Abstract
We have developed a novel multiplex reverse transcription-PCR ligase detection reaction (RT-PCR/LDR) assay for the detection of West Nile virus (WNV) in both clinical and mosquito pool samples. The method relies on the amplification of three different genomic regions, one in the coding sequence of nonstructural protein NS2a and two in nonstructural protein NS5, to minimize the risk of detection failure due to genetic variation. The sensitivity of the PCR is complemented by the high specificity of the LDR step, and the detection of the LDR products can be achieved with capillary electrophoresis (CE) or a universal DNA microarray. We evaluated the limit of detection by both one-step and two-step multiplex RT-PCR/LDR/CE approaches, which reached, respectively, 0.005 and 0.017 PFU. The assay demonstrated 99% sensitivity when mosquito pool samples were tested and 100% sensitivity with clinical samples when the one-step approach was used. The broad strain coverage was confirmed by testing 34 WNV isolates belonging to lineages 1 and 2, and the high specificity of the assay was determined by testing other flaviviruses, as well as negative mosquito pool and clinical samples. In summary, the multiplex RT-PCR/LDR assay could represent a valuable complement to WNV serological diagnosis, especially in early symptomatic patients. In addition, the multiplexing capacity of the technique, which can be coupled to universal DNA microarray detection, makes it an amenable tool to develop a more comprehensive assay for viral pathogens.
West Nile virus (WNV) was first isolated in 1937 from a patient in Uganda (32) and has since become the most widespread member of the Japanese encephalitis virus (JEV) complex (2). It made its first appearance in the western hemisphere in 1999, when 62 cases of WNV encephalitis and seven fatalities were reported in New York City (10, 22, 26). A surveillance program of mosquito pools was initiated by the New York City Department of Health and Mental Hygiene in 2000 (36, 37), and blood donor screening was introduced in the United States in June 2003 after the report of WNV transmission to 23 individuals from 16 viremic donors in the preceding year (8, 33). Since its introduction into the United States, the virus has rapidly spread and it is now endemic in 41 states and the District of Columbia (11).
WNV strains belong primarily to two different lineages, which exhibit considerable genomic diversity, thus posing challenges to their identification and detection in both clinical and environmental samples. Lineage 1 comprises strains from several continents, and it can be subdivided into at least three clades, clade 1a, which includes strains from Europe, Africa, the United States and Israel; clade 1b, which is specific for Kunjin isolates from Australia; and clade 1c, which includes two strains from India. Lineage 2 comprises WNV strains from sub-Saharan regions of Africa and from Madagascar (6, 20). The genetic distance between lineage 2 isolates is particularly great (75.7 to 76.8%) compared to that observed in lineage 1 strains (95.2 to 99.9%) (20).
In addition to this inherent diversity, the spread of the various lineages of WNV and the isolation of new viral strains are constantly being recognized. A lineage 2 strain was recently reported in central Europe (2), and two new isolates from central Europe and southern Russia have recently been characterized which, based on their genetic distances, can be considered either separate lineages of WNV (lineages 3 and 4) or new members of the JEV group (1). In addition, phylogenetic analyses of North American isolates have shown an accumulation of mutations in the genome of prototype New York strain WN-NY99 leading to new genetic variants (5, 13, 20).
Screening of both mosquito pools and the blood supply is performed primarily by reverse transcription (RT)-PCR-based nucleic acid amplification testing (NAT) methods (31, 33). Despite the genomic diversity, a recent proficiency study revealed that less than 40% of the participating laboratories could detect lineage 2 strains (28). Molecular detection methods which are sensitive yet capable of detecting both lineages, as well as the numerous and emerging strains of WNV, are therefore needed (34).
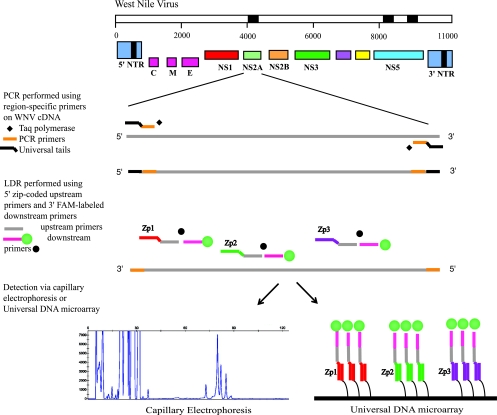
In this report, we describe the development of a new, sensitive assay for the detection of both lineages of WNV, based on multiplex RT-PCR and the ligase detection reaction (LDR) (Fig. 1). LDR was originally developed for discriminating single-base mutations or polymorphisms (3, 4). The technique is amenable to multiplexing and has been used to detect mutations and single nucleotide polymorphisms in cancer genes (14-16, 18, 19) and recently to detect and identify bacteria in clinical blood samples (29).
FIG. 1.
Schematic of the multiplex RT-PCR/LDR assay for WNV. Multiple PCR primer pairs are designed to amplify three distinct regions of the WNV cDNA (in black). Each PCR primer contains between one and three degenerate positions to accommodate minor sequence variation at the primer binding sites. For simplicity, only one PCR amplicon is shown. The presence of each PCR amplicon is detected by LDR primer pairs specific for three regions within each amplicon. The 5′ upstream LDR primers bear zip code complements, while the 3′ LDR downstream primers bear a 6-carboxyfluorescein (FAM) fluorescent label. Ligation of an LDR primer pair results in fluorescently labeled products of different lengths that can be detected either by CE or by hybridization to a universal DNA microarray. NTR, nontranslated region.
We describe the validation of the technique for WNV detection and identification with mosquito pool samples, clinical isolates, and national, as well as international, strains. The high sensitivity and broad strain coverage of the multiplex RT-PCR/LDR assay could render it a valuable complement to WNV serological diagnosis, especially in early symptomatic patients. In addition, the multiplexing capacity of the technique makes it amenable to the development of a more comprehensive assay for viral pathogens.
MATERIALS AND METHODS
Viruses.
The WNV strains used in this study (Table 1) from Uganda, France, Israel, and New York (NY99) belonged to the European Network for Diagnostics of Imported Viral Diseases and were provided by M. Niedrig at the Robert Koch Institute, Berlin, Germany (28). All of the other strains were obtained from the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch in Galveston (7).
TABLE 1.
WNV strains used in this study
| Designation | Strain | Origin/yr | Lineage-clade(s) (reference) |
|---|---|---|---|
| CAR67 | ArB-310/67 | Central African Republic/1967 | 1-b |
| NIG65 | IbAn7019 | Nigeria/1965 | 1-b |
| SEN79 | ArD-27875 | Senegal/1979 | 1-b |
| USA99a | 31A | United States/1999 | 1-b |
| IND68 | 68856 | India/1968 | 1-a |
| EGY51 | Egypt101 | Egypt/1951 | 1-a |
| ETH76 | EthAn4766 | Ethiopia/1976 | 1-a |
| AUS60 | MRM16(Kunjin) | Australia/1960 | KUN 1-b |
| AUS91 | K6453(Kunjin) | Australia/1991 | KUN 1-b |
| IND57 | IG-15578 | India/1957 | IND 1-c; 5 (9)a |
| IND80 | 804994 | Bangalore, India/1980 | IND 1-c; 5 (9)a |
| SEN920 | ArD-76104 | Senegal/1990 | 2 |
| CAR82 | ArB3573/82 | Central African Republic/1982 | 2 |
| SA58 | SAH-442 | South Africa/1958 | 2 |
| SA89 | SPU116-89 | South Africa/1989 | 2 |
| MAD-88 | ArMg-979 | Madagascar/1988 | 2 |
| MAD78 | DakAnMg78 | Madagascar/1978 | 2 |
| CYP68 | Q3574-5 | Cyprus/1968 | 2 |
| RabV | 97-103 | Czech Republic/1997 | 3 (1)b |
| WNV-Ug37 | B 956 (WNFCG) | Uganda/1937 | 2 |
| New York99 | NY99 | United States/1999 | 1 |
| PaAn001 | M12294 Fr00 | France/2000 | 1 |
| WNV 0043 | WNV 0043 | Israel/2000 | 1 |
| MXH 442 | TVP-9497 | Sonora, Mexico/2004 | 1 |
| IA 377-03 | TVP 9115 | Tabasco, Mexico/2003 | 1 |
| TX 2676 | TVP 10267 | Texas/2006 | 1 |
| AR 2771 | TVP 8833 | Quebec, Canada/2002 | 1 |
| 15476-04 | TVP 9241 | Nebraska/2003 | 1 |
| IL-10861 | TVP 9536 | Illinois/2004 | 1 |
| CT 8495-04 | TVP 10030 | Connecticut/2004 | 1 |
| TX 5784 | TVP10122 | Texas/2006 | 1 |
| I01-2032 | TVP 8852 | Florida/2001 | 1 |
| LA 02-01 | TVP 9177 | California/2004 | 1 |
| GRLA 1260 | TVP 9744 | California/2003 | 1 |
Classified also as a new distinct fifth lineage (clade 5; see Discussion).
Classified either as a new third lineage or as a novel flavivirus within the JEV group.
For sensitivity testing, a WNV load panel containing plasma samples spiked with defined dilutions of NY99 virus was kindly provided by R. Lanciotti from the Centers for Disease Control and Prevention (CDC) Division of Vector-Borne Infectious Diseases in Fort Collins, CO (21). The panel titers ranged from 180,000 to 0.15 PFU/ml (quantified by standard plaque assay). Other flaviviruses used to determine the specificity of the assay were obtained from various sources. The St. Louis encephalitis, Murray Valley fever, Powassan encephalitis, and yellow fever viruses were obtained from R. Lanciotti (CDC, Fort Collins); JEV and tick-borne encephalitis virus were obtained from M. Niedrig (Robert Koch Institute, Berlin, Germany); and dengue virus serotypes I to IV were obtained from J. Muñoz (CDC Dengue Branch in San Juan, Puerto Rico) (Table 2).
TABLE 2.
Other flaviviruses used in this study
| Flavivirus | Strain | Origin/yr | Titer |
|---|---|---|---|
| Dengue virus serotypes I, II, III, IV | NDa | Puerto Rico/2004-2006 | ND |
| St. Louis encephalitis virus | MSI-7 | Mississippi/1977 | 3 × 108 PFU/ml |
| Murray Valley fever virus | OR2 | Victoria, Australia/1951 | ND |
| Powassan encephalitis virus | M11665 | Ontario, Canada/1965 | ND |
| Yellow fever virus | 17D | Ghana/1927 | 5 × 105 PFU/ml |
| JEV | SA14-14-2 | China/1954 | 2 × 107 copies/ml |
| Tick-borne encephalitis virus | K23 | Reference 27 | 1 × 107 copies/ml |
ND, not determined.
Mosquito pools and clinical samples.
Ninety-eight positive and 20 negative mosquito pool samples were obtained from the New York City Department of Health and Mental Hygiene (NYC DOHMH), which collects and tests trapped mosquitoes from different areas in New York City as part of its WNV surveillance program (36, 37).
Plasma samples from 50 NAT-positive blood donors and 92 NAT-negative donors were tested. The positive plasma samples were provided by the Gulf Coast Regional Blood Center in Houston, TX; the negative samples were obtained from the CDC Dengue branch in Puerto Rico. Twenty additional plasma samples which tested positive for dengue virus and negative for WNV were also provided by the CDC in Puerto Rico. Additional samples for initial primer validation were obtained from S. Stramer at the American Red Cross National Testing and Reference Laboratories, Gaithersburg, MD.
RNA extraction.
Samples from mosquito pool homogenates were clarified by centrifugation at 2,000 × g for 3 min. Viral RNA was extracted from the mosquito pool sample supernatants and clinical samples, as well as from viral seeds, with the QIAamp viral RNA mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. RNA was eluted in 60 μl and stored at −70°C until used. One negative and one positive extraction control were processed along with each group of 10 samples subjected to RNA extraction.
Selection of target regions for multiplex PCR/LDR.
Thirty-nine WNV complete genomic sequences available from the GenBank database (accessed in January 2005) were aligned by using the ClustalW algorithm to select optimal primer binding regions. These regions were characterized by a higher degree of conservation among different WNV strains so as to achieve maximum strain coverage. Primer sets with partially overlapping sequences were designed to simultaneously amplify three different regions (Integrated DNA Technology, Coralville, IA). One region was in the coding sequence of nonstructural protein NS2a, and two regions were in nonstructural protein NS5 (respectively, 6, 8, and 8 PCR primers were used, for a total of 22 primers). Each of the three amplicons was ∼500 bp. Each primer sequence contained no more that two degenerate positions and had a melting temperature of around 80°C (Table 3).
TABLE 3.
Primers used for PCR and RT-PCR
| Target region | Forward primer sequencea (5′-3′) | Tm (°C)b | Reverse primer sequencea (5′-3′) | Tm (°C) | Amplicon length (bp) |
|---|---|---|---|---|---|
| 1 (NS2a) | WNV_PCR_F5, GCCAACTACCGCAACACAGYTGGGCCTTMTGGTCGTGTTC | 81-83 | WNV_PCR_R7A, CCAACTACCGCAACCACTTTGATGAGGCTTCCAACTCCAACCAT | 81 | 540-580 |
| WNV_PCR_F6, GCCAACTACCGCAACACTGGCCACCCAGGAGGTCCTTC | 83 | WNV_PCR_R7B, CCAACTACCGCAACCACCCTGATCAARCTGCCTATTCCRACCAT | 81-83 | ||
| WNV_PCR_R8A, CCAACTACCGCAACCAATGAGGCAAGCTCCTTTCTTTTTTGC | 79 | ||||
| WNV_PCR_R8B, CCAACTACCGCAACCAARCAGACTKGCTCCTTTCTTYTTTGC | 78-81 | ||||
| 2 (NS5) | WNV_PCR_F9A, GCCAACTACCGCAACACGATGTGGAAGAGGCGGYTGGTGTTA | 82-83 | WNV_PCR_R11A, CCAACTACCGCAACCACCCAGAAGCACCTGGCTWGTCAT | 81 | 460-510 |
| WNV_PCR_F9B, GCCAACTACCGCAACACGGTGTGGTAGAGGCGGCTGGTGCTA | 85 | WNV_PCR_R11B, CCAACTACCGCAACCACCTARGAGYACCTGGCTGGTCAT | 80-82 | ||
| WNV_PCR_F10A, GCCAACTACCGCAACACAGAAGAGGGTRCAGGAAGTGAAAGGGTACAC | 83-84 | WNV_PCR_R12A, CCAACTACCGCAACCAGGTCCCTTCCAKGTYTTCTTYTCCAT | 79-82 | ||
| WNV_PCR_F10B, GCCAACTACCGCAACACAAAAAAGAGTYCAAGAAGTCAGAGGGTACAC | 81 | WNV_PCR_R12B, CCAACTACCGCAACCAGGTCCCTTCCAGGTCCTYTTYTCCAT | 81-83 | ||
| 3 (NS5) | WNV_PCR_F13A, GCCAACTACCGCAACACGCGAACCACCCCTACAGGACCTGGAAC | 85 | WNV_PCR_R15A, CCAACTACCGCAACCAGGYTTTTTCTCYCTCTTTCCCATC | 78-80 | 420-560 |
| WNV_PCR_F13B, GCCAACTACCGCAACACGAGAACCACCCATATAGAACCTGGAAC | 82 | WNV_PCR_R15B, CCAACTACCGCAACCAGGTTTCTTYTCTCTCTTSCCCATC | 79-80 | ||
| WNV_PCR_F14A, GCCAACTACCGCAACACCAATGTGACCACGATGGCCATGAC | 82 | WNV_PCR_R16A, CCAACTACCGCAACCACAGAAAGCGAGCTCCKAGCCAC | 81-82 | ||
| WNV_PCR_F14B, GCCAACTACCGCAACACGAATGTYACCACMATGGCCATGAC | 80-82 | WNV_PCR_R16B, CCAACTACCGCAACCACAGGAAGCGRGCYCCCAGCCAC | 83-85 |
The universal tail sequences appended to the 5′ ends of all primers are underlined.
Tm, melting temperature.
A major barrier to high-sensitivity multiplexed PCR amplification in other systems has been the exponential increase in potential false amplicons and primer dimers that results from using too many PCR primers in the same reaction mixture. We circumvent this potential pitfall by using PCR primers containing 5′ universal tail sequences on the forward (5′-GCCAACTACCGCAACAC-3′) and reverse (5′-CCAACTACCGCAACCA-3′) primers (Table 3). Primer dimers and short aberrant amplicons do not amplify efficiently because such products form panhandle structures. The correct PCR product is just the right size for efficient amplification (300 to 600 bp), while false longer amplicons do not amplify as well. We have successfully applied this strategy where standard multiplexed PCR has failed (12, 15, 17).
LDR primers were chosen in three different conserved regions within each of the three PCR amplicons and, just as for the PCR primers, designed with the intent of achieving the highest strain coverage. Each LDR primer pair was composed of an upstream probe bearing a 20-mer zip code complement sequence at its 5′ end and a downstream probe bearing a 6-carboxyfluorescein fluorophore at its 3′ end. The zip code complements are unique 20-mer oligonucleotide sequences complementary to the zip code addresses spotted on the universal DNA microarray (18).
During LDR, both upstream and downstream probes hybridize to the template sequence and ligation occurs only when there is perfect complementarity at the ligation junction (Fig. 1). Forty-nine LDR primers targeting a total of nine regions (three LDR primer pairs per PCR product) were designed, with the aim of detecting as many strains as possible and potential new variants (Table 4) .
TABLE 4.
Primers used for LDR
| Target region | Downstream primer, sequence (5′-3′) | Tm (°C)b | Allele-specific (upstream) primer,a sequence (5′-3′) | Tm (°C) | LDR product length (bp) |
|---|---|---|---|---|---|
| 1 (NS2a) | WNV831aCOM, (Phos)CTATCATGCTTGCACTCCTAGTCCTAGTGTTTGG(6-FAM) | 74 | WNV831GZp52, (NH2)TCCGTTCCGCCAGAGGGTGAGTGGACRGCCAAGATCAGCATKCCAG | 85-87 | 80 |
| WNV831bCOM, (Phos)CTATACTGATTGCTCTGCTAGTYCTGGTGTTTGG(6-FAM) | 73-74 | ||||
| WNV831cCOM, (Phos)CCATACTGATTGCCCTGCTAGTTCTAGTGTTTGG(6-FAM) | 74 | ||||
| WNV939aCOM, (Phos)GAGACGTCG,TGCACTTGGCACTTATGG(6-FAM) | 71 | WNV939aGZp53, (NH2)TCGCCGTCCGCTGTCTTTGGCGTTTGCTGAAGCRAACTCAGGAG | 84 | 72 | |
| WNV939bCOM, (Phos)GAGACGTGGTACACTTGGCGCTCATGG(6-FAM) | 73 | WNV939bGZp53, (NH2)TCGCCGTCCGCTGTCTTTGGCTTTCGCAGAATCYAATTCRGGAG | 82-84 | ||
| WNV939cCOM, (Phos)GAGATGTGGTGCATCTGGCGCTCATGG(6-FAM) | 73 | WNV939cGZp53, (NH2)TCGCCGTCCGCTGTCTTTGGCCTTTGCAGAATCMAACTCAGGAG | 83-84 | ||
| WNV1021aCOM, (Phos)GACCAACCAAGAGAGTATTTTGCTCATGCTTG(6-FAM) | 71 | WNV1021aGZp54, (NH2)GACCAGGCCTCGACCCACCCGGTGGCTTCCTTTTTGAAGGCAAGGTG | 87 | 79 | |
| WNV1021bCOM, (Phos)GACCAACCAGGAGAACATYTTGTTGATGTTGG(6-FAM) | 71-73 | WNV1021bGZp54, (NH2)GACCAGGCCTCGACCCACCCGGTGGCATCGTTTCTCAAAGCGAGATG | 87 | ||
| WNV1021cCOM, (Phos)GACCAACCAAGAAAACATTCTGCTGATGTTGG(6-FAM) | 71 | WNV1021cGZp54, (NH2)GACCAGGCCTCGACCCACCCGGTAGCATCATTTCTCAAGGCGAGATG | 86 | ||
| 2 (NS5) | WNV5341aCOM, (Phos)GTTGGAATATTGTTACCATGAAGAGTGGAGTCGACGTC(6-FAM) | 76 | WNV5341aGZp55, (NH2)GCCACGCTGCCAGGACTCCGATGAAGAACCACAACTGGTGCAGAGCTATG | 86 | 88 |
| WNV5341bCOM, (Phos)GATGGAACATTGTCACCATGAAGAGYGGRGTGGATGTG(6-FAM) | 77-79 | WNV5341bGZp55, (NH2)GCCACGCTGCCAGGACTCCGATGAAGAGCCCCAACTRGTGCAAAGTTATG | 85-86 | ||
| WNV5341cGZp55, (NH2)GCCACGCTGCCAGGACTCCGATGAAGAGCCCCAGCTAGTGCAGAGCTATG | 88 | ||||
| WNV5427aCOM, (Phos)AGTCATCGTCAAGTGCCGAGGTAGAAGAACACCGC(6-FAM) | 78 | WNV5427aGZp56, (NH2)TCCGGTCTTGGTCGCTTCGCGCGAGCGACACACTGCTCTGTGACATTGGAG | 88 | 86 | |
| WNV5427bCOM, (Phos)AGTCYTCRTCAAGTGCTGAGGTTGAAGAGCATAGG(6-FAM) | 74-76 | WNV5427bGZp56, (NH2)TCCGGTCTTGGTCGCTTCGCTGYTGCGAYACYCTCCTTTGTGACATCGGAG | 85-88 | ||
| WNV5427cCOM, (Phos)AGTCCTCATCAAGTGCTGAAGTTGAAGAACATAGA(6-FAM) | 72 | WNV5427cGZp56, (NH2)TCCGGTCTTGGTCGCTTCGCTGTTGTGACACTCTCCTTTGTGATATCGGAG | 85 | ||
| WNV5548aCOM, (Phos)CCAAAGTGATTGAGAAGATGGAAACACTCC(6-FAM) | 69 | WNV5548aCZp57, (NH2)GTCTTCGCGGTGGGTGCCTGCTGCATCAAAGTGCTATGCCCTTACATGC | 86 | 79 | |
| WNV5548bCOM, (Phos)CRAAAGTCATAGAGAAGATGGAGCTRCTCC(6-FAM) | 69-72 | WNV5548bCZp57, (NH2)GTCTTCGCGGTGGGTGCCTGTTGYGTGAAGGTRCTSTGCCCCTACATGC | 86-88 | ||
| WNV5548cCOM, (Phos)CAAAGGTCATAGAAAAGATGGAGCTGCTCC(6-FAM) | 71 | ||||
| 3 (NS5) | WNV6094aCOM, (Phos)GAGTCAAATACGTCCTCAATGAGACCACGAACTGGCTG(6-FAM) | 78 | WNV6094aGZp58, (NH2)GCGTTTGGTTGGCTGCGGACCACAAAGGCTCCAGAGCCTCCAGAAG | 86 | |
| WNV6094bCOM, (Phos)GAGTGAAGTATGTGCTCAAYGAAACCACCAAYTGGTTG(6-FAM) | 75-77 | WNV6094bGZp58, (NH2)GCGTTTGGTTGGCTGCGGACCACGAAAGCTCCBGAACCGCCAGAAG | 86-87 | 84 | |
| WNV6094cCOM, (Phos)GAGTGAAGTACGTGCTCAAYGAGACCACCAAYTGGTTG(6-FAM) | 77-79 | ||||
| WNV6094dCOM, (Phos)GAGTTAAGTATGTGCTCAATGAAACCACCAATTGGTTG(6-FAM) | 73 | ||||
| WNV6168aCOM, (Phos)TGYTCCCGGGAGGAATTTATYGGAAAAGTCAACAG(6-FAM) | 73-76 | WNV6168aGZp59, (NH2)GGACCTCGGCCACGCTCTGCTTAGCCCGCGAYAARAAACCCAGGATG | 86-88 | 82 | |
| WNV6168bCOM, (Phos)TGCTCGCGAGAGGAATTTATAAGGAAGGTCAATAG(6-FAM) | 73 | WNV6168bGZp59, (NH2)GGACCTCGGCCACGCTCTGCCTGGCACGAGAAAAGCGTCCCAGAATG | 88 | ||
| WNV6168cCOM, (Phos)TGCTCTCGAGAGGARTTCATAAGAAAGGTCAACAG(6-FAM) | 73-74 | WNV6168cGZp59, (NH2)GGACCTCGGCCACGCTCTGCCTGGCCAGAGAAAAACGTCCCAGAATG | 87 | ||
| WNV6244aCOM, (Phos)GGAAGAACGCCCGGGAAGCTGTAGAGGATCC(6-FAM) | 77 | WNV6244aTZp60, (NH2)GCCACTCGTCCGTCCGCCACAGGAGCGATGTTTGAAGAACAGAACCAAT | 85 | 80 | |
| WNV6244bCOM, (Phos)GGAGGAGTGCTAGAGAAGCGGTTGAAGATCC(6-FAM) | 75 | WNV6244bTZp60, (NH2)GCCACTCGTCCGTCCGCCACGGGTGCCATGTTTGAAGARCAGAAYCAAT | 85-86 | ||
| WNV6244cCOM, (Phos)GGAGGAGCGCCAGAGAAGCAGTTGAAGATCC(6-FAM) | 76 |
The zip code complements on the upstream primers are in bold.
Tm, melting temperature.
LDR products ranged from 72 to 88 bp in length. The oligonucleotide probes (Integrated DNA Technology, Coralville, IA) were designed to have similar thermodynamic features and to avoid hairpin and self-dimer formation.
Two-step multiplex RT-PCR.
RT reactions were performed in a 60-μl volume with the Superscript First Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) and random hexamers. Briefly, 20 μl of RNA extracted from mosquito pools or from clinical samples was mixed with 6 μl of 50 ng/ml random hexamers, 3 μl of a 10 mM deoxynucleoside triphosphate (dNTP) mixture, and 1 μl of water and denatured at 65°C for 5 min. After cooling on ice, 1× RT buffer (20 mM Tris-HCl, 50 mM KCl), 5 mM MgCl2, 0.01 M dithiothreitol, and 2 U of RNaseOUT were added for a 2-min incubation at 25°C. Six units of Superscript II RT was added, and the mixture was incubated first at 25°C for 10 min and then at 42°C for 50 min. The reaction was terminated by heating at 70°C for 15 min. Degradation of residual RNA or cleavage of RNA-DNA hybrids was achieved by incubation with 6 U of RNase H for 20 min at 37°C.
Five microliters of newly synthesized cDNA was subjected to multiplex PCR amplification in a final volume of 25 μl which contained 1× GeneAmp PCR Gold buffer, 1.5 mM MgCl2, 200 μM each dNTP, 5 pmol of each PCR primer, and 1 U of Taq polymerase (AmpliTaq Gold; Applied Biosystems, Foster City, CA). After 10 min of incubation at 95°C for Hot Start Taq activation, a total of 40 cycles were performed, each consisting of a denaturation step at 94°C for 30 s, an annealing step at 60°C for 1 min, and a extension step at 72°C for 1 min, with a final extension step at 72°C for 10 min. A nontemplate negative control and a WNV cDNA positive control were included in each round. All PCR thermal cycling was performed in a Perkin-Elmer GeneAmp PCR System 9700 Thermal Cycler (Applied Biosystems, Foster City, CA). The amplification products were visualized by electrophoresis in a 2% agarose gel.
One-step multiplex RT-PCR.
Alternatively, the RNA extracted from clinical samples was subjected to a one-step multiplex RT-PCR (OneStep RT-PCR kit; Qiagen, Valencia, CA). Briefly, 15 μl of RNA was added to a 50-μl final volume containing 1× OneStep RT-PCR buffer, 0.4 mM dNTPs, 0.6 μM each PCR primer, and 2 μl of OneStep RT-PCR Enzyme Mix. RT (at 50°C for 30 min) was followed by a denaturation step at 95°C for 15 min and 45 cycles of amplification (94°C for 30 s, 60°C for 1 min, and 72°C for 1 min) with a final extension step at 72°C for 10 min.
Multiplex LDR.
LDRs were carried out in a final volume of 20 μl containing 5 μl of amplified DNA, 1× LDR buffer (20 mM Tris [pH 7.6], 10 mM MgCl2, 100 mM KCl), 1 mM NAD, 1 mM dithiothreitol, 250 fmol of each LDR primer, and 0.01 μM AK16D DNA ligase (4, 25).
LDR mixtures were thermally cycled for 20 cycles of 30 s at 94°C and 4 min at 64°C. Prior to LDRs, a mixture containing 7.5 pmol of each LDR primer was phosphorylated in a 30-μl kinase reaction mixture containing 1× T4 ligase buffer (50 mM Tris-HCl, 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, 25 μg/ml bovine serum albumin) and 10 U of T4 kinase (New England BioLabs, Ipswich, MA). The mixture was incubated at 37°C for 60 min, followed by 10 min of incubation at 65°C and storage at 4°C.
Capillary electrophoresis (CE).
After the LDR, the mixture was diluted 1:10 in water and 2 μl was added to 8 μl of a CE master mixture containing Hi-Di Formamide (Applied Biosystems, Foster City, CA) and 0.3 μl of GeneScan 500 LIZ size standard (Applied Biosystem, Foster City, CA). The CE mixture was denatured at 94°C for 2 min and chilled on ice. The LDR products were analyzed on a 3730 DNA analyzer (Applied Biosystems, Foster City, CA). A sample was considered WNV positive when a minimum of two LDR products in any of the three PCR amplicons was detected by CE.
Universal DNA microarray spotting and hybridization conditions.
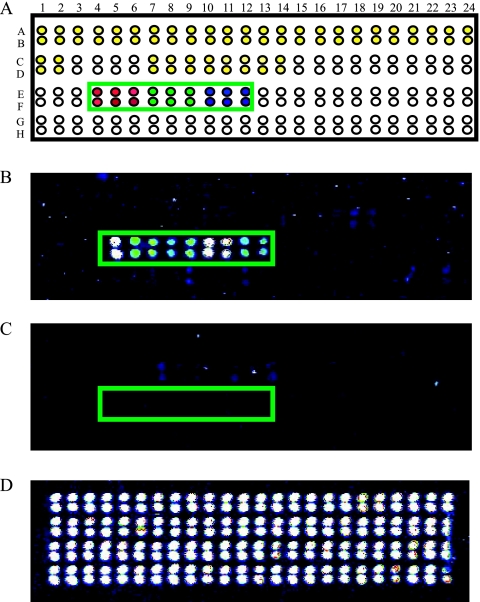
Universal microarrays were prepared by spotting unique 20-mer zip code oligonucleotides (12, 14, 15, 18) on activated Codelink slides (GE Healthcare, Piscataway, NJ) with a QArrayMini robotic array printer (Genetix, Boston, MA) according to the manufacturer's instructions. The zip code addresses were plated into a 384-well microplate in 50 mM sodium phosphate (pH 8.5) at a final concentration of 25 μM. A 1 μM concentration of a fiducial oligonucleotide was added to the printing mixture in each well and cospotted with each zip code address. A carboxy-X-rhodamine-labeled fiducial complement was included in the hybridization mixture to determine the position and quality of each spot. Robotic printing was carried out at 10°C and 50 to 60% humidity. Printed slides were incubated in a saturated NaCl chamber overnight and then treated with a blocking solution (0.1 M Tris, 50 mM ethanolamine [pH 9.0]) to block residual reactive carboxyl groups. The slides were washed with 4× SSC (20× SSC is 3 M sodium chloride and 0.3 M sodium citrate, pH 7.0)-0.1% sodium dodecyl sulfate (SDS) and spin dried. Each printing layout contained a total of 96 zip code addresses spotted in duplicate (see Fig. 4D). Nine zip code addresses were designed to hybridize the zip code complements appended to the WNV-specific upstream LDR primers. The other zip codes on the array were complementary to zip code complements on LDR primers specific for other blood-borne viral pathogens (dengue virus and other hemorrhagic fever viruses). LDR products from a subset of nine mosquito pool samples and from positive and negative controls were denatured at 94°C for 3 min and chilled on ice prior to hybridization to the arrays. The hybridization solution consisted of the entire volume of the LDR products, 5× SSC, 0.1% SDS, 0.1 mg/ml salmon sperm DNA (Fisher Scientific), and a 5 nM concentration of the fiducial complement in a total volume of 30 μl. The hybridization solution was applied to the microarray slide with a multichamber ProPlate Slide Module (Grace Bio-Labs, Bend, OR). The hybridizations were carried out in the dark on a rocking platform within a hybridization oven (Lab-line; VWR West Chester, PA) at 60°C for 2 h. The slides were rinsed with 5× SSC and washed with 1× SSC-0.1% SDS at 60°C for 15 min. Two more washes followed, first with 0.2× SSC at 22°C for 1 min and then with 0.1× SSC at 22°C for 1 min. The slides were spun dry and scanned on a ProScanArray microarray scanner (Perkin-Elmer, Wellesley, MA).
FIG. 4.
Representative WNV detection with a universal DNA microarray. (A) Schematic of the microarray. The green box includes the area where the nine WNV-specific zip codes were spotted in duplicate (E4 to E12 and F4 to F12), with red, green, and blue spots indicating the three sets of zip codes corresponding to LDR products for amplicon regions 1, 2, and 3, respectively. Other zip codes on the array are designated for detecting dengue virus (yellow spots) and other hemorrhagic fever viruses (white spots). (B) LDR products from a representative WNV-positive mosquito pool sample revealing hybridization of LDR products from each region to the correct zip codes. The colors represent the fluorescence intensity at each spot, white being the strongest and blue being the weakest. (C) LDR products from a WNV negative control mosquito pool sample. (D) Hybridization of a carboxy-X-rhodamine-labeled fiducial complement internal control to verify uniform spotting of zip codes.
LOD.
The limit of detection (LOD) of the assay was measured with a WNV load panel containing plasma samples spiked with defined dilutions of NY99 virus. The panel titers ranged from 180,000 to 0.15 PFU/ml (quantified by standard plaque assay). Dilutions ranging from 1,800 PFU/ml to 0.07 PFU/ml were tested. For each dilution, RNA was extracted from a 140-μl aliquot with the QIAamp viral RNA mini kit (Qiagen, Valencia, CA) as described above. The RNA was subjected to either the one-step or the two-step multiplex RT-PCR as described above, followed by multiplexed LDR and CE as described earlier. Thus, a dilution of 1,800 PFU/ml corresponds to a LOD of 63 PFU for the one-step method (a 140-μl aliquot was extracted into a final volume of 60 μl of RNA, of which 15 μl was used for the RT-PCR) and 2.8 PFU was used for the two-step method (a 140-μl aliquot was extracted into a final volume of 60 μl of RNA, of which 20 μl was used in the RT step in a total volume of 60 μl. Of this 60-μl volume of cDNA, 2 μl was used in the PCR step).
RESULTS
PCR/LDR/CE primer selection and validation.
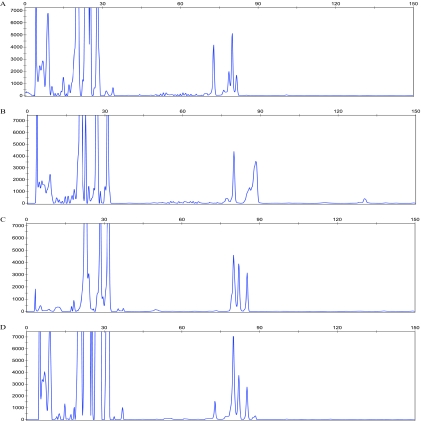
A multiplex PCR/LDR/CE assay was developed for detecting WNV based on the simultaneous screening of three WNV genomic regions (Fig. 1). The three target regions were initially tested separately by performing PCR/LDR/CE assays with primers specific for each region (Fig. 2A to C). In this way, it was possible to evaluate the primer performance for each region and to ensure signal detection from each of the expected total of nine LDR products. In the initial evaluation phase, performed with WNV cultures, as well as WNV-positive plasma samples obtained from the American Red Cross, the primers which failed to produce either the PCR amplicon or one of the LDR products were discarded and replaced with newly designed primers (data not shown).
FIG. 2.
Representative CE profiles of ligation products from the PCR/LDR. (A to C) CE profiles from a uniplex PCR/LDR for each of the three regions of WNV. (A) Peaks at 72, 79, and 80 bases represent LDR products from regions WNV939, WNV1021, and WNV831, respectively. (B) Peaks at 79, 86, and 88 bases represent LDR products from regions WNV5548, WNV5427, and WNV5341, respectively. (C) Peaks at 80, 82, and 84 bases represent LDR products from regions WNV6244, WNV6168, and WNV6094, respectively. (D) CE cumulative profile obtained after a multiplex PCR/LDR when all three regions are amplified and detected together. Note that the peaks at 79 and 80 bases merge together to give a single peak at 80 bases and that the peak at 86 bases merges with the peak at 88 bases, giving a small peak at around 86 bases. Fluorescence intensity is indicated on the y axis, and the number of bases is indicated on the x axis.
Region-specific LDR products for each region produced peaks at 72, 79, and 80 bases for region 1 (along with a minor peak at 81 bases arising from ligation of a primer with a single base difference at one of the ligation sites); 79, 86, and 88 bases for region 2; and 80, 82, and 84 bases for region 3 (Fig. 2A to C). Figure 2D shows the CE profile obtained when the LDR was multiplexed for all three WNV regions. LDR products with identical lengths but different sequences (due to the use of degenerate oligonucleotides) can migrate at separate positions on CE, resulting in broadened peaks that may overlap nearby peaks. The algorithm for the identification of a positive sample requires detecting the presence of at least two LDR products from any one amplicon or one LDR product from any two amplicons, i.e., at least two separate peaks. No specific peak need be present. The cumulative profile from multiplexed LDR was sufficient to positively identify a sample.
LOD.
To evaluate the LOD of the WNV multiplex PCR/LDR/CE assay, we tested a viral load panel containing plasma samples spiked with serial dilutions of WNV. The LOD was determined after performing RNA extraction, RT, and multiplex PCR/LDR/CE, and it was calculated both for the assay that used the two-step approach and for the one-step multiplex RT-PCR.
In the two-step approach, the RT was performed with random hexamers, followed by PCR amplification with region-specific primers. This approach allowed the detection of the sample dilutions from the viral load panel containing 11 PFU/ml. This corresponds to a LOD of 0.017 PFU. Previous studies using preparations of the NY99 virus strain grown in Vero cells have consistently shown that 1 PFU represents 500 copies (30; R. Lanciotti personal communication). Therefore, based upon these calculations, PCR/LDR has an LOD of approximately eight genome copies.
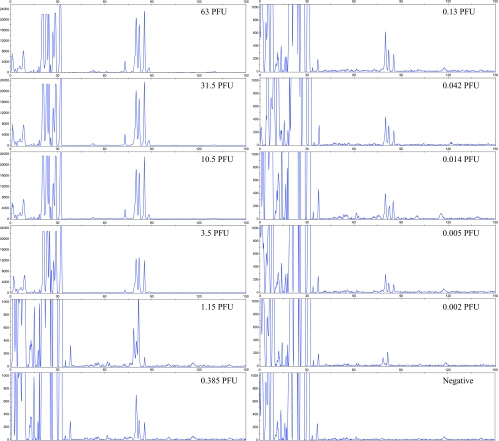
The one-step multiplex RT-PCR approach allowed the detection of the viral load panel dilutions containing 0.15 PFU/ml, about 70 times less concentrated than that detected by the two-step method. The LOD with this method corresponds to 0.005 PFU or 2.5 genome copies (Fig. 3). This compares favorably to other detection systems, including those that detect both lineages 1 and 2, such as the FDA-licensed PROCLEIX System (21, 23, 24).
FIG. 3.
Determination of the sensitivity of the one-step multiplex PCR/LDR/CE assay. CE profiles were obtained after RNA extraction and one-step multiplex RT-PCR/LDR of the WNV load panel. CE signals below a threshold of 200 fluorescence units were considered negative. Fluorescence intensity is indicated on the y axis, and the number of bases is indicated on the x axis.
Sensitivity and specificity of multiplex PCR/LDR/CE.
The sensitivity of the multiplex PCR/LDR/CE system was determined with WNV cultures and environmental and clinical samples. A sample was considered WNV positive when a minimum of two LDR products in any of the three PCR amplicons was detected. WNV cultures included 34 strains from 19 countries (Table 1) which belonged to both lineages 1 and 2, as well as the Kunjin and Rabensburg viruses. All of the strains tested positive, except for the Rabensburg virus and two Indian isolates. Both the Rabensburg virus and the two Indian isolates nevertheless produced positive PCR amplification products visible after gel electrophoresis, indicating that the LDR did not work.
To evaluate the specificity of the method, seven other flaviviruses were tested, as listed in Table 2. Although four of them (St. Louis encephalitis virus, yellow fever virus, Murray valley fever virus, and JEV) produced PCR amplification products detected by agarose gel electrophoresis, no ligation products were obtained after the LDR. Ninety-eight pooled mosquito homogenates which had previously tested positive according to the NYC DOHMH were subjected to the two-step multiplex PCR/LDR/CE assay. All but one sample produced a positive signal, for a sensitivity of about 99%. Twenty WNV-negative mosquito pools were also tested, and no false positives were found.
Fifty WNV-positive plasma samples with a representative range of concentrations (a minimum of 100 copies/ml) were obtained from the Gulf Coast Regional Blood Center in Texas. These samples were subjected to RNA extraction and were tested in parallel by both the two-step and one-step multiplex PCR/LDR/CE methods. While the one-step approach detected WNV RNA in all 50 samples with 100% sensitivity, the two-step protocol displayed 82% sensitivity (41 out of 50 positive samples detected). Ninety-two additional WNV-negative plasma samples, together with another 20 dengue virus-positive but WNV-negative samples (obtained from CDC, Puerto Rico), were included in the analysis. No false positives were detected from any of the total of 112 WNV-negative plasma samples, providing 100% specificity.
Universal DNA microarray.
A subset of nine WNV-positive mosquito pool samples was tested with the universal DNA microarray as an alternative readout system. Successful ligation of the LDR primers results in the formation of LDR products that bear a zip code complement at the 5′ end and a fluorescent label at the 3′ end. The universal DNA microarray permits the detection of the ligation products via hybridization of the zip code complements to zip codes spotted on the array. The results obtained with these samples showed that the universal array could detect a fluorescent signal from each of the nine different LDR products which correctly hybridized to their designated addresses on the array (Fig. 4). This indicates that the assay can be performed by using either CE or a universal array as the final readout.
DISCUSSION
In this report, we describe the development of a new WNV detection method based on multiplex RT-PCR followed by LDR. Our detection strategy was based on finding regions in the WNV genome that were most invariant among the different strains belonging to both lineages. We then designed PCR primers that had the required specificity to amplify WNV-specific RNA (after RT) while tolerating sequence variation without amplifying the far more abundant human RNA. Likewise, the LDR primers were designed to specifically ligate, even if the target sequence varied in up to three positions. The high sensitivity of the initial RT-PCR step, with degenerate primers, allows some tolerance to mismatches, which is complemented by the high specificity of the LDR step. LDR uses an exquisitely specific thermostable AK16D DNA ligase that permits ligation only when the sequence at the junction between the paired oligonucleotides is complementary to the template sequence. This type of assay is ideal for multiplexing, since several primer sets can ligate along a template without the interference encountered in polymerase-based assays (18, 19).
The multiplex RT-PCR/LDR/CE test was evaluated with both mosquito pools and clinical samples, with the clinical samples being subjected to both one-step and two-step RT-PCR protocols. The sensitivity obtained with the mosquito pool samples was 98%; the only sample which gave a negative result was retested at the NYC DOHMH, where it was confirmed as negative, suggesting possible sample degradation.
The WNV-positive clinical samples tested with the one-step protocol, which uses target-specific primers for RT and PCRs, were detected with a sensitivity of 100%. On the other hand, with the two-step method, where random hexamers are used for the RT step, 82% of the samples gave a positive result. The higher sensitivity of the one-step versus the two-step approach may result from the better performance of the former over the latter method, which was demonstrated during LOD testing (Fig. 3). Although the sensitivity obtained by the two-step approach with mosquito pools did not seem to be affected by this drawback, it has been reported that for low mRNA levels (like those expected to be found in clinical samples versus mosquito pools), gene-specific priming provides a more sensitive method (35).
When the test was evaluated on 34 different WNV strains, the LDR failed to detect the Rabensburg strain and the two Indian isolates belonging to clade 1c or, as new evidence shows, forming a distinct fifth lineage (9). Genomic sequences for these isolates were not available when the LDR primers were designed. Alignment of the Rabensburg and IND 804994 strain sequences, which have since been published, reveal that they are too divergent to successfully anneal with the initial primers. Since LDRs can be highly multiplexed without compromising the ligation efficiency (14, 29), primers permitting the detection of these isolates may easily be incorporated into future versions of the assay. The flexibility of the technique will permit the expansion of the assay to include emerging new WNV strains in a similar manner.
Over the past few years, the PCR/LDR approach has been used for several applications in our laboratories (12, 14-16, 19). The use of LDR primers with specific sequences appended, termed “zip code” complements, has enabled the detection of LDR products through a universal DNA microarray containing designated addressable zip codes (16, 18). A universal DNA microarray offers the advantage of being completely programmable and permits the inclusion of new genomic target sequences without redesigning the array. In addition, different pathogens can be detected simultaneously since the hybridization event is mediated by the spotted zip code and zip code complements on the LDR primers in place of the actual pathogen's genomic sequence. By uncoupling pathogen detection from pathogen identification, the same type of array can be used simultaneously for different organisms without changing the spotted probes.
Our group recently demonstrated the utility of PCR/LDR/CE in the multiplexed detection of blood-borne bacterial infectious agents (29). Due to the frequently nonspecific clinical symptoms of viral infections and the overlap of different arboviruses in the same geographic area, we envision a similar approach to the detection of blood-borne viral pathogens, both in clinical specimens and for environmental surveillance. The use of a multiplex RT-PCR/LDR for detection and a universal DNA microarray for identification represents a convenient tool given the frequent sequence variation in RNA viruses which may necessitate additions to the detection primers used. This approach can assist epidemiologists in rapidly tracking unknown and emerging strains of HFV. We have designed the same type of test for the detection and serotype determination of dengue virus in clinical samples (unpublished data), as well as other hemorrhagic fever viruses, paving the way for a comprehensive viral detection method.
Acknowledgments
We thank Pius Brzoska at Applied Biosystems for providing us with genomic sequence alignments for WNV and M. Niedrig from the Robert Koch Institute, Berlin, Germany, for supplying the viruses from the European Network for Diagnostics of Imported Viral Diseases.
This work was supported by Public Health Service grant UC1-AI062579 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 21 May 2008.
REFERENCES
- 1.Bakonyi, T., Z. Hubalek, I. Rudolf, and N. Nowotny. 2005. Novel flavivirus or new lineage of West Nile virus, central Europe. Emerg. Infect. Dis. 11225-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakonyi, T., E. Ivanics, K. Erdelyi, K. Ursu, E. Ferenczi, H. Weissenbock, and N. Nowotny. 2006. Lineage 1 and 2 strains of encephalitic West Nile virus, central Europe. Emerg. Infect. Dis. 12618-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barany, F. 1991. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc. Natl. Acad. Sci. USA 88189-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barany, F., and D. H. Gelfand. 1991. Cloning, overexpression and nucleotide sequence of a thermostable DNA ligase-encoding gene. Gene 1091-11. [DOI] [PubMed] [Google Scholar]
- 5.Beasley, D. W., C. T. Davis, H. Guzman, D. L. Vanlandingham, A. P. Travassos da Rosa, R. E. Parsons, S. Higgs, R. B. Tesh, and A. D. Barrett. 2003. Limited evolution of West Nile virus has occurred during its southwesterly spread in the United States. Virology 309190-195. [DOI] [PubMed] [Google Scholar]
- 6.Beasley, D. W., C. T. Davis, M. Whiteman, B. Granwehr, R. M. Kinney, and A. D. Barrett. 2004. Molecular determinants of virulence of West Nile virus in North America. Arch. Virol. Suppl. 1835-41. [DOI] [PubMed] [Google Scholar]
- 7.Beasley, D. W., L. Li, M. T. Suderman, and A. D. Barrett. 2001. West Nile virus strains differ in mouse neurovirulence and binding to mouse or human brain membrane receptor preparations. Ann. N. Y. Acad. Sci. 951332-335. [DOI] [PubMed] [Google Scholar]
- 8.Biggerstaff, B. J., and L. R. Petersen. 2003. Estimated risk of transmission of the West Nile virus through blood transfusion in the US, 2002. Transfusion 431007-1017. [DOI] [PubMed] [Google Scholar]
- 9.Bondre, V. P., R. S. Jadi, A. C. Mishra, P. N. Yergolkar, and V. A. Arankalle. 2007. West Nile virus isolates from India: evidence for a distinct genetic lineage. J. Gen. Virol. 88875-884. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 1999. Outbreak of West Nile-like viral encephalitis—New York, 1999. MMWR Morb. Mortal. Wkly. Rep. 48845-849. [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2006. West Nile virus activity—United States, January 1-November 7, 2006. MMWR Morb. Mortal. Wkly. Rep. 551204-1205. [PubMed] [Google Scholar]
- 12.Cheng, Y.-W., C. Shawber, D. Notterman, P. Paty, and F. Barany. 2006. Multiplexed profiling of candidate genes for CpG island methylation status using a flexible PCR/LDR/Universal Array assay. Genome Res. 16282-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, C. T., D. W. Beasley, H. Guzman, R. Raj, M. D'Anton, R. J. Novak, T. R. Unnasch, R. B. Tesh, and A. D. Barrett. 2003. Genetic variation among temporally and geographically distinct West Nile virus isolates, United States, 2001, 2002. Emerg. Infect. Dis. 91423-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Favis, R., and F. Barany. 2000. Mutation detection in K-ras, BRCA1, BRCA2, and p53 using PCR/LDR and a universal DNA microarray. Ann. N. Y. Acad. Sci. 90639-43. [DOI] [PubMed] [Google Scholar]
- 15.Favis, R., J. P. Day, N. P. Gerry, C. Phelan, S. Narod, and F. Barany. 2000. Universal DNA array detection of small insertions and deletions in BRCA1 and BRCA2. Nat. Biotechnol. 18561-564. [DOI] [PubMed] [Google Scholar]
- 16.Favis, R., N. P. Gerry, Y. W. Cheng, and F. Barany. 2005. Applications of the universal DNA microarray in molecular medicine. Methods Mol. Med. 11425-58. [DOI] [PubMed] [Google Scholar]
- 17.Favis, R., J. Huang, N. P. Gerry, A. Culliford, P. Paty, T. Soussi, and F. Barany. 2004. Harmonized microarray/mutation scanning analysis of TP53 mutations in undissected colorectal tumors. Hum. Mutat. 2463-75. [DOI] [PubMed] [Google Scholar]
- 18.Gerry, N. P., N. E. Witowski, J. Day, R. P. Hammer, G. Barany, and F. Barany. 1999. Universal DNA microarray method for multiplex detection of low abundance point mutations. J. Mol. Biol. 292251-262. [DOI] [PubMed] [Google Scholar]
- 19.Khanna, M., P. Park, M. Zirvi, W. Cao, A. Picon, J. Day, P. Paty, and F. Barany. 1999. Multiplex PCR/LDR for detection of K-ras mutations in primary colon tumors. Oncogene 1827-38. [DOI] [PubMed] [Google Scholar]
- 20.Lanciotti, R. S., G. D. Ebel, V. Deubel, A. J. Kerst, S. Murri, R. Meyer, M. Bowen, N. McKinney, W. E. Morrill, M. B. Crabtree, L. D. Kramer, and J. T. Roehrig. 2002. Complete genome sequences and phylogenetic analysis of West Nile virus strains isolated from the United States, Europe, and the Middle East. Virology 29896-105. [DOI] [PubMed] [Google Scholar]
- 21.Lanciotti, R. S., A. J. Kerst, R. S. Nasci, M. S. Godsey, C. J. Mitchell, H. M. Savage, N. Komar, N. A. Panella, B. C. Allen, K. E. Volpe, B. S. Davis, and J. T. Roehrig. 2000. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J. Clin. Microbiol. 384066-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 2862333-2337. [DOI] [PubMed] [Google Scholar]
- 23.Linke, S., H. Ellerbrok, M. Niedrig, A. Nitsche, and G. Pauli. 2007. Detection of West Nile virus lineages 1 and 2 by real-time PCR. J. Virol. Methods 146355-358. [DOI] [PubMed] [Google Scholar]
- 24.Linnen, J. M., M. L. Deras, J. Cline, W. Wu, A. S. Broulik, R. E. Cory, J. L. Knight, M. M. Cass, C. S. Collins, and C. Giachetti. 2007. Performance evaluation of the PROCLEIX West Nile virus assay on semi-automated and automated systems. J. Med. Virol. 791422-1430. [DOI] [PubMed] [Google Scholar]
- 25.Luo, J., D. E. Bergstrom, and F. Barany. 1996. Improving the fidelity of Thermus thermophilus DNA ligase. Nucleic Acids Res. 243071-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mostashari, F., M. L. Bunning, P. T. Kitsutani, D. A. Singer, D. Nash, M. J. Cooper, N. Katz, K. A. Liljebjelke, B. J. Biggerstaff, A. D. Fine, M. C. Layton, S. M. Mullin, A. J. Johnson, D. A. Martin, E. B. Hayes, and G. L. Campbell. 2001. Epidemic West Nile encephalitis, New York, 1999: results of a household-based seroepidemiological survey. Lancet 358261-264. [DOI] [PubMed] [Google Scholar]
- 27.Niedrig, M., U. Klockmann, W. Lang, J. Roeder, S. Burk, S. Modrow, and G. Pauli. 1994. Monoclonal antibodies directed against tick-borne encephalitis virus with neutralizing activity in vivo. Acta Virol. 38141-149. [PubMed] [Google Scholar]
- 28.Niedrig, M., S. Linke, H. Zeller, and C. Drosten. 2006. First international proficiency study on West Nile virus molecular detection. Clin. Chem. 521851-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pingle, M. R., K. Granger, P. Feinberg, R. Shatsky, B. Sterling, M. Rundell, E. Spitzer, D. Larone, L. Golightly, and F. Barany. 2007. Multiplexed identification of blood-borne bacterial pathogens by use of a novel 16S rRNA gene PCR-ligase detection reaction-capillary electrophoresis assay. J. Clin. Microbiol. 451927-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi, P. Y., E. B. Kauffman, P. Ren, A. Felton, J. H. Tai, A. P. Dupuis II, S. A. Jones, K. A. Ngo, D. C. Nicholas, J. Maffei, G. D. Ebel, K. A. Bernard, and L. D. Kramer. 2001. High-throughput detection of West Nile virus RNA. J. Clin. Microbiol. 391264-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi, P. Y., and L. D. Kramer. 2003. Molecular detection of West Nile virus RNA. Expert Rev. Mol. Diagn. 3357-366. [DOI] [PubMed] [Google Scholar]
- 32.Smithburn, K. C., T. P. Hughes, A. W. Burke, and J. H. Paul. 1940. A neurotropic virus isolated from the blood of a native of Uganda. Am. J. Trop. Med. s1-20471-492. [Google Scholar]
- 33.Stramer, S. L., C. T. Fang, G. A. Foster, A. G. Wagner, J. P. Brodsky, and R. Y. Dodd. 2005. West Nile virus among blood donors in the United States, 2003 and 2004. N. Engl. J. Med. 353451-459. [DOI] [PubMed] [Google Scholar]
- 34.Tang, Y., C. Anne Hapip, B. Liu, and C. T. Fang. 2006. Highly sensitive TaqMan RT-PCR assay for detection and quantification of both lineages of West Nile virus RNA. J. Clin. Virol. 36177-182. [DOI] [PubMed] [Google Scholar]
- 35.Wacker, M. J., and M. P. Godard. 2005. Analysis of one-step and two-step real-time RT-PCR using SuperScript III. J. Biomol Tech. 16266-271. [PMC free article] [PubMed] [Google Scholar]
- 36.White, D. J. 2001. Vector surveillance for West Nile virus. Ann. N. Y. Acad. Sci. 95174-83. [DOI] [PubMed] [Google Scholar]
- 37.White, D. J., L. D. Kramer, P. B. Backenson, G. Lukacik, G. Johnson, J. A. Oliver, J. J. Howard, R. G. Means, M. Eidson, I. Gotham, V. Kulasekera, and S. Campbell. 2001. Mosquito surveillance and polymerase chain reaction detection of West Nile virus, New York State. Emerg. Infect. Dis. 7643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]