Abstract
Vancomycin-resistant enterococci (VRE) are becoming widespread worldwide, and the rapid identification of VRE carriers from surveillance cultures is crucial for the efficient control of their spread. We assessed a new selective chromogenic medium, chromID VRE (bioMérieux, France), that enhanced the isolation and presumptive identification of VRE directly from rectal swabs and reduced unnecessary confirmatory and time-consuming tests.
Enterococcus species are members of the normal intestinal flora (14), but over the last two decades they have also emerged as important nosocomial pathogens (14, 22, 23). Since their initial discovery from patients in France and the United Kingdom in 1988 (12, 20), vancomycin-resistant enterococci (VRE) have been reported worldwide (3, 5-7, 9-11, 15, 17). The resistance phenotype VanA is the most common and features high-level resistance to both vancomycin and teicoplanin. Hospital outbreaks of VRE have been reported extensively in the United States (14, 22) and in recent years have been increasingly reported in European hospitals, with observed prevalences of 10.4% in the United Kingdom and up to 19.6% in Italy (8, 22). The most notable consequences of VRE infection are increases in mortality and the length and cost of hospital stays (2, 19, 21).
Management of a VRE outbreak requires strategies to contain cases and decrease rates of transmission, including isolation of VRE-infected or colonized patients (14, 23). VRE colonization can be monitored by screening cultures of stool or rectal swabs using differential and/or selective media. To date, the widely used medium for VRE screening is bile-esculin azide agar supplemented with 6 μg of vancomycin/ml (EVA) (16, 23). Although reasonably sensitive, this approach requires additional confirmatory tests to identify isolates and confirm glycopeptide resistance. Alternatively, molecular methods have been developed that allow identification of the glycopeptide resistance genotype (18, 23), with the drawbacks that they identify either antimicrobial resistance genes in the absence of a viable organism or resistance determinants carried by an organism other than the targeted bacterium (1).
Recent progress in the use of highly specific chromogenic substrates with sufficient sensitivity to identify VRE in a selective agar medium led to the development of chromID VRE (bioMérieux, Marcy l'Etoile, France). The aim of the present study was to evaluate the sensitivity and specificity of this selective chromogenic agar medium in the course of an ongoing outbreak faced by several wards at the Hôpital Bicêtre since August 2004 and caused by the vancomycin-resistant E. faecium strain BCT-1 expressing a heterogeneous VanD phenotype vanA genotype (15).
(This study was presented in part at the 17th European Congress of Clinical Microbiology and Infectious Diseases, Munich, Germany, 31 March to 3 April 2007.)
A total of 498 rectal swabs (351 patients) were tested in routine conditions over a 6-week period. Due to a low-level expression of the glycopeptide resistance, the swabs were first incubated for 18 h in a bile broth (AES, Bruz, France) supplemented with 3 μg of vancomycin/ml, 5 μg of colistin/ml, and 50 μg of amphotericin B/ml at 37°C prior to plating on either the conventional BD BBL Enterococcosel vancomycin agar (EVA; BD Diagnostics, Marcy l'Etoile, France) or the chromID VRE medium. Plates were incubated at 37°C under aerobic atmosphere.
Colonies on chromID VRE were screened for purple VR E. faecium (VREfm) or blue-green VR E. faecalis (VREfs) colors (Fig. 1A and B). Purple or blue-green colonies were easily differentiated from each other and from other flora, even in mixed cultures and when present at low colony counts. EVA plates were screened for colonies causing a blackening of the medium around the colony. EVA has no differential capabilities, making it impossible to distinguish VREfm from VREfs without further testing. Gram staining and catalase reactions were performed on characteristic colonies. Only gram-positive cocci and catalase-negative colonies were retained for further identification. Confirmatory identification with the VITEK 2 instrument and VITEK 2 GP cards (bioMérieux) was done only on isolates that were positive on PYRase test strips (Oxoid, Dardilly, France).
FIG. 1.
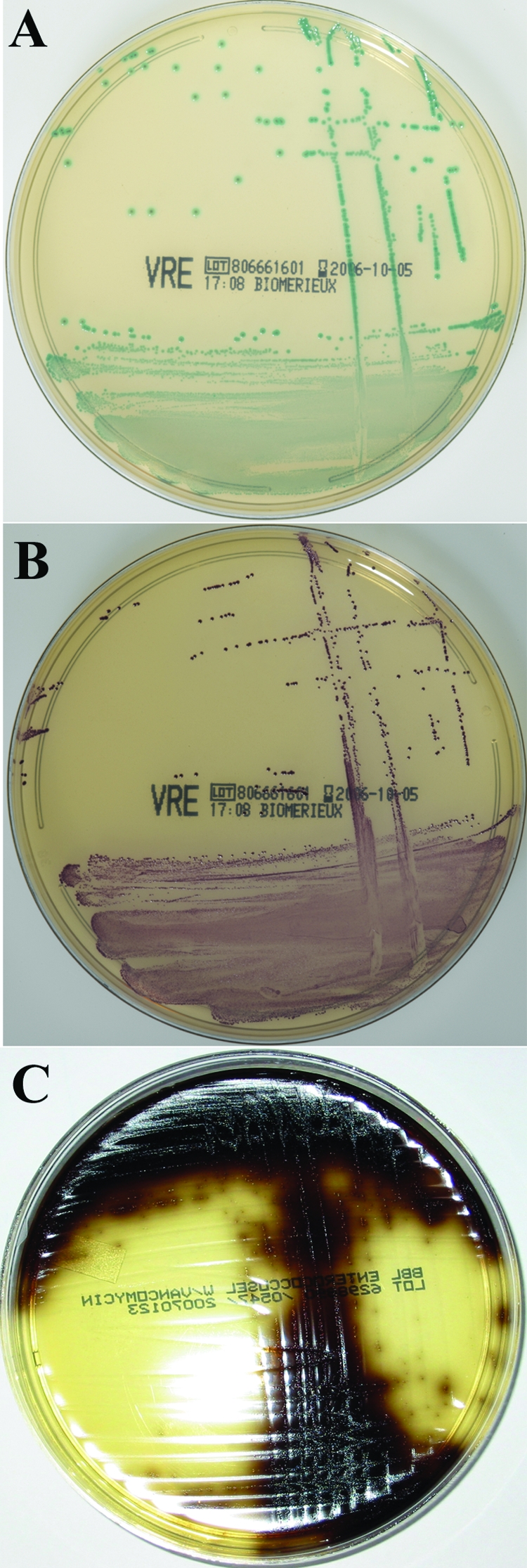
ChromID VRE medium plated with E. faecalis (A) or E. faecium (B) and EVA medium with E. faecium (C). On chromID VRE colonies of E. faecalis appear blue-green and colonies of E. faecium appear purple, and on EVA colonies of E. faecium dark brown. For the purposes of the present study, rectal swabs were plated to chromID VRE and EVA after a 24-h enrichment culture, and the plates were observed after 48 h of growth.
Interfering flora (IF), defined as colonies with characteristic color but catalase-positive or rod/yeast Gram stain shapes and as colonies without any characteristic color, was evaluated for both media. Among the 86 specimens that yielded IF on EVA, 77 yielded growth of non-VRE colonies presenting a typical appearance of VRE. On chromID VRE, 66 specimens yielded IF, but only 3 of them yielded growth of non-VRE colonies presenting the typical color of VRE. These false positives recovered from chromID VRE were two strains of E. faecalis and one strain of Lactococcus gergoviae, all being susceptible to glycopeptides. False positives on EVA medium were predominantly naturally vancomycin-resistant species such as E. gallinarum, E. casseliflavus, and Lactobacillus spp. These strains had typical aspects of VRE and needed to be subcultured for further confirmation tests (except for Lactobacillus, where the Gram stain was sufficient).
All VRE isolates had the same resistance phenotype, as revealed by disk diffusion antibiograms and interpreted according to CLSI guidelines (4). The MICs of vancomycin and teicoplanin were determined by the E-test method (AB Biodisk, Solna, Sweden). Although a few isolates displayed a true VanA phenotype (vancomycin MIC of >256 μg/ml, teicoplanin MIC of 48 μg/ml), most of the isolates had a VanD phenotype (vancomycin MICs of 12 to 24 μg/ml, teicoplanin MIC of 4 to 8 μg/ml). All VRE isolates were of the vanA genotype, as revealed by PCR (18), and pulsed-field gel electrophoresis showed that all E. faecium isolates were clonally related to the epidemic E. faecium clone BCT-1.
Among the screened patients, 6.6% were found to be colonized with VRE, which reflects the actual prevalence of VRE in the affected wards of the Hôpital Bicêtre. The two main human enterococcal species were isolated, but E. faecium was the most prevalent species (n = 31; 93.4%). Overall, 33 VRE isolates were recovered from the 498 specimens (6.6%) on at least one of the two media: 32 on chromID VRE (sensitivity, 96.9%) and 31 on EVA (sensitivity, 93.9%) (Table 1). One strain could not be recovered on chromID VRE, probably because the plate was covered by Pseudomonas aeruginosa growth.
TABLE 1.
Sensitivity analysis of chromID VRE and EVA for E. faecium or E. faecalis at 48 h
| Medium | Organism(s) | No. of resultsa
|
Sensitivity
|
||
|---|---|---|---|---|---|
| TP | FN | % | 95% CI | ||
| chromID VRE | E. faecium | 30 | 1 | 96.8 | 83.4-99.4 |
| E. faecalis | 2 | 0 | 100 | 33.3-100 | |
| E. faecium + E. faecalis | 32 | 1 | 96.9 | 84.3-99.5 | |
| EVA | E. faecium | 29 | 2 | 93.6 | 78.9-98.3 |
| E. faecalis | 2 | 0 | 100 | 33.3-100 | |
| E. faecium + E. faecalis | 31 | 2 | 93.9 | 80.0-98.4 | |
A true positive (TP) is defined as a blue-green or purple colony on chromID VRE or a brown to black colony on EVA that was identified as VRE by VITEK 2 and confirmed by PCR. A false negative (FN) is defined as an isolate that was confirmed as a VRE on one medium but did not grow on the other medium.
The specificities after 48 h of incubation were 99.4 and 83.4%, for chromID VRE and EVA, respectively (Table 2). chromID VRE yielded positive predictive values (PPVs) of 96.8% for VREfm and 50.0% for VREfs at 48 h. Taken together, the PPV of chromID VRE was 91.4%. By comparison, the PPV of EVA for all VRE was 28.7% at 48 h (Table 3). These results are difficult to compare to those obtained in other studies because an enrichment step is not always performed (13). Moreover, a recent study indicated that specificity was found to be better after only 24 h of incubation (5). However, the epidemic E. faecium strain BCT-1 required with both media a prolonged incubation of 48 h in order to have the best sensitivity.
TABLE 2.
Sensitivity and specificity analysis of chromID VRE and EVA for VRE at 48 h
| Medium | No. of resultsa
|
Sensitivity
|
Specificity
|
|||
|---|---|---|---|---|---|---|
| TP | FN | % | 95% CI | % | 95% CI | |
| chromID VRE | 32 | 1 | 96.9 | 84.3-99.5 | 99.4 | 98.1-99.8 |
| EVA | 31 | 2 | 93.9 | 80.1-98.4 | 83.4 | 79.1-86.6 |
A true positive (TP) is defined as a blue-green or purple colony on chromID VRE or a brown to black colony on EVA that was identified as VRE by VITEK 2 and confirmed by PCR. A false negative (FN) is defined as an isolate that was confirmed as a VRE on one medium but did not grow on the other medium.
TABLE 3.
PPVs and NPVs of chromID VRE and EVA for VRE at 48 h
| Medium | No. of resultsa
|
PPV
|
NPV
|
|||||
|---|---|---|---|---|---|---|---|---|
| TP | FP | TN | FN | % | 95% CI | % | 95% CI | |
| chromID VRE | 32 | 3 | 462 | 1 | 91.4 | 77.3-97.1 | 99.8 | 98.8-99.9 |
| EVA | 31 | 77 | 388 | 2 | 28.7 | 20.9-38.1 | 99.5 | 98.1-99.8 |
A true positive (TP) is defined as a blue-green or purple colony on chromID VRE or a brown to black colony on EVA that was identified as VRE by VITEK 2 and confirmed by PCR. A false positive (FP) is defined as an isolate that exhibited typical coloration on the respective medium but was not identified as VRE by VITEK 2 or confirmed by PCR. A true negative (TN) is defined as the lack of a typically colored colony. A false negative (FN) is defined as an isolate that was confirmed as a VRE on one medium but did not grow on the other medium.
The cost of the chromID VRE was only slightly greater than that of EVA (additional cost for 498 samples, $90). In contrast, because of the false positives, VRE detection using EVA required numerous subcultures, supplementary identifications, and susceptibility tests (additional material, $990; additional technician time, $135). Therefore, the use of chromID VRE allowed a saving of at least $2 per sample for a VRE prevalence of 6.6%.
This chromogenic medium, chromID VRE, provides an interesting tool for screening patients for gut colonization with VRE. This culture medium is able to identify and differentiate VREfm from VREfs, while inhibiting the growth of vancomycin-susceptible Enterococcus spp. It proved to be more specific than conventional EVA and reduced the need for additional biochemical analysis or antimicrobial susceptibility testing in the clinical laboratory. Moreover, BCT-1 strains that may appear susceptible to glycopeptides on routine disk diffusion antibiograms were efficiently detected on ChromID VRE. These VanD- or VanB-type phenotypes with vanA genotype have been observed now in several countries and might be efficiently detected using this medium (6, 7, 9, 10, 15).
Acknowledgments
This study was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, Paris, France.
Footnotes
Published ahead of print on 7 May 2008.
REFERENCES
- 1.Ballard, S. A., K. K. Pertile, M. Lim, P. D. Johnson, and M. L. Grayson. 2005. Molecular characterization of vanB elements in naturally occurring gut anaerobes. Antimicrob. Agents Chemother. 491688-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carmeli, Y., G. Eliopoulos, E. Mozaffari, and M. Samore. 2002. Health and economic outcomes of vancomycin-resistant enterococci. Arch. Intern. Med. 282223-2228. [DOI] [PubMed] [Google Scholar]
- 3.Christiansen, K. J., J. D. Turnidge, J. M. Bell, N. M. George, J. C. Pearson, et al. 2007. Prevalence of antimicrobial resistance in Enterococcus isolates in Australia, 2005: report from the Australian Group on Antimicrobial Resistance. Commun. Dis. Intell. 31392-397. [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Delmas, J., F. Robin, C. Schweitzer, O. Lesens, and R. Bonnet. 2007. Evaluation of a new chromogenic medium, chromID VRE, for detection of vancomycin-resistant enterococci in stool samples and rectal swabs. J. Clin. Microbiol. 452731-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eom, J. S., I. S. Hwang, B. Y. Hwang, J. G. Lee, Y. J. Lee, H. J. Cheong, Y. H. Park, S. C. Park, and W. J. Kim. 2004. Emergence of vanA genotype vancomycin-resistant enterococci with low or moderate levels of teicoplanin resistance in Korea. J. Clin. Microbiol. 421785-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ergani-Ozcan, A., T. Naas, B. O. Baysan, D. Inan, D. Colak, and P. Nordmann. 2008. Nosocomial outbreak of vancomycin-resistant Enterococcus faecium in a paediatric unit at a Turkish university hospital. J. Antimicrob. Chemother. doi: 10.1093/jac/dkn066. [DOI] [PubMed]
- 8.Goossens, H., D. Jabes, R. Rossi, C. Lammens, G. Privitera, and P. Courvalin. 2003. European survey of vancomycin-resistant enterococci in at-risk hospital wards and in vitro susceptibility testing of ramoplanin against these isolates. J. Antimicrob. Chemother. 51iii5-iii12. [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto, Y., K. Tanimoto, Y. Ozawa, T. Murata, and Y. Ike. 2000. Amino acid substitutions in the VanS sensor of the VanA-type vancomycin-resistant Enterococcus strains result in high-level vancomycin resistance and low-level teicoplanin resistance. FEMS Microbiol. Lett. 185247-254. [DOI] [PubMed] [Google Scholar]
- 10.Lauderdale, T. L., L. C. McDonald, Y. R. Shiau, P. C. Chen, H. Y. Wang, J. F. Lai, and M. Ho. 2002. Vancomycin-resistant enterococci from humans and retail chickens in Taiwan with unique VanB phenotype-vanA genotype incongruence. Antimicrob. Agents Chemother. 46525-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leavis, H. L., M. J. M. Bonten, and L. J. L. Willems. 2006. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr. Opin. Microbiol. 9454-460. [DOI] [PubMed] [Google Scholar]
- 12.Leclercq, R., E. Derlot, J. Duval, and P. Courvalin. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium strain. N. Engl. J. Med. 319157-161. [DOI] [PubMed] [Google Scholar]
- 13.Ledeboer, N. A., R. J. Tibbetts, and W. M. Dunne. 2007. A new chromogenic agar medium, chromID VRE, to screen for vancomycin-resistant Enterococcus faecium and Enterococcus faecalis. Diagn. Microbiol. Infect. Dis. 59477-479. [DOI] [PubMed] [Google Scholar]
- 14.Moellering, R. C. 2005. Enterococcus species, Streptococcus bovis, and Leuconostoc species, p. 2411-2421. In G. L Mandell, J. R. Bennett, and R. Dolin (ed.), Principles and practices of infectious diseases, 6th ed., vol. 2. Elsevier/Churchill Livingstone, Philadelphia, PA. [Google Scholar]
- 15.Naas, T., N. Fortineau, R. Snanoudj, C. Spicq, A. Durrbach, and P. Nordmann. 2005. First nosocomial outbreak of vancomycin-resistant Enterococcus faecium expressing a VanD-like phenotype associated with a vanA genotype. J. Clin. Microbiol. 433642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novicki, T. J., J. M. Schapiro, B. K. Ulness, A. Sebeste, L. Busse-Johnston, K. M. Swanson, S. R. Swanzy, W. Leisenring, and A. P. Limaye. 2004. Convenient selective differential broth for isolation of vancomycin-resistant enterococcus from fecal material. J. Clin. Microbiol. 421637-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ofner-Agostini, M., B. L. Johnston, A. E. Simor, J. Embil, A. Matlow, M. Mulvey, D. Ormiston, J. Conly, et al. 2008. Vancomycin-resistant enterococci in Canada: results from the Canadian Nosocomial Infection Surveillance Programme, 1999-2005. Infect. Control. Hosp. Epidemiol. 29271-274. [DOI] [PubMed] [Google Scholar]
- 18.Sloan, L. M., J. R. Uhl, E. A. Vetter, C. D. Schleck, W. S. Harmsen, J. Manahan, R. L. Thompson, J. E. Rosenblatt, and F. R. Cockerill III. 2004. Comparison of the Roche LightCycler vanA/vanB detection assay and culture for detection of vancomycin-resistant enterococci from perianal swabs. J. Clin. Microbiol. 422636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song, X., A. Srinivasan, D. Plaut, and T. M. Perl. 2003. Effect of nosocomial vancomycin-resistant enterococcal bacteremia on mortality, length of stay, and costs. Infect. Control Hosp. Epidemiol. 24251-256. [DOI] [PubMed] [Google Scholar]
- 20.Uttley, A. H., C. H. Collins, J. Naidoo, and R. C. George. 1988. Vancomycin-resistant enterococci. Lancet i57-58. [DOI] [PubMed] [Google Scholar]
- 21.Webb, M., L. W. Riley, and R. B. Roberts. 2001. Cost of hospitalization for and risk factors associated with vancomycin-resistant Enterococcus faecium infection and colonization. Clin. Infect. Dis. 33445-452. [DOI] [PubMed] [Google Scholar]
- 22.Willems, R. J. L., J. Top, M. Van Santen, D. A. Robinson, T. M. Coque, F. Baquero, H. Grundmann, and M. J. M. Bonten. 2005. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg. Infect. Dis. 11821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zirakzadeh, A., and R. Patel. 2006. Vancomycin-resistant enterococci: colonization, infection, detection, and treatment. Mayo Clin. Proc. 81529-536. [DOI] [PubMed] [Google Scholar]


