Abstract
Comparative genomics analysis of the Tamil Nadu strain of Mycobacterium leprae has uncovered several polymorphic sites with potential as epidemiological tools. In this study we compared the stability of two different markers of genomic biodiversity of M. leprae in several biopsy samples isolated from the same leprosy patient. The first type comprises five different variable-number tandem repeats (VNTR), while the second is composed of three single nucleotide polymorphisms (SNP). Contrasting results were obtained, since no variation was seen in the SNP profiles of M. leprae from 42 patients from 7 different locations in Mali whereas the VNTR profiles varied considerably. Furthermore, since variation in the VNTR pattern was seen not only between different isolates of M. leprae but also between biopsy samples from the same patient, these VNTR may be too dynamic for use as epidemiological markers for leprosy.
Leprosy remains a public health problem, and on average, 400,000 new cases have been reported annually during the last 5 years (2), although there is no known reservoir for the etiologic agent, Mycobacterium leprae, other than human beings. In order to understand better the transmission and epidemiology of leprosy, several investigators have searched for polymorphic markers within the genome of M. leprae with the aim of developing robust molecular typing systems for epidemiology. Variable-number tandem repeats (VNTR), such as runs of di- or trinucleotides, have been examined as potential typing markers and found to vary in copy number between strains of M. leprae (7, 12, 16, 19, 24), thus arousing interest in their application as epidemiological tools. Other workers have investigated single nucleotide polymorphisms (SNP) and found that despite the reductive evolution and genome decay undergone by the leprosy bacillus (3), the genome sequence is highly conserved and SNP are rare. Three SNP were found to be informative and have been used to reconstitute the evolution and global spread of M. leprae (12). While the SNP are useful in studies of long-range transmission of leprosy, the VNTR, owing to their more dynamic nature, appear more appropriate for monitoring the spread of M. leprae over shorter epidemiological distances, such as within a region or a large city. A number of reports have appeared that present the findings of VNTR studies of leprosy bacilli from different Asian countries (20, 25) and in one case from patients belonging to the same family (24). Here we present the findings of a study aimed at comparing the performance of three SNP and five VNTR markers in typing strains present in different biopsy samples from the same leprosy patients living in Mali.
MATERIALS AND METHODS
Biopsy samples.
The skin biopsy samples were isolated from 45 patients presenting at the Centre National d'Appui à la Lutte Contre la Maladie (CNAM) in Bamako, Mali. Details of the sex, age, type of leprosy, prior treatment history, and biopsy site may be found in Table 1. Generally, three biopsies were taken from each patient, two on the day of arrival at the leprosy clinic (D0) and the third after 1 month of treatment with moxifloxacin (D31) prior to commencement of standard multidrug therapy. At least two biopsies were used in this study (Table 1). All human biopsies were obtained in accordance with a defined protocol, which had been approved by the appropriate institutional review boards, and after informed patient consent.
TABLE 1.
Patients' detailsa
| Patient no. | Sex | Yr of Birth | Town of origin | Leprosy type | Diagnosis | Treatment | Biopsy site 1 | Biopsy site 2 | PCR result |
|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 1940 | Koulikoro | LL | RE | D | L elbow | L elbow | + |
| 2 | M | 1950 | Sikasso | LL | RE | D | L lumbar region | L lumbar region | + |
| 3 | M | 1951 | Ségou | LL | RE | D | Abdomen | Back | + |
| 4 | M | 1975 | Kayes | LL | N | U | L scapula | L scapula | + |
| 5 | F | 1955 | Segou | LL | RE | D | L flank | L flank | − |
| 6 | F | 1982 | Ségou | LL | N | U | L scapula | R Lumbar region | + |
| 7 | M | 1977 | Mopti | LL | N | U | L scapula | R lumbar region | + |
| 8 | M | 1955 | Ségou | LL | RE | D | R scapula | R scapula | + |
| 9 | M | 1980 | Ségou | LL | N | U | R back | L arm | + |
| 10 | M | 1938 | Mopti | LL | N | U | L back | R lumbar region | − |
| 11 | F | 1971 | Bamako | LL | N | U | L lumbar region | L lumbar region | + |
| 12 | M | 1960 | Bamako | LL | RE | D | R back | R lumbar region | + |
| 13 | M | 1940 | Gao | LL | RE | D | R lumbar region | R arm | + |
| 14 | F | 1945 | Mopti | LL | N | U | R lumbar region | L back | + |
| 15 | M | 1938 | Mauritania | LL | N | U | R lumbar region | R lumbar region | + |
| 16 | F | 1970 | Koulikoro | LL | N | U | R back | L lumbar region | + |
| 17 | F | 1960 | Ségou | LL | N | U | R back | R lumbar region | + |
| 18 | M | 1972 | Koulikoro | LL | N | U | L back | L back | + |
| 19 | F | 1960 | Bamako | LL | N | U | L lumbar region | L lumbar region | + |
| 20 | F | 1983 | Bamako | LL | N | U | L back | L back | + |
| 21 | F | 1975 | Sikasso | LL | N | U | L back | L back | + |
| 22 | F | 1944 | Koulikoro | LL | RE | D | R back | R lumbar region | + |
| 23 | M | 1943 | Koulikoro | LL | RE | D | L lumbar region | L lumbar region | + |
| 24 | M | 1980 | Kayes | LL | N | U | L lumbar region | L Lumbar region | + |
| 25 | F | 1940 | Sikasso | LL | RE | D | R lumbar region | L lumbar region | + |
| 26 | F | 1980 | Bamako | LL | N | U | L lumbar region | R lumbar region | + |
| 27 | M | 1975 | Kayes | LL | N | U | L back | L lumbar region | + |
| 28 | M | 1941 | Kayes | LL | RE | D | R breast | R breast | + |
| 29 | M | 1951 | Kayes | LL | N | U | R lumbar region | L lumbar region | + |
| 30 | F | 1951 | Guinea Rep | LL | RE | D | L back | L back | − |
| 31 | M | 1976 | Ségou | LL | N | U | R lumbar region | R lumbar region | + |
| 32 | M | 1972 | Kolokani | LL | N | U | R lumbar region | L lumbar region | + |
| 33 | F | 1976 | Kayes | LL | N | U | L lumbar region | L lumbar region | + |
| 34 | M | 1953 | Kayes | LL | N | U | L lumbar region | L lumbar region | + |
| 35 | F | 1983 | Mopti | LL | N | U | R lumbar region | R forearm | + |
| 36 | M | 1971 | Mopti | BL | N | U | L elbow | R elbow | + |
| 37 | M | 1938 | Koulikoro | LL | N | U | R arm | R arm | + |
| 38 | M | 1978 | Ségou | BL | N | U | L arm | L arm | + |
| 39 | M | 1965 | Koulikoro | LL | RE | D | L back | L back | + |
| 40 | M | 1983 | Kayes | BL | N | U | R back | L back | + |
| 41 | M | 1971 | Sikasso | LL | N | U | L arm | R lumbar region | + |
| 42 | M | 1986 | Mopti | LL | N | U | R elbow | L elbow | + |
| 43 | M | 1956 | Mauritania | LL | N | U | R lumbar region | R lumbar region | + |
| 44 | M | 1965 | Kayes | LL | N | U | R lumbar region | R lumbar region | + |
| 45 | M | 1946 | Sikasso | LL | N | U | L lumbar region | L lumbar region | + |
M, male; F, female; LL, lepromatous leprosy; BL, borderline leprosy; N, new case; RE, relapse; U, unknown; R, right; L, left.
SNP and VNTR analysis.
Details of the three SNP and five VNTR sites examined and the primers used for their amplification by PCR may be found in Table 2. PCR fragments were generated from M. leprae DNA present in human biopsies, processed by standard procedures (22), and then sequenced using an AB3100 DNA sequencer (Applied Biosystems, Foster City, CA).
TABLE 2.
Primers used in this study
| Polymorphism | Locusa | Primer sequence |
|---|---|---|
| SNP-14676-F | ML0009-ML0010 IG | AATGGAATGCTGGTGAGAGC |
| SNP-14676-R | ML0009-ML0010 IG | CAATGCATGCTAGCCTTAATGA |
| SNP-1642875-F | ML1378 | TGCTAGTTTAACCGAGTACTGCTA |
| SNP-1642875-R | ML1378 | GTAGTAGTCTTCCAAGTTGTGGTG |
| SNP-2935685-F | ML2462 | ATCTGGTCCGGGTAGGAATC |
| SNP-2935685-R | ML2462 | ACCGGTGAGCGCACTAAG |
| VNTR 21_TTC-F | ML2345 | ACTCGATCGAAGAACCAACC |
| VNTR 21_TTC-R | ML2345 | GGACCTAAACCATCCCGTTT |
| VNTR 9_GTA-F | ML2172-ML2173 IG | CTCGATTAGTGCGATCAACG |
| VNTR 9_GTA-R | ML2172-ML2173 IG | GAGCCAGCGGTAGTACTGGA |
| VNTR 14_AT-F | ML0235-ML0236 IG | AGCGTTATGAGCCGTAAGGA |
| VNTR 14_AT-R | ML0235-ML0236 IG | CGAACCACTAACCTGGCAAC |
| VNTR 15_AT-F | ML079-ML0799 IG | TGATCAATATGCGGGTTGG |
| VNTR 15_AT-R | ML0798-ML0799 IG | GGTTATGTTCGGCATCCATC |
| VNTR 17_AT-F | ML2183 | TTGAGCGAAAGAAAGCAGGT |
| VNTR 17_AT-R | ML2183 | TGCATTTAGCAGGACGATTG |
IG, intergenic region.
Briefly, PCR was carried out using 1.25 U of Taq DNA polymerase (Q-Biogene, MP Biomedicals) in a 25-μl volume containing 2.5 μl of prepared biopsy samples and 200 nM of custom primers (Table 2) with other reagents supplied by the manufacturer. The mixture was denatured at 94°C for 3 min, followed by 45 PCR cycles (1 min at 94°C, 1 min at 55°C, extension at 72°C for 2 min), with a final extension at 72°C for 10 min in a thermocycler (PTC-100; MJ Research, Inc.). We purified amplified DNA by enzymatic procedures; 8 μl of PCR products were incubated with 2.5 U of exonuclease I (USB Corp., Cleveland, OH) and 0.25 U of shrimp alkaline phosphatase (USB Corp., Cleveland, OH), with a final volume of 10 μl, at 37°C for 15 min before enzyme inactivation at 80°C for 15 min. Then, we added 2 μl of BigDye v3.1, 4 μl of BigDye v3.1 buffer (Applied Biosystems, Foster City, CA), and 200 nM of primer for a final volume of 20 μl. The sequencing mixture was denatured at 96°C for 1 min, followed by 40 cycles of denaturation at 96°C for 30 s, annealing at 56°C for 15 s, and extension at 60°C for 4 min.
In some instances, involving AT-rich regions, PCR products were purified using a QIAquick PCR purification kit (Qiagen, Inc., Valencia, CA) and cloned into the pGEM-T Easy Vector systems (Promega, Madison, WI) prior to sequencing. The complete results of SNP and VNTR analysis are summarized in the text and Table 3.
TABLE 3.
Alleles present at VNTR loci in M. leprae from patients in Mali
| Locus | Allele | No. of isolates | Frequency (%) | Total no. of isolates for locus |
|---|---|---|---|---|
| 21_TTC | 10 | 4 | 9.5 | 42 |
| 11 | 5 | 11.9 | ||
| 12 | 6 | 14.3 | ||
| 13 | 6 | 14.3 | ||
| 14 | 11 | 26.2 | ||
| 15 | 5 | 11.9 | ||
| 16 | 2 | 4.8 | ||
| 17 | 3 | 7.1 | ||
| 9_GTA | 8 | 6 | 14.3 | 42 |
| 9 | 18 | 42.9 | ||
| 10 | 9 | 21.4 | ||
| 11 | 5 | 11.9 | ||
| 12 | 2 | 4.8 | ||
| 13 | 1 | 2.4 | ||
| 14 | 1 | 2.4 | ||
| 14_AT | 8 | 1 | 2.4 | 42 |
| 12 | 2 | 4.8 | ||
| 13 | 4 | 9.5 | ||
| 14 | 11 | 26.2 | ||
| 15 | 5 | 11.9 | ||
| 16 | 4 | 9.5 | ||
| 17 | 2 | 4.8 | ||
| 18 | 3 | 7.1 | ||
| 19 | 2 | 4.8 | ||
| 21 | 2 | 4.8 | ||
| 22 | 3 | 7.1 | ||
| 24 | 1 | 2.4 | ||
| 25 | 2 | 4.8 | ||
| 15_AT | 11 | 2 | 5 | 40 |
| 12 | 6 | 15 | ||
| 13 | 10 | 25 | ||
| 14 | 5 | 12.5 | ||
| 15 | 4 | 10 | ||
| 16 | 3 | 7.5 | ||
| 17 | 1 | 2.5 | ||
| 18 | 2 | 5 | ||
| 19 | 1 | 2.5 | ||
| 20 | 1 | 2.5 | ||
| 23 | 1 | 2.5 | ||
| >25 | 4 | 10 | ||
| 17_AT | 9 | 1 | 2.4 | 42 |
| 10 | 1 | 2.4 | ||
| 11 | 7 | 16.7 | ||
| 12 | 11 | 26.2 | ||
| 13 | 12 | 28.6 | ||
| 14 | 5 | 11.9 | ||
| 15 | 2 | 4.8 | ||
| 16 | 2 | 4.8 | ||
| 20 | 1 | 2.4 |
Bioinformatics.
Sequences were compiled and analyzed using Gap4 (http://staden.sourceforge.net/manual/gap4_unix_2.html) and Artemis (13), as previously described (3). Phylogenetic trees were constructed using the QuickTree software (8), which employs the neighbor-joining method (14) to calculate distances that correspond to the square of the number of VNTR differences.
RESULTS
Setting and characteristics of study population.
Mali is a sparsely inhabited West African country (<9 inhabitants per km2) with a well-functioning leprosy control program that reached the goal of leprosy elimination in April 2001. As part of a chemotherapy trial to test the efficacy of a new regimen including moxifloxacin (5), a group of 45 leprosy patients (Table 1) was constituted at the CNAM. This comprised 29 males and 16 females, and they came from 7 different towns, 5 of which are located on the Niger river; Bamako and Gao are separated by 1,213 km and are the towns that are furthest apart (Fig. 1).
FIG. 1.
Map of Mali showing major towns and cities. The patient's town of origin is colored and boxed; the same color scheme applies to Fig. 2.
In the current study, leprosy was diagnosed clinically on the basis of skin lesions with impairment of sensation and skin smear positivity. With three exceptions, classed as borderline lepromatous (BL), all patients were diagnosed with lepromatous leprosy (LL). Thirty-two of the patients were new cases with no known history of antileprosy treatment, and 13 patients had relapsed, having previously been treated with dapsone monotherapy. At the time of the study, the patients ranged in age from 21 to 69 years old, with an average age of 44 years old. The average age of relapsed cases was 58 years old. Skin biopsies were taken from lesions at D0 before treatment began and again at D31. The biopsy site was noted, and in the case of large lesions, both D0 and D31 biopsies were taken from the same site. In six cases, two different sites were biopsied at D0 but not at D31.
Amplification of M. leprae DNA by PCR.
To prepare DNA suitable for sequencing purposes, as part of the SNP and VNTR analysis, a variety of PCR primers were used to amplify the eight corresponding loci (Table 2). Samples from three relapsed patients (patients no. 5, 10, and 30) consistently yielded no products (Table 1) despite multiple efforts, including the highly sensitive RLEP amplification procedure (4, 21, 22), suggesting that either they contained no M. leprae DNA or inhibitors of Taq polymerase were present. The great majority of samples from both the D0 and D31 biopsies generated PCR samples suitable for DNA sequencing, and in most cases the sequencing reaction was successful, yielding unambiguous results. To eliminate possible technical errors, all experiments were performed in duplicate by three different experimentalists with the same result.
SNP analysis.
On sequence interrogation of the three polymorphic genomic positions, which are the basis of the SNP typing system, it was found that all 42 PCR-positive M. leprae strains belonged to the same group, SNP type 4 (12). This SNP type is characterized by the nucleotides T, T, and C at genome positions 14676, 1642875, and 2935685, respectively, and has been found mainly in West Africa but also in the Americas, where it was believed to have been introduced during the era of the slave trade. Since these isolates could not be distinguished by SNP typing, five VNTR loci were amplified, sequenced, and compared.
VNTR analysis.
PCR was used to amplify fragments suitable for sequencing from five genomic loci known to harbor VNTR, namely, two trinucleotide repeats (21_TTC and 9_GTA) and three dinucleotide repeats (14_AT, 15_AT, and 17_AT), as described in Table 2. These repeats were chosen since, like the three SNP, they are located in pseudogenes or intergenic regions and thus should not be functionally constrained. All loci were readily amplified and sequenced well, with the exception of 15_AT, which occasionally yielded fragments that could not be sequenced directly. Some of the larger 15_AT-repeat-bearing fragments were cloned prior to sequencing. In these cases the same repeat copy number was obtained from all clones examined, but in some instances, when the copy number was >25, the exact number of repeats could not be established.
In our study population, eight alleles of the 21_TTC repeat were found, and the number of TTC repeats ranged from 10 to 17, with 14 being the most common (Table 3). Likewise, seven alleles of the 9_GTA repeat were detected, and the range was from 8 to 14, with 9 the most common type. Most allelic diversity was seen at the 14_AT locus, where 13 different versions were observed, ranging from 8 to 25 copies, with 14 being by far the most abundant form. The numbers of alleles at the 15_AT and 17_AT loci were 12 and 9, respectively, and in both cases 13 was the most commonly occurring repeat unit. Full details of the frequency of the alleles at these five VNTR loci are reported in Table 3. The sigA (rpoT) locus (11) of the Malian strains was also examined for variability in its hexanucleotide repeat region, but none was found (data not shown).
Phylogenetic analysis.
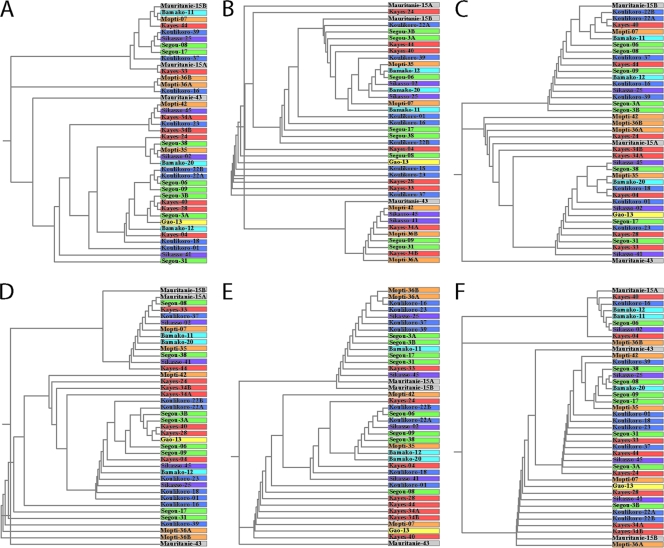
To determine whether any links could be established between the M. leprae strains and the town of origin of the 33 corresponding patients for which complete data sets were available, we used the neighbor-joining method (14) to produce phylogenetic trees. In no case was the same tree obtained, nor was convincing evidence for clustering associated with the geographical origin of the strain detected (Fig. 2), irrespective of whether VNTR loci were used individually (Fig. 2B to F) or collectively (Fig. 2A).
FIG. 2.
Phylogenetic trees generated by the neighbor-joining method. Trees were obtained using all VNTR (A), 21_TTC (B), 9_GTA (C), 14_AT (D), 15_AT (E), or 17_AT (F). Note the widely different arrangements between all trees. Samples labeled A and B correspond to different specimens from the same patient.
Intrapatient variability in VNTR type.
To establish whether there was any copy number variation at selected VNTR loci taken from different sites on the same patient or from the same site at two different times, we screened our biopsy collection using PCR and sequencing of the four most-robust sites. Owing to its lower reliability in PCR, the 15_AT locus was excluded. This comparative analysis revealed no variation in 36 of the 42 M. leprae isolates where 2 samples were available. However, in six cases, reproducible copy number differences were seen (Table 4); four strains varied at a single locus, one at three loci, and the remaining isolate at all four. Greatest variability was seen at the 21_TTC locus (5 out of 11), followed by the 17_AT repeat (3 out of 11). Most of the differences were due to a loss or gain of a single repeat unit (9 out of 11), with strain 15 showing two and three unit differences at the 14_AT and 9_GTA loci, respectively.
TABLE 4.
Allelic variation at VNTR loci in M. leprae isolated from the same patient
| Patient no. | Site(s) of biopsy | Allele at VNTR locusa
|
|||
|---|---|---|---|---|---|
| 21_TTC | 9_GTA | 14_AT | 17_AT | ||
| 3 | Abdomen, back | 12, 12 | 10, 10 | 22, 22 | 13, 12 |
| 34 | L lumbar region, L lumbar region | 16, 15 | 9, 9 | 16, 16 | 12, 12 |
| 36 | L elbow, R elbow | 15, 16 | 9, 9 | 15, 15 | 12, 12 |
| 26 | L lumbar region, R lumbar region | 15, 14 | 10, 10 | 14, 14 | 12, 12 |
| 21 | L back, L back | 14, 13 | 10, 10 | 14, 13 | 12, 13 |
| 15 | R lumbar region, R lumbar region | 14, 12 | 9, 12 | 12, 14 | 11, 12 |
The left number corresponds to VNTR copy number in the biopsy sample at day 0, the right at day 31, with allele numbers shown in bold differing between them.
When SNP analysis was performed on both samples from all of the 42 M. leprae specimens, no variation was seen at any of the three sites examined.
DISCUSSION
The aim of this study was to compare the performances of selected SNP and VNTR as potential epidemiological markers for transmission of M. leprae within a defined setting by taking advantage of a set of serial biopsy samples from a patient cohort participating in a trial of a new regimen for the treatment of leprosy. Although the SNP analysis was technically easier than VNTR analysis, since only interrogation of a single position is required, this failed to detect any differences between the M. leprae isolates, all of which belong to SNP type 4. This finding was not unexpected in light of the known stability of this type of marker and the confinement of the study to a single country. By contrast, extensive variation was seen at all five VNTR loci that were examined in this work, both between patient isolates and within samples from the same patient. Consistent with the results of earlier studies (7, 19, 25), we observed greatest allelic variation in the dinucleotide repeats (in descending order of allelic variation, 14_AT, 15_AT, and 17_AT) and then the trinucleotide repeats (21_TTC followed by 9_GTA).
Other workers have compared the stability of these VNTR in strains of M. leprae that have been passaged through animals (19, 25) and in clinical specimens taken directly from patients (24, 25). On the basis of the animal passage work, it was concluded that with the exception of strain Thai-53, the VNTR genotypes were stable during serial passaging, which augured well for their use in epidemiological studies of humans (19). However, the situation changes when the results of the relatively few studies pertaining to leprosy patients belonging to the same household are examined. In a study of 3 different households involving 11 patients, no variation was seen at 9 VNTR loci from 9 patients, whereas the 2 samples from the third household differed in the 17_AT and 9_GTA loci by 1 repeat unit (25). Similar findings of VNTR differences were reported by others when skin and nerve biopsies from the same patient were compared (24).
The present work involves the largest number of serial biopsy samples (42) yet examined by VNTR analysis and thus has considerably greater statistical significance than previous studies involving only three to four patients. We find that as many as 15% (6/42) of our paired samples differ in 1 or more VNTR loci when they come from two distinct biopsy sites or two separate sites within the same large lesion (Table 4). VNTRs are known to be highly prone to slipped-strand mispairing during replication (10), and the finding of copy number differences in two separate lesions probably reflects the fact that following infection with an isolate of M. leprae with a particular VNTR genotype, the bacilli are transported within circulating phagocytes to different niches in the body, where they replicate their DNA and multiply in number independently. We consider that in five of these six cases, the variability corresponds to clonal instability within VNTR rather than coinfection with two different strains, for the following reasons. First, although the allelic range is quite large, the copy number difference is only a single repeat unit. Second, the time frame of 31 days between sampling is too short to generate diversity since it corresponds to only two generations in the life of M. leprae (9). In the sixth case, patient 15 (Table 4), infection with two different strains of M. leprae appears more probable because four VNTR differ simultaneously and three of these by two, two, and three units, respectively. While coinfection with different strains has been reported in pulmonary tuberculosis (15), it has not been described previously for leprosy, to our knowledge.
An interesting observation was made when comparing the allelic diversity of four of the five VNTR surveyed in this work with that of their counterparts in 68 strains of M. leprae isolated from patients in the Yunnan Province of the People's Republic of China (20). The Chinese strains were all members of SNP type 3 and displayed far greater diversity at the trinucleotide VNTR loci than those from Mali. For instance, there were 22 and 24 alleles at the 21_TTC and 9_GTA loci, respectively, compared to 8 and 9 in the Malian population. Furthermore, the median repeat length was longer in type 3 than in type 4, with 14 versus 19 copies at the 21_TTC locus and 13 versus 16 copies at the 15_AT locus, while the other two VNTR (9_GTA and 17_AT) that could be compared showed the same median repeat length in both populations. This apparent shortening of the VNTR and the resultant decrease in allelic diversity are consistent with the finding that SNP type 4 strains are descended from type 3 and might therefore be expected to have undergone further genome reduction. This hypothesis could be tested by performing large-scale analysis of VNTR loci in strains of SNP types 1 and 2, which in turn precede types 3 and 4 in the evolutionary scheme for the leprosy bacillus (12).
From the different phylogenetic analyses conducted here, it was not possible to establish any convincing relationships between the M. leprae strains, even those isolated from patients living in the same town, where one might have expected a common source of infection to occur. Other workers were also unable to establish a link between the VNTR genotype and the geographic origin of M. leprae or its hosts (19). It is possible, however, that by sampling a larger number of VNTR or additionally by developing new algorithms to interpret the findings, more-robust phylogeographic lineages may be established, as has been done for Mycobacterium tuberculosis (1, 17).
While our work was in review, Young and coworkers (23) published the findings of a VNTR study of M. leprae in which eight short tandem repeats were surveyed (of which only one was common to the set used in our study) in serial biopsy samples from the same four patients. These investigators found that none of the collective profiles, based on five VNTR, from the same patient were identical over time and observed that genotypic differences were more likely between bacilli from different tissues. This is reminiscent of our own findings, especially the variation in the VNTR profiles of leprosy bacilli isolated from different lesions on the same patient. Taken together, these 2 studies indicate that the 12 VNTR examined may not be suitable for epidemiological purposes owing to their excessive variability. Indeed, in a phylogeographic study of M. tuberculosis, Filliol et al. found that microsatellites, such as the mycobacterial interspersed repetitive units (18), another kind of VNTR, are less sturdy and informative as phylogenetic markers than SNP (6). It is ironic to note that, by contrast, the SNP in M. leprae are too stable for short-range studies of leprosy transmission, meaning that further genome mining will be required in order to find appropriate sites of biodiversity for epidemiological tool development.
Acknowledgments
We thank the staff and patients of the CNAM for their participation.
This study received financial support from the Institut Pasteur, the Association Française Raoul Follereau, and the National Institutes of Health, National Institute of Allergy and Infectious Diseases (grant RO1-AI47197 and contract NO1-AI25469).
Footnotes
Published ahead of print on 21 May 2008.
REFERENCES
- 1.Allix-Beguec, C., M. Fauville-Dufaux, and P. Supply. 2008. Three-year population-based evaluation of standardized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 461398-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anon, W. 2007. Global leprosy situation. 2007. Wkly. Epidemiol. Rec 82225-232. [PubMed] [Google Scholar]
- 3.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honoré, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, S. Duthoy, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, A. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 4091007-1011. [DOI] [PubMed] [Google Scholar]
- 4.Cole, S. T., P. Supply, and N. Honoré. 2001. Repetitive sequences in Mycobacterium leprae and their impact on genome plasticity. Lepr. Rev. 72449-461. [PubMed] [Google Scholar]
- 5.Consigny, S., A. Bentoucha, P. Bonnafous, J. Grosset, and B. Ji. 2000. Bactericidal activities of HMR 3647, moxifloxacin, and rifapentine against Mycobacterium leprae in mice. Antimicrob. Agents Chemother. 442919-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbon, M. Bobadilla del Valle, J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. Leon, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendon, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groathouse, N. A., B. Rivoire, H. Kim, H. Lee, S. N. Cho, P. J. Brennan, and V. D. Vissa. 2004. Multiple polymorphic loci for molecular typing of strains of Mycobacterium leprae. J. Clin. Microbiol. 421666-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howe, K., A. Bateman, and R. Durbin. 2002. QuickTree: building huge Neighbour-Joining trees of protein sequences. Bioinformatics 181546-1547. [DOI] [PubMed] [Google Scholar]
- 9.Levy, L. 1976. Studies of the mouse footpad technique for cultivation of Mycobacterium leprae. III. Doubling time during logarithmic multiplication. Lepr. Rev. 47103-106. [DOI] [PubMed] [Google Scholar]
- 10.Lovett, S. T. 2004. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 521243-1253. [DOI] [PubMed] [Google Scholar]
- 11.Matsuoka, M., S. Maeda, M. Kai, N. Nakata, G. T. Chae, T. P. Gillis, K. Kobayashi, S. Izumi, and Y. Kashiwabara. 2000. Mycobacterium leprae typing by genomic diversity and global distribution of genotypes. Int. J. Lepr. Other Mycobact. Dis. 68121-128. [PubMed] [Google Scholar]
- 12.Monot, M., N. Honore, T. Garnier, R. Araoz, J. Y. Coppee, C. Lacroix, S. Sow, J. S. Spencer, R. W. Truman, D. L. Williams, R. Gelber, M. Virmond, B. Flageul, S. N. Cho, B. Ji, A. Paniz-Mondolfi, J. Convit, S. Young, P. E. Fine, V. Rasolofo, P. J. Brennan, and S. T. Cole. 2005. On the origin of leprosy. Science 3081040-1042. [DOI] [PubMed] [Google Scholar]
- 13.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16944-945. [DOI] [PubMed] [Google Scholar]
- 14.Saitou, N., and M. Nei. 1987. The neighbour-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 15.Shamputa, I. C., L. Jugheli, N. Sadradze, E. Willery, F. Portaels, P. Supply, and L. Rigouts. 2006. Mixed infection and clonal representativeness of a single sputum sample in tuberculosis patients from a penitentiary hospital in Georgia. Respir. Res. 799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin, Y. C., H. Lee, H. Lee, G. P. Walsh, J. D. Kim, and S. N. Cho. 2000. Variable numbers of TTC repeats in Mycobacterium leprae DNA from leprosy patients and use in strain differentiation. J. Clin. Microbiol. 384535-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Supply, P., C. Allix, S. Lesjean, M. Cardoso-Oelemann, S. Rusch-Gerdes, E. Willery, E. Savine, P. de Haas, H. van Deutekom, S. Roring, P. Bifani, N. Kurepina, B. Kreiswirth, C. Sola, N. Rastogi, V. Vatin, M. C. Gutierrez, M. Fauville, S. Niemann, R. Skuce, K. Kremer, C. Locht, and D. van Soolingen. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 444498-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36762-771. [DOI] [PubMed] [Google Scholar]
- 19.Truman, R., A. B. Fontes, A. B. De Miranda, P. Suffys, and T. Gillis. 2004. Genotypic variation and stability of four variable-number tandem repeats and their suitability for discriminating strains of Mycobacterium leprae. J. Clin. Microbiol. 422558-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weng, X., Z. Wang, J. Liu, M. Kimura, W. C. T. Black, P. J. Brennan, H. Li, and V. D. Vissa. 2007. Identification and distribution of Mycobacterium leprae genotypes in a region of high leprosy prevalence in China: a 3-year molecular epidemiological study. J. Clin. Microbiol. 451728-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woods, S. A., and S. T. Cole. 1990. A family of dispersed repeats in Mycobacterium leprae. Mol. Microbiol. 41745-1751. [DOI] [PubMed] [Google Scholar]
- 22.Woods, S. A., and S. T. Cole. 1989. A rapid method for the detection of potentially viable Mycobacterium leprae in human biopsies: a novel application of PCR. FEMS Microbiol. Lett. 53305-309. [DOI] [PubMed] [Google Scholar]
- 23.Young, S. K., J. M. Ponnighaus, S. Jain, S. Lucas, S. Suneetha, D. N. Lockwood, D. B. Young, and P. E. Fine. 2008. Use of short tandem repeat sequences to study Mycobacterium leprae in leprosy patients in Malawi and India. PLoS Negl. Trop. Dis. 2e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young, S. K., G. M. Taylor, S. Jain, L. M. Suneetha, S. Suneetha, D. N. Lockwood, and D. B. Young. 2004. Microsatellite mapping of Mycobacterium leprae populations in infected humans. J. Clin. Microbiol. 424931-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, L., T. Budiawan, and M. Matsuoka. 2005. Diversity of potential short tandem repeats in Mycobacterium leprae and application for molecular typing. J. Clin. Microbiol. 435221-5229. [DOI] [PMC free article] [PubMed] [Google Scholar]