation, multiplex-PCR, PCR product labeling, microarray production, microarray hybridization, and microarray washing. The equipment and labor costs were not included. We calculated a price of 2.9 Chinese yuan ($0.41 [United States]) per SNP for the microarray method; this cost was lower than that for pyrosequencing ($2.28 per SNP), conventional sequencing ($3.66 per SNP), and RFLP ($6.58 per SNP) (37).
Filter paper samples used in the present study offered considerable advantages for field collection, transportation, and storage over frozen liquid samples, but eight samples collected from Hainan province in 2001 failed to be analyzed by microarray. We found that these samples were not stored in a dry environment and became moldy before tests were conducted. Thus, it is probable that the genomic DNA of the parasite in these samples was destroyed or degraded. Therefore, it is important to store samples properly to ensure high-quality DNA. In addition, since false microarray determinations were mainly caused by nonspecific signals and poor probe spotting, the microarray silylated slides should be carefully kept from dust contamination and more efforts should be made to further improve the spotting quality.
The testing results of the 20 combinations of the synthesized oligonucleotides show that the microarray is able to identify all of the possible genotypes at the loci studied. Furthermore, it indicates that the technique can also differentiate the SNPs of mixed infections with P. falciparum strains of different genotypes, which allows the application of microarrays in various regions for different genetic background of P. falciparum.
As a limitation, the microarray could only be used to analyze the known SNPs in the resistance-related genes. Further studies are needed to develop a microarray assay for detection of the Pfmdr1 copy number, which has been associated with resistance against chloroquine, mefloquine, and probably artemisinins. Since new SNPs associated with antimalarial drug resistance continue to be discovered, the present microarray should be regularly updated by adding new probes.
It should be noted that microarray technology requires a microarray spotter and a microarray scanner, which may not be available in ordinary laboratories. We hereby suggest that a microarray laboratory be established in one or several countries where P. falciparum is endemic so that every site within the area can send filter paper samples to that laboratory for testing. Moreover, in the next phase, we will try to adopt chemiluminescence in the microarray technology so that the microarray assay can be performed in more laboratories.
In conclusion, mPCR-based microarray provides a promising tool for resistance-associated SNP detection in P. falciparum. This approach not only saves time and cost but also permits high-throughput detection of the SNPs. However, we consider that molecular techniques, including microarray, will complement rather than replace conventional in vivo or in vitro testing. Conventional resistance testing will remain indispensable for the detection of resistance because the molecular results do not always correlate with the clinical response associated with P. falciparum. Therefore, microarray-based analysis will be most useful in combination with conventional methods. We expect that the method developed here will considerably facilitate the molecular surveillance of antimalarial resistance in countries and regions where malaria is endemic.
Acknowledgments
This study was supported by the Key Science-Technology Project of the National Tenth Five-Year Plan of China (no. 2004BA718B13).
We are grateful to Yi Zhong Duan and Ying Xue Lin for providing P. falciparum samples. We thank Yao Yu Feng and Yi Chang Ni for critically reading the manuscript. We also thank the Malaria Research and Reference Reagent Resource Center for providing the genomic DNA for P. falciparum 3D7, HB3, and Dd2.
Footnotes
Published ahead of print on 30 April 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Abdel-Muhsin, A. M., L. C. Ranford-Cartwright, A. R. Medani, S. Ahmed, S. Suleiman, B. Khan, P. Hunt, D. Walliker, and H. A. Babiker. 2002. Detection of mutations in the Plasmodium falciparum dihydrofolate reductase (dhfr) gene by dot blot hybridization. Am. J. Trop. Med. Hyg. 6724-27. [DOI] [PubMed] [Google Scholar]
- 2.Alifrangis, M., S. Enosse, R. Pearce, C. Drakeley, C. Roper, I. F. Khalil, W. M. Nkya, A. M. Ronn, T. G. Theander, and I. C. Bygbjerg. 2005. A simple, high-throughput method to detect Plasmodium falciparum single nucleotide polymorphisms in the dihydrofolate reductase, dihydropteroate synthase, and P. falciparum chloroquine resistance transporter genes using polymerase chain reaction- and enzyme-linked immunosorbent assay-based technology. Am. J. Trop. Med. Hyg. 72155-162. [PubMed] [Google Scholar]
- 3.Antwerpen, M. H., M. Schellhase, E. Ehrentreich-Forster, F. Bier, W. Witte, and U. Nubel. 2007. DNA microarray for detection of antibiotic resistance determinants in Bacillus anthracis and closely related Bacillus cereus. Mol. Cell Probes 21152-160. [DOI] [PubMed] [Google Scholar]
- 4.Booth, S. A., M. A. Drebot, I. E. Martin, and L. K. Ng. 2003. Design of oligonucleotide arrays to detect point mutations: molecular typing of antibiotic resistant strains of Neisseria gonorrhoeae and hantavirus-infected deer mice. Mol. Cell Probes 1777-84. [DOI] [PubMed] [Google Scholar]
- 5.Bruant, G., C. Maynard, S. Bekal, I. Gaucher, L. Masson, R. Brousseau, and J. Harel. 2006. Development and validation of an oligonucleotide microarray for detection of multiple virulence and antimicrobial resistance genes in Escherichia coli. Appl. Environ. Microbiol. 723780-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djimde, A., O. K. Doumbo, J. F. Cortese, K. Kayentao, S. Doumbo, Y. Diourte, A. Dicko, X. Z. Su, T. Nomura, D. A. Fidock, T. E. Wellems, C. V. Plowe, and D. Coulibaly. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 344257-263. [DOI] [PubMed] [Google Scholar]
- 7.Djimde, A., A. Dolo, A. Ouattara, S. Diakite, C. V. Plowe, and O. K. Doumbo. 2004. Molecular diagnosis of resistance to antimalarial drugs during epidemics and in war zones. J. Infect. Dis. 190853-855. [DOI] [PubMed] [Google Scholar]
- 8.Duraisingh, M. T., J. Curtis, and D. C. Warhurst. 1998. Plasmodium falciparum: detection of polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes by PCR and restriction digestion. Exp. Parasitol. 891-8. [DOI] [PubMed] [Google Scholar]
- 9.Duraisingh, M. T., and P. Refour. 2005. Multiple drug resistance genes in malaria: from epistasis to epidemiology. Mol. Microbiol. 57874-877. [DOI] [PubMed] [Google Scholar]
- 10.Durand, R., J. Eslahpazire, S. Jafari, J. F. Delabre, A. Marmorat-Khuong, J. P. di Piazza, and J. Le Bras. 2000. Use of molecular beacons to detect an antifolate resistance-associated mutation in Plasmodium falciparum. Antimicrob. Agents Chemother. 443461-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckstein-Ludwig, U., R. J. Webb, I. D. Van Goethem, J. M. East, A. G. Lee, M. Kimura, P. M. O'Neill, P. G. Bray, S. A. Ward, and S. Krishna. 2003. Artemisinins target the SERCA of Plasmodium falciparum. Nature 424957-961. [DOI] [PubMed] [Google Scholar]
- 12.Ehrenreich, A. 2006. DNA microarray technology for the microbiologist: an overview. Appl. Microbiol. Biotechnol. 73255-273. [DOI] [PubMed] [Google Scholar]
- 13.Gregson, A., and C. V. Plowe. 2005. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol. Rev. 57117-145. [DOI] [PubMed] [Google Scholar]
- 14.Happi, C. T., G. O. Gbotosho, O. A. Folarin, D. O. Akinboye, B. O. Yusuf, O. O. Ebong, A. Sowunmi, D. E. Kyle, W. Milhous, D. F. Wirth, and A. M. Oduola. 2005. Polymorphisms in Plasmodium falciparum dhfr and dhps genes and age related in vivo sulfadoxine-pyrimethamine resistance in malaria-infected patients from Nigeria. Acta Trop. 95183-193. [DOI] [PubMed] [Google Scholar]
- 15.Hyde, J. E. 2005. Drug-resistant malaria. Trends Parasitol. 21494-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jambou, R., E. Legrand, M. Niang, N. Khim, P. Lim, B. Volney, M. T. Ekala, C. Bouchier, P. Esterre, T. Fandeur, and O. Mercereau-Puijalon. 2005. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet 3661960-1963. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, D. J., D. A. Fidock, M. Mungthin, V. Lakshmanan, A. B. Sidhu, P. G. Bray, and S. A. Ward. 2004. Evidence for a central role for PfCRT in conferring Plasmodium falciparum resistance to diverse antimalarial agents. Mol. Cell 15867-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung, M., H. Kim, K. Y. Nam, and K. T. No. 2005. Three-dimensional structure of Plasmodium falciparum Ca2+-ATPase(PfATP6) and docking of artemisinin derivatives to PfATP6. Bioorg Med. Chem. Lett. 152994-2997. [DOI] [PubMed] [Google Scholar]
- 19.Kublin, J. G., F. K. Dzinjalamala, D. D. Kamwendo, E. M. Malkin, J. F. Cortese, L. M. Martino, R. A. Mukadam, S. J. Rogerson, A. G. Lescano, M. E. Molyneux, P. A. Winstanley, P. Chimpeni, T. E. Taylor, and C. V. Plowe. 2002. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J. Infect. Dis. 185380-388. [DOI] [PubMed] [Google Scholar]
- 20.Lakshmanan, V., P. G. Bray, D. Verdier-Pinard, D. J. Johnson, P. Horrocks, R. A. Muhle, G. E. Alakpa, R. H. Hughes, S. A. Ward, D. J. Krogstad, A. B. Sidhu, and D. A. Fidock. 2005. A critical role for PfCRT K76T in Plasmodium falciparum verapamil-reversible chloroquine resistance. EMBO J. 242294-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ndiaye, D., J. P. Daily, O. Sarr, O. Ndir, O. Gaye, S. Mboup, and D. F. Wirth. 2005. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase genes in Senegal. Trop. Med. Int. Health. 101176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearce, R. J., C. Drakeley, D. Chandramohan, F. Mosha, and C. Roper. 2003. Molecular determination of point mutation haplotypes in the dihydrofolate reductase and dihydropteroate synthase of Plasmodium falciparum in three districts of northern Tanzania. Antimicrob. Agents Chemother. 471347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perreten, V., L. Vorlet-Fawer, P. Slickers, R. Ehricht, P. Kuhnert, and J. Frey. 2005. Microarray-based detection of 90 antibiotic resistance genes of gram-positive bacteria. J. Clin. Microbiol. 432291-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plowe, C. V. 2003. Monitoring antimalarial drug resistance: making the most of the tools at hand. J. Exp. Biol. 2063745-3752. [DOI] [PubMed] [Google Scholar]
- 25.Reed, M. B., K. J. Saliba, S. R. Caruana, K. Kirk, and A. F. Cowman. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403906-909. [DOI] [PubMed] [Google Scholar]
- 26.Sisowath, C., J. Stromberg, A. Martensson, M. Msellem, C. Obondo, A. Bjorkman, and J. P. Gil. 2005. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem). J. Infect. Dis. 1911014-1017. [DOI] [PubMed] [Google Scholar]
- 27.Tagelsir, N., Z. Ibrahim, A. Medani, O. Salih, A. Hamad, H. Giha, A. El-Agib, B. Khan, N. Saeed, and M. Ibrahim. 2006. High frequency of Plasmodium falciparum PfCRT K76T and PfpghN86Y in patients clearing infection after chloroquine treatment in the Sudan. Acta Trop. 9719-25. [DOI] [PubMed] [Google Scholar]
- 28.Veiga, M. I., P. E. Ferreira, A. Bjorkman, and J. P. Gil. 2006. Multiplex PCR-RFLP methods for pfcrt, pfmdr1, and pfdhfr mutations in Plasmodium falciparum. Mol. Cell Probes 20100-104. [DOI] [PubMed] [Google Scholar]
- 29.Vora, G. J., C. E. Meador, M. M. Bird, C. A. Bopp, J. D. Andreadis, and D. A. Stenger. 2005. Microarray-based detection of genetic heterogeneity, antimicrobial resistance, and the viable but nonculturable state in human pathogenic Vibrio spp. Proc. Natl. Acad. Sci. USA 10219109-19114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walliker, D., I. A. Quakyi, T. E. Wellems, T. F. McCutchan, A. Szarfman, W. T. London, L. M. Corcoran, T. R. Burkot, and R. Carter. 1987. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science 2361661-1666. [DOI] [PubMed] [Google Scholar]
- 31.Wilson, P. E., A. P. Alker, and S. R. Meshnick. 2005. Real-time PCR methods for monitoring antimalarial drug resistance. Trends Parasitol. 21278-283. [DOI] [PubMed] [Google Scholar]
- 32.Wooden, J., S. Kyes, and C. H. Sibley. 1993. PCR and strain identification in Plasmodium falciparum. Parasitol. Today 9303-305. [DOI] [PubMed] [Google Scholar]
- 33.Woodrow, C. J., and S. Krishna. 2006. Antimalarial drugs: recent advances in molecular determinants of resistance and their clinical significance. Cell Mol. Life Sci. 631586-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Health Organization. 2005. Susceptibility of Plasmodium falciparum to antimalarial drugs: report on global monitoring, 1996-2004. World Health Organization, Geneva, Switzerland.
- 35.World Health Organization and UNICEF. 2005. World malaria report 2005. World Health Organization, Geneva, Switzerland.
- 36.Zhang, F., S. Hu, J. Huang, H. Wang, Z. Wen, G. Yongyao, and S. Wang. 2006. Development and clinical evaluation of oligonucleotide microarray for HLA-AB genotyping. Pharmacogenomics 7973-985. [DOI] [PubMed] [Google Scholar]
- 37.Zhou, Z., A. C. Poe, J. Limor, K. K. Grady, I. Goldman, A. M. McCollum, A. A. Escalante, J. W. Barnwell, and V. Udhayakumar. 2006. Pyrosequencing, a high-throughput method for detecting single nucleotide polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum. J. Clin. Microbiol. 443900-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zolg, J. W., G. X. Chen, and J. R. Plitt. 1990. Detection of pyrimethamine resistance in Plasmodium falciparum by mutation-specific polymerase chain reaction. Mol. Biochem. Parasitol. 39257-265. [DOI] [PubMed] [Google Scholar]