Abstract
To identify host proteins interacting with Tomato bushy stunt virus (TBSV) replication proteins in a genome-wide scale, we have used a yeast (Saccharomyces cerevisiae) proteome microarray carrying 4,088 purified proteins. This approach led to the identification of 58 yeast proteins that interacted with p33 replication protein. The identified host proteins included protein chaperones, ubiquitin-associated proteins, translation factors, RNA-modifying enzymes, and other proteins with yet-unknown functions. We confirmed that 19 of the identified host proteins bound to p33 in vitro or in a split-ubiquitin-based two-hybrid assay. Further analysis of Cdc34p E2 ubiquitin-conjugating enzyme, which is one of the host proteins interacting with p33, revealed that Cdc34p is a novel component of the purified viral replicase. Downregulation of Cdc34p expression in yeast, which supports replication of a TBSV replicon RNA (repRNA), reduced repRNA accumulation and the activity of the tombusvirus replicase by up to fivefold. Overexpression of wild-type Cdc34p, but not that of an E2-defective mutant of Cdc34p, increased repRNA accumulation, suggesting a significant role for the ubiquitin-conjugating enzyme function of Cdc34p in TBSV replication. Also, Cdc34p was able to ubiquitinate p33 in vitro. In addition, we have shown that p33 becomes ubiquitinated in vivo. We propose that ubiquitination of p33 likely alters its function or affects the recruitment of host factors during TBSV replication.
The host cell plays a major role during the replication of small plus-stranded RNA viruses with limited coding capacity. A yet-unknown number of host proteins, termed host factors, are likely exploited by viruses to facilitate their replication and spread. Indeed, viruses likely coopt or alter many cellular processes, such as translation and RNA and protein degradation, via their coded proteins (1, 27, 29). The host cells also activate antiviral defense mechanisms, such as gene silencing, to destroy viral RNAs (20). Recent systematic genome-wide screens of a single-gene knockout library of yeast (Saccharomyces cerevisiae) conducted with Brome mosaic virus (BMV) (19) and Tomato bushy stunt virus (TBSV) (17, 35) revealed that their replication is affected by ∼100 mostly unique host genes. Additional genome-wide screens revealed that 32 host factors affected tombusvirus evolution via stimulating or inhibiting RNA recombination (50, 51). However, the above-described genome-wide screens could have missed the identification of those host genes, which are part of multimember gene families with overlapping functions. Therefore, additional screens might lead to the identification of new host genes affecting virus replication.
Genomic RNA of TBSV and the closely related Cucumber necrosis virus (CNV) codes for two replication proteins, termed p33 and p92pol, and three additional proteins involved in encapsidation, cell-to-cell movement, and suppression of gene silencing (63). p92pol is the RNA-dependent RNA polymerase (28, 31, 34, 37), whereas p33 is an essential replication cofactor involved in RNA template selection/recruitment (26, 33, 41) and in the assembly of the viral replicase (38), and it is an integral part of the tombusvirus replicase complex (45, 49). Interestingly, yeast cells expressing tombusvirus p33 and p92pol replication proteins can efficiently replicate a TBSV-derived replicon RNA (repRNA), which is a defective interfering (DI) RNA identified in TBSV-infected plants (34, 39). The tombusvirus repRNA serves not only as a template for replication but also as an assembly platform for the viral replicase complex (38, 39). Altogether, yeast serves as a useful model host to study replication and recombination of tombusviruses, allowing the utilization of powerful genomics and proteomics tools developed for yeast.
The p33 replication protein likely interacts with many host proteins that could affect various steps during tombusvirus infections. Host proteins are likely involved in intracellular trafficking of p33 and in posttranslational modification of p33, such as phosphorylation, which has been shown to affect the ability of p33 to bind to the viral RNA (55, 58). To identify those host proteins that interact with the tombusvirus p33 replication cofactor, we took a global approach based on the yeast proteome microarray (66, 67). Previous studies using the yeast proteome microarray have identified numerous yeast proteins involved in protein-protein interactions, lipid binding, DNA binding, and small-substrate binding, thus demonstrating the usefulness of the global analysis approach (14, 15, 56, 66, 67). In addition, a yeast protoarray approach was used successfully to identify many host proteins interacting with a 3′ fragment of the BMV RNA (68). Two of those host proteins were confirmed to play a role during BMV infection in plants.
This work has led to the identification of 58 host proteins that bound to the purified recombinant p33, whereas 11 bound to the unique C-terminal portion of p92pol. The identified proteins include helicases, ubiquitin (Ub) ligases and proteases, translation factors, RNA-modifying enzymes, and proteins with unknown functions. Additional data from in vitro binding experiments and the split-Ub two-hybrid assay confirmed many of the host protein-p33 interactions. Further detailed work with Cdc34p Ub-conjugating enzyme demonstrated that it is present within the viral replicase complex. Downregulation of Cdc34p decreased the accumulation of TBSV repRNA in yeast and reduced the in vitro activity of the tombusvirus replicase. Ubiquitination of p33 has been performed in vitro with purified Cdc34p, which likely plays a regulatory role in TBSV replication. In addition, we have shown that p33 becomes mono- and biubiquitinated in yeast cells. Overall, the presence of Cdc34p within the tombusvirus replicase validates the usefulness of the protoarray approach.
MATERIALS AND METHODS
Yeast and Escherichia coli plasmids.
To study the effect of overexpression of selected yeast proteins on viral RNA replication and recombination, we used the yeast open reading frame (ORF) collection from Open Biosystems. In this yeast ORF collection, each ORF is expressed from 2μ plasmid BG1805 under the control of the GAL1 promoter and fused to a tandem affinity tag that includes a hemagglutinin tag and the zz domain of protein A at the C terminus. For the replication assay, the parental strain (BY4741) was cotransformed with three separate plasmids: (i) pHisGBK-His33/DI72-CUP1 [coexpressing CNV p33 from the ADH1 promoter and DI72(+) RNA from the CUP1 promoter], (ii) pGAD-His92-CUP1 (containing the CNV p92pol gene behind the CUP1 promoter), and (iii) one of the individual yeast ORF clones (Open Biosystems) or 2μ plasmid pYES-NT-C (Invitrogen) as a control. For the recombination assay, pHisGBK-His33-CUP1/DI-AU-FP-GAL1 (coexpressing CNV p33 from the CUP1 promoter and DI-AU-FP from the GAL1 promoter) (7) was used together with pGAD-His92-CUP1 and one of the yeast ORF plasmids. For FLAG purification of virus replicase from yeast, the yeast plasmids pGBK-33HF and pGAD-92HF, expressing His6- and FLAG-tagged p33 and p92, and pGBK-His33 and pGAD-His92, expressing only His6-tagged p33 and p92, were described previously (49).
The full-length CDC34 and the regions of CDC34 corresponding to the N-terminal 170-amino-acid (aa) and C-terminal 125-aa sequences were amplified by PCR (the sequences of the primers are available upon request). The PCR products were treated by BamHI and XhoI and ligated into pYES-NT-C or pYC2/CT (a centromeric, low-copy-number plasmid) (Invitrogen) digested with the same enzymes. The PCR products were also cloned into pGEX-2T (Novagen) to construct protein expression plasmids in E. coli. The CDC34 C95S mutation was constructed by use of a QuikChange XL site-directed mutagenesis kit (Stratagene), changing the Cys95 TGT codon to TCT (serine). The mutated DNA was cloned into pYES-NT-C, pYC2/CT, and pGEX-2T as described above.
To construct the expression vector for the glutathione S-transferase (GST)/Ub fusion, we amplified the yeast Ub sequence with PCR using primers 2227 (GGCGGGATCCATGCAGATTTTCGTCAAGACTTTG) and 2228 (GGCCCTCGAGTTAACCACCTCTTAGCCTTAGCACAAG). The PCR product was digested with BamHI and XhoI and cloned into pGEX-2T at the BamHI/XhoI sites.
Yeast strains and culturing.
Saccharomyces cerevisiae strains BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and InvSc1 were obtained from Invitrogen. The CDC34 strain (pCDC34::Kanr Tet07 TATA URA3::CMV-tTA MATa his3-1 leu2-0 met15-0) with the titratable yeast Tet promoters from the Hughes collection (yTHC) was obtained from Open Biosystems. Yeast cultures were grown at 29°C in YPD (1% yeast extract, 2% peptone, and 2% glucose) or defined synthetic medium (SC) supplemented with appropriate amino acids and 2% glucose or 2% galactose as a carbon source. Yeast transformation was done by using the standard lithium acetate-single-stranded DNA-polyethylene glycol method, and transformants were selected by complementation of auxotrophic markers (34, 36). The URA3 gene in the CDC34 yTHC strain was mutated by transforming yeast with an SdaI-truncated ura3 fragment and then growing yeast on SC medium with 2% glucose in the presence of 1 g/liter 5-fluororotic acid. The 5-fluororotic acid-resistant colonies were selected and the colonies were further checked by their inability to grow on SC medium lacking uracil (SC-U−).
Expression and purification of recombinant tombusvirus replication proteins.
The sequence of a v5 epitope tag was added at the C terminus of TBSV p33 in plasmid pMAL-p33 (44) by use of PCR with p33-specific primers 788 (GAGGGATCCGAGACCATCAAGAGAATG) and 1621 (CGCGTCTAGATTTGACACCCAGGGACTCCTGTGA) and v5-specific primers 1619 (CGCGTCTAGAGGGCCCTTCGAAGGTAAGCCT) and 1620 (CGGGCTGCAGTCAATGGTGATGGTGATGATGACCGGT). The obtained PCR products were digested with BamHI-XbaI and with XbaI-PstI, respectively. The two PCR products were cloned simultaneously into BamHI-PstI-digested pMAL-c2 vector (NEB). The obtained plasmid was used to express the maltose binding protein (MBP)-p33-v5 fusion protein in E. coli (44, 46), followed by the addition of 100 μg of RNase A to the bacterial pellet prior to resuspension in 50 mM HEPES, pH 7.4, and 100 mM NaCl. Affinity purification was performed on maltose binding resin in 50 mM HEPES and 100 mM NaCl according to the manufacturer's instructions (NEB). The MBP tag of the MBP-p33-v5 fusion protein was cleaved off with 1 μl factor Xa at 4°C in 1× protoarray buffer plus 0.5 M NaCl and 0.5% Triton X-100 (46). The final concentration of p33-v5 was 0.7 μg/μl. The v5 epitope tag was added to the C terminus of pMAL-p33N82 and pMAL-p92C (Fig. 1A) (43) by using a strategy as described above for pMAL-p33.
FIG. 1.
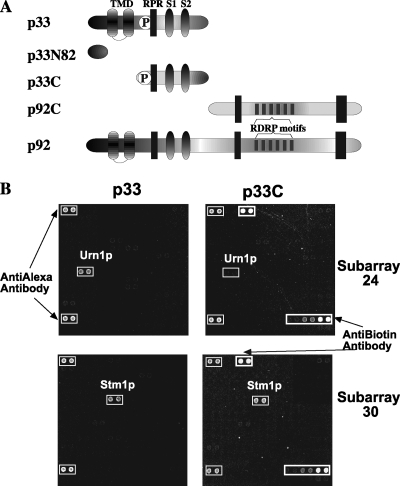
Yeast protoarray approach to identify host proteins interacting with TBSV replication proteins. (A) Schematic representation of the overlapping TBSV p33 and p92pol replication proteins and their derivatives expressed in E. coli. TMD, predicted transmembrane domains; RPR, arginine-proline-rich RNA-binding region; RDRP, RNA-dependent RNA polymerase; S1 and S2, p33:p33 interaction subdomains. Note that the sequence in the N-terminal region in p92pol is identical with the p33 sequence due to the gene expression strategy of tombusviruses. (B) Identification of host proteins binding to p33 and a truncated p33 (termed p33C) (panel A) replication protein based on the protoarray. The whole chip contains 4,088 purified yeast proteins in duplicate. For the binding assay, we used E. coli-expressed v5-tagged p33 and biotinylated p33C cleaved from the MBP tag. Two representative subarrays are shown to illustrate the binding of host proteins to p33 but not to p33C (top) or to both p33 and p33C (bottom). Note that the protoarray contains various amounts of yeast proteins (the actual values are supplied by Invitrogen), and this information was used to calculate the relative binding of each host protein to p33/p33C (Table 1).
Biotinylation of p33C for protoarray analysis.
Twenty microliters of the affinity-purified MBP-p33C (2.5 μg/μl) (44) in 50 mM HEPES, pH 7.4, and 100 mM NaCl was biotinylated by the addition of 1.6 μl of 5 nmol/μl biotin-sulfosuccinimidyl ester sodium salt and 2 μl of 1 M NaHCO3, pH 8.4. The biotinylation reaction was performed at room temperature for 60 min, followed by cleaning through a gel filtration minicolumn (exclusion limit, 6,000 Da). The final protein concentration was 0.5 μg/μl. The quality of the protein biotinylation was assessed by Western blotting using streptavidin-alkaline phosphatase conjugate and a fluorescent substrate, CDP-Star, according to the manufacturer's instructions (Invitrogen).
Yeast protoarray analysis.
The protein array slide (Invitrogen) was blocked for 1 h at 4°C in the blocking buffer (1× phosphate-buffered saline, 1.0% bovine serum albumin, and 0.1% Tween 20). Then, 120 μl of the p33-v5 probe (5 ng/μl) or the biotinylated p33C probe (50 ng/μl) in the probing buffer (1× phosphate-buffered saline, 5 mM MgCl2, 0.5 mM dithiothreitol [DTT], 0.05% Triton X-100, 5% glycerol, and 1.0% bovine serum albumin) was pipetted on each protoarray slide. The slide was then covered and incubated at 4°C for 1.5 h. After three 1-min washes with the cold probing buffer, the protein array was incubated in the dark on ice with 0.1 μg/ml streptavidin-Alexa Fluor 647 for the detection of the biotinylated protein probe or with anti-v5-Alexa Fluor 647 conjugate for the v5-tagged p33 probe. The unbound Alexa Fluor 647 conjugates were washed off three times with 1-min washes in the probing buffer. The array was centrifuged for 4 min at 800 × g and left in the slide holder to air dry for 30 min. Scanning and quantification were performed with a ScanArray 4000 (Packard Bioscience, Billerica, MA) scanner and QuantArray V3.0 software. We scored those host proteins that had a relative value of higher than 150, as calculated on the basis of the actual amounts of yeast proteins spotted on the protoarray and the measured signal in repeated experiments, as shown in Table 1.
TABLE 1.
The names and functions of yeast proteins bound to TBSV p33
| Genea | Binding of corresponding protein tob:
|
Protein functionc | |||
|---|---|---|---|---|---|
| p33 | p33N | p33C | p92C | ||
| ALA1 | ++ | − | − | − | Cytoplasmic alanyl-tRNA synthetase |
| ARP8* | ++ | − | + | − | Nuclear actin-related protein involved in chromatin remodeling |
| BFR1 | ++ | − | − | − | Component of mRNP complexes associated with polyribosomes |
| BUD21 | ++ | − | − | − | Component of SSU processosome |
| CDC34 | ++ | − | ++++ | − | Ub-conjugating enzyme or E2 |
| DDR48 | ++ | ++ | ++ | ++ | DNA damage-responsive protein; expression is increased in response to heat shock stress |
| EFB1* | ++ | − | − | − | Translation elongation factor 1 beta |
| ELF1* | ++ | − | ++ | ++ | A conserved zinc finger containing transcription elongation factor; deletion inhibits BMV gene expression |
| EMI2 | + | + | − | − | Protein of unknown function |
| ERB1 | ++++ | ++ | ++ | ++ | Protein required for maturation of the 25S and 5.8S ribosomal RNAs; constituent of 66S preribosomal particles; homologous to mammalian Bop1 |
| ERR2 | ++ | + | − | − | Protein of unknown function; has similarity to enolases |
| FOX2 | ++++ | − | − | − | Multifunctional enzyme of the peroxisomal fatty acid beta-oxidation pathway |
| FMP40 | + | + | − | − | Protein of unknown function |
| GIM3 | ++ | − | − | − | Subunit of the heterohexameric cochaperone prefoldin complex, which binds specifically to cytosolic chaperonin and transfers target proteins to it |
| GPH1 | ++ | ++ | + | − | Glycogen phosphorylase |
| GSY2* | + | + | ++ | ++ | Glycogen synthase |
| HBS1* | ++ | − | ++ | − | GTP binding protein with sequence similarity to the elongation factor class of G proteins, EF-1alpha and Sup35p |
| HOR2 | ++ | + | − | − | dl-Glycerol-3-phosphatase |
| IPI3 | ++ | − | − | − | Protein of unknown function; a possible role in assembly of the ribosomal large subunit |
| ISN1 | ++ | − | − | − | IMP-specific 5′ nucleotidase |
| IWR1* | ++++ | − | ++++ | ++++ | Protein of unknown function |
| JJJ1* | ++ | − | ++ | ++++ | Protein that may function as a cochaperone |
| JJJ3 | ++ | ++ | − | − | Protein of unknown function, contains a J-domain |
| MAM33 | ++ | − | + | − | Acidic protein of the mitochondrial matrix involved in oxidative phosphorylation |
| NAP1* | ++ | + | ++++ | − | Protein that interacts with mitotic cyclin Clb2p; required for the regulation of microtubule dynamics |
| PDI1 | ++ | + | − | − | Protein disulfide isomerase, multifunctional protein resident in the endoplasmic reticulum lumen; essential for the formation of disulfide bonds in secretory and cell surface proteins |
| PLP2 | ++ | − | − | − | Similarity to phosducins, which are GTPase inhibitors |
| POL30 | + | − | − | − | PCNA |
| PYC1 | ++ | − | + | − | Pyruvate carboxylase isoform |
| QCR6 | ++ | − | ++ | ++ | Subunit 6 of the ubiquinol cytochrome-c reductase complex; highly acidic protein |
| RIB2 | + | − | − | − | DRAP deaminase; cytoplasmic tRNA pseudouridine synthase |
| RPL8A* | ++ | − | ++++ | ++++ | Ribosomal protein L4 of the large (60S) ribosomal subunit |
| RSP5 | + | − | − | − | Ub-protein ligase involved in Ub-mediated protein degradation |
| RTT106 | ++ | − | ++ | − | Protein of unknown function |
| SAS10* | ++++ | − | ++++ | ++++ | Component of the SSU processosome |
| SHO1 | ++ | − | − | − | Transmembrane osmosensor |
| STM1* | + | − | ++ | ++ | Protein that binds G4 quadruplex and purine motif triplex nucleic acid |
| SPT16* | ++++ | + | ++++ | ++++ | Pol II transcription elongation factor activity |
| TIF1* | + | − | − | − | Translation initiation factor eIF4A; identical to Tif2p; DEA(D/H)-box RNA helicase |
| TIF11* | + | − | − | + | Translation initiation factor eIF1A; essential protein that forms a complex with Sui1p (eIF1) and the 40S ribosomal subunit and scans for the start codon |
| TOM71 | ++ | − | ++ | ++ | Mitochondrial outer membrane protein; minor component of the TOM |
| TRM1* | ++++ | − | ++++ | ++++ | tRNA methyltransferase; localizes to both nucleus and mitochondrion to produce the modified base N2,N2-dimethylguanosine in tRNAs in both compartments |
| TRZ1 | + | − | + | − | tRNase Z; involved in RNA processing; has two putative nucleotide triphosphate-binding motifs (P-loop) and a conserved histidine motif; homolog of the human candidate prostate cancer susceptibility gene ELAC2 |
| TSR2 | ++++ | − | − | − | Protein with a potential role in pre-rRNA processing |
| UBA1 | + | + | − | − | Ub-activating enzyme involved in Ub-mediated protein degradation |
| UBP10* | + | − | ++ | ++ | UBP that deubiquitinates Ub-protein moieties |
| UBP15* | ++++ | ++ | ++ | − | UBP that may play a role in Ub precursor processing |
| URN1 | ++ | + | − | ++ | Pre-mRNA splicing factor associated with the U2-U5-U6 snRNPs, the RES complex, and the Prp19-associated complex (NTC) |
| YCR016W | ++ | − | ++++ | ++++ | Unknown; involved in ribosome biogenesis |
| YDR161W | ++ | − | − | − | Unknown; involved in ribosome biogenesis |
| YGL242C | ++ | − | ++++ | ++ | Unknown |
| YGR017W | ++ | + | ++ | ++ | Unknown |
| YGR027W | ++++ | − | − | − | Retrotransposon TYA Gag gene |
| YHL013C/OTU2 | + | − | − | + | member of the OTU superfamily of predicted cysteine proteases |
| YHR009C | + | − | + | − | Unknown |
| YLR125W | + | − | − | − | Unknown |
| YOR251C | + | + | − | − | Thiosulfate sulfurtransferase activity |
Genes corresponding to proteins binding specifically to various regions of TBSV p33 and p92. Boldface data indicate yeast proteins that interacted with recombinant p33C in an in vitro protein-binding assay (Fig. 2). Host proteins that interacted with p33 in the split-Ub assay (Fig. 3) are marked with asterisks.
Binding of the yeast protein to p33, p33N (the N-terminal 82-aa region), p33C, and p92C was calculated according to the following formula: (signal − background signal/amount of the selected protein on the array) × 10,000. Relative binding values are indicated as follows: ++++, above 1,000; ++, between 350 and 1000; +, between 150 and 350; −, below 150. The values were derived from repeated experiments.
Documented functions of the yeast proteins are based on the Saccharomyces Genome Database at http://www.yeastgenome.org/. SSU, small ribosomal subunit; PCNA, proliferating cell nuclear antigen; Pol II, polymerase II; TOM, translocase of outer membrane; OTU, ovarian tumor-like; IMP, inosine 5′-monophosphate.
Pulldown assay.
The recombinant TBSV p33C was expressed as a fusion with MBP in E. coli Epicurian BL21-CodonPlus (DE3)-RIL (Stratagene) as described previously (43). E. coli cells were sonicated in MBP column buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol), and the fusion protein was immobilized on amylose resin (100 μl). The unbound material was drained and washed with MBP column buffer twice. Yeast strains expressing individual GST-His6-tagged proteins were selected from a GST-His6 ORF library (a generous gift from Brenda Andrews) (57). Yeast cells were first pregrown in SC-U− with 2% glucose at 29°C and then grown in SC-U− with 2% galactose until reaching mid-logarithmic phase (optical density at 600 nm, 0.8 to 1.0). Yeast cells were suspended in binding buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% Nonidet P-40 [NP-40], 10 mM β-mercaptoethanol, and 1% [vol/vol] yeast protease inhibitor mix) and were broken by glass beads in a Genogrinder. The total yeast lysate was cleared by centrifugation at 21,000 × g for 10 min at 4°C (performed twice) and loaded on the affinity column with immobilized MBP-p33C or MBP. The binding reactions were performed at 4°C for 1 h with continuous rotation. After the binding, the beads were washed five times with 1 bed volume of binding buffer containing 200 mM NaCl. Proteins bound to the beads were eluted by incubation in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer at 85°C for 10 min. Samples (10 μl) were loaded onto SDS-PAGE gels. Standard Western blotting was performed with anti-His/anti-mouse immunoglobulin G antibodies (39).
Host protein overexpression studies.
For inducible launching of TBSV replication, we used the copper-regulatable CUP1 promoter. The CUP1 promoter was amplified form the yeast genomic DNA with primers 1779 (CGCGGAATTCGACATTTGGGCGCTATACGTGCATATGT) and 1780 (CGCGCTCGAGTACAGTTTGTTTTTCTTAATATCTATTTCGA), followed by digestion of the PCR product with XhoI/EcoRI. The sequence of DI72 in combination with the 3′-located ribozyme was amplified with primers 471 (CCCGCTCGAGGGAAATTCTCCAGGATTTCTC) and 1069 (CCGGTCGAGCTCTACCAGGTAATATACCACAACGTGTGT) and digested with XhoI/SacI. Both fragments were cloned simultaneously into pHisGBK-His33-DI72 (17) digested with EcoRI/SacI to yield pHisGBK-His33-CUP-DI72.
Yeast strain BY4741 was transformed with the selected ORFs under the GAL1 promoter (Open Biosystems) along with pGAD-His92 and pHisGBK-His33-CUP-DI72. Selection was done in synthetic complete medium lacking Ura, Leu, and His (SC-ULH− medium), whereas the culturing of yeast transformants was done in 1.5 ml of SC-ULH plus 2% galactose for 20 h at 30°C to produce the particular host protein. Then, TBSV repRNA replication was launched by adding 50 μM CuSO4 to the media, followed by additional culturing for 24 h at 30°C. The final optical density was ∼0.7 to 1.0. For the detection of the expressed host and viral proteins, we performed Western blotting as described previously (39).
Split-Ub yeast two-hybrid assay.
The split-Ub assay was based on the Dualmembrane kit 3 (Dualsystems Biotech). The His6-tagged CNV p33 was amplified from pGBK-His33 (39) with primers 2261 (GTCGCTGCAGTACTAGTAGGCCTGGAGGTTCTCATCATCATC) and 2262 (GTCGCCATGGAGGCCTCTATTTCACACCAAGGGAC) and cloned into PstI/NcoI-digested pBT3-N (Dualsystems). From the resulting plasmid, the expression cassette comprising CYC1pro-LexA-VP16-Cub-6xHis-p33 was amplified by PCR using primers 2236 (CGGCCTGCAGGCTCATTTGGCGAGCGTTGG) and 2262 and cloned into SdaI/NcoI-digested pGAD-H (40), creating pGAD-BT2-N-His33. pPR3-N and pPR3-C (Dualsystems) were modified by the addition of a synthetic polylinker. To do that, CNV p33 was amplified from pGBK-His33 by use of primers 2181 (GTGGGATCCGAATTCAGATCTGGGCCCGGGATGGATACCATCAAGAGGATG) and 2182 (GTCGTCGACTTAATCGATGCTAGCCCATGGCCCGGGTTTCACACCAAGGGACTC). The resulting PCR product was digested with BamHI and SalI and ligated into similarly digested pPR3-N (Dualsystems) to generate pPR3-N-p33. Alternatively, the PCR product was digested with BamHI and ClaI and ligated into similarly digested pPR3-C (Dualsystems) to generate pPR3-C-p33. Plasmids pPR3-N-p33 and pPR3-C-p33 were digested with XmaI and gel purified to excise the portion containing p33 and religated to create pPR-N-RE and pPR-C-RE. Host genes were amplified by PCR using specific primers and cloned into pPR-N-RE or pPR-C-RE in frame with the hemagglutinin-NubG cassette. Yeast strain NMY51 [MATa his3Δ200 trp1-901 leu2-3,112 ade2 LYS2::(lexAop)4-HIS3 ura3::(lexAop)8-lacZ ade2::(lexAop)8-ADE2 GAL4; Dualsystems] was transformed with pGAD-BT2-N-His33 and pPR-N-RE (NubG) or one of the pPR host gene constructs and plated onto Trp−/Leu− minimal medium plates. Transformed colonies were picked with a loop, resuspended in water, and streaked onto Trp−/Leu−/His−/Ade− plates to test for p33-host protein interactions.
Replicase purification.
Yeast SC1 strain transformed with expression plasmids for His6- and FLAG-tagged p33 and p92 and DI72 were grown at 29°C until reaching an optical density at 600 nm of 0.8 to 1.0. Replicase purification was performed as described earlier with modification (49). Briefly, 200 mg of yeast cells was broken in TG buffer (50 mM Tris-HCl [pH 7.5], 10% glycerol, 15 mM MgCl2, and 10 mM KCl) supplemented with 0.5 M NaCl, 0.1% NP-40, and 1% (vol/vol) yeast protease inhibitor cocktail. The enriched membrane fraction was solubilized in 1 ml TG buffer with 0.5 M NaCl, 1% NP-40, 5% caprylyl sulfobetaine (SB3-10; Sigma) via gentle rotation for 2 h at 4°C. Then, the tube was incubated at 37°C for 5 min and centrifuged at 21,000 × g at 4°C for 15 min. Supernatant was incubated with 50 μl anti-FLAG M2-agarose affinity gel (Sigma) for 2 h at 4°C. The unbound materials were removed by centrifugation at 500 × g, and the resin was washed three times with 1 ml TG buffer with 0.5 M NaCl, 1% NP-40, and 1% SB3-10 and twice with 1 ml TG buffer with 150 mM NaCl and 0.1% NP-40. Proteins bound to affinity beads were eluted by incubating in SDS-PAGE loading buffer at 85°C for 10 min and then subjected to SDS-PAGE on a 10% gel by use of anti-His (Amersham) and anti-cdc34 (generous gift from Mark Goebl) antibodies.
In vitro ubiquitination.
The yeast strain expressing UBA1 was from the GST-His6 ORF library (57). The purification of GST-His6 UBA1 was performed from yeast, while the GST-CDC34 and GST/Ub was obtained from E. coli by use of glutathione Sepharose beads (Novagen). The obtained GST fusion proteins were dialyzed in 40 mM Tris-HCl (pH 7.5), 10% glycerol, 50 mM NaCl, 2 mM MgCl2, and 0.5 mM DTT overnight at 4°C (two changes of 0.5 liter buffer each). The purified His6-tagged human Ub was obtained from Boston Biochem. In vitro ubiquitination was carried out in 20 μl ubiquitination buffer (40 mM Tris-HCl [pH 7.5], 50 mM NaCl, 5 mM MgCl2, 2 mM ATP, 1 mM DTT, 10 mM creatine phosphate [Roche], and 1 U creatine kinase [Roche]) containing 50 nM GST-His6-UBA1, 500 nM GST-CDC34, 100 μM His6-Ub, or 30 μM GST/Ub and purified recombinant 0.5 μg MBP-p33 or MBP as substrates. The ubiquitination assays were performed at 30°C for 60 min and then terminated by boiling for 5 min with SDS sample buffer containing 0.1 M DTT, followed by Western blot analysis with anti-MBP antibody.
Analysis of p33 ubiquitination in yeast.
To construct pGBK-FLAG-p33, we used primers 2450 (GGCAAGCTTACCATGGGTCGGGATTACAAGGAC) and 992B (GAGCTGCAGCTATTTCACACCAAGGGA) and pGBK-33HFH as a template. The PCR product was digested with NcoI and PstI and ligated into similarly digested pGBK-33HFH. S. cerevisiae strain Sc1 was transformed with pYES2/NT-C and YEp105 (8), which expresses a copper-inducible c-Myc-tagged Ub and pGBK-33HFH or pGBK-FLAG-p33. Transformed yeast cells were pregrown in medium lacking Ura, Trp, and His (UTH− minimal medium) supplemented with 2% glucose for 24 h at 29°C and then pelleted and grown in UTH− minimal medium plus 2% galactose and 50 mM CuSO4 for an additional 24 h. Cultures were centrifuged and washed in 20 mM Tris-HCl, pH 8.0. Four hundred microliters of yeast pellet was resuspended in 600 μl of ice-cold TG buffer supplemented with 0.5 M NaCl, 0.25% yeast protease inhibitor cocktail, and 10 mM N-ethylmaleimide. Cells were broken with glass beads in a homogenizer for 6 min at 1,500 strokes/min. Two volumes of cold extraction buffer was added and the extracts were transferred to Eppendorf tubes and centrifuged at 21,000 × g for 15 min at 4°C. The supernatant was discarded and the pellet was resuspended in 1 ml ice-cold TG buffer plus 1% NP-40 and 5% SB3-10. This mixture was incubated for 1 h at 4°C with rotation and 5 min at 37°C to solubilize membranes, followed by centrifugation at 21,000 × g for 15 min at 4°C. The supernatant containing the solubilized membrane proteins was loaded onto columns containing 25 μl of anti-FLAG M2 agarose from mouse (Sigma) (equilibrated in TG buffer plus 0.5 M NaCl and 0.1% NP-40) and incubated for 2 h at 4°C with rotation. Columns were drained and filled with 1 ml cold washing buffer I (TG buffer plus 0.5 M NaCl and 1% NP-40) and rotated for 20 min at 4°C. Columns were drained and washed three additional times with washing buffer I and two times with washing buffer II (TG buffer plus 0.05 M NaCl and 0.1% NP-40). After the last wash, columns were centrifuged at 1,000 × g for 1 min to remove excess liquid and proteins were eluted with 50 μl of SDS-PAGE loading buffer without β-mercaptoethanol at 85°C. Columns were centrifuged for a short period to recover the proteins, and then 2.5 μl of β-mercaptoethanol was added to each sample and samples were boiled for 5 min and subjected to SDS-PAGE and Western blotting using monoclonal anti-FLAG M2 antibody from mouse (1:5,000 dilution; Sigma) and alkaline phosphatase-conjugated anti-mouse (1:5,000) followed by Nitro Blue Tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate) detection. c-Myc-tagged Ub was detected using anti-c-Myc antibody from rabbit (1:10,000 dilution; Bethyl) and alkaline phosphatase-conjugated anti-rabbit (1:10,000) followed by Nitro Blue Tetrazolium-BCIP detection.
RESULTS
Yeast protoarray-based identification of host proteins that bind to the tombusvirus p33 replication protein.
The viral replication proteins likely interact with many host proteins to facilitate viral replication. Also, some host proteins could interact with and inhibit the function of the viral replication proteins as an antiviral mechanism. To determine host proteins interacting with the viral replication proteins on a proteome-wide scale, we have utilized a protoarray-based approach with purified host proteins. Since TBSV repRNA replicates efficiently in yeast (34, 39), we took advantage of the yeast proteome microarray carrying 4,088 purified yeast proteins (∼70% coverage of all the yeast genes). To probe the yeast proteome microarray for the identification of host proteins binding to the tombusvirus p33 replication cofactor protein, we purified recombinant p33 fused to MBP from E. coli by use of affinity chromatography as described previously (44). After purification, the N-terminal MBP fusion was cleaved off from p33 by use of factor Xa protease. Importantly, p33 also carried a C-terminal v5 tag to facilitate its detection. The highly purified p33 preparation was applied (in the presence of an excess amount of bovine serum albumin as a nonspecific competitor) onto the yeast protoarray to promote protein-protein interactions, followed by thorough washing. The bound p33 was detected via v5-specific antibody conjugated with Alexa Fluor dye and visualized with a microchip scanner (Fig. 1B). It is worth noting that the v5 tag in p33 could interfere with binding to host proteins and/or the binding of a host protein to this region of p33 could block the detection of the v5 tag on the array.
We found that 58 host proteins bound to the recombinant p33 efficiently and reliably under the in vitro conditions employed (see Materials and Methods). Among these host proteins, there are 3 protein chaperones (Gim3p, Jjj1p, and Jjj3p), 5 proteins involved in protein ubiquitination (Cdc34p, Rsp5p, Uba1p, Ubp10p, and Ubp15p), 6 translation factors involved in mRNA translation (Bfr1p, Efb1p, Hbs1p, Rpl8Ap, Tif1p, and Tif11p), and 10 proteins involved in RNA processing and metabolism (Ala1p, Bud21p, Erb1, Rib2p, Sas10p, Stm1p, Trm1p, Trz1p, Tsr2p, and Urn1p) (Table 1). Additional proteins are known to be involved in various cellular processes and the functions of nine proteins are not yet defined (Table 1).
To obtain further insight into p33-host protein interactions, we also performed similar binding experiments with purified recombinant p33N82 (v5 tagged), which contains only the N-terminal 82 aa of p33, as well as with p33C (Fig. 1A) with defined RNA-binding and protein interaction domains (43-45) by use of the yeast protoarray. However, for p33C, we used biotin labeling followed by detection via streptavidin conjugated with Alexa Fluor dye and visualization with a microchip scanner. Also, because p33C is more soluble than full-length p33, which carries long hydrophobic stretches (Fig. 1A), we could use p33C at a concentration ∼10 times higher than that used for p33 to increase the sensitivity in the protoarray experiments (see Materials and Methods). Interestingly, 29 host proteins (50% of the 58 bound to the full-length p33 on the chip) bound to p33C, and 12 (21%) bound to p33N82, whereas 8 (14%) bound to both p33C and p33N82 (Table 1). The remaining nine (15%) host proteins did not bind to p33N82 and p33C, suggesting that this group of proteins might bind to the hydrophobic central domain in p33, which is missing from both p33N82 and p33C (Fig. 1A).
Yeast protoarray-based identification of host proteins that bind to the unique C-terminal fragment of replication protein p92pol.
To identify host proteins binding specifically to the unique, nonoverlapping portion of p92pol, we also performed protoarray-based binding experiments with purified recombinant p92C (v5 tagged), which contains the RNA-dependent RNA polymerase motifs in the unique C-terminal segment of p92pol (Fig. 1A). We found that 21 host proteins bound both to p33 and to p92C (Table 1), whereas 11 host proteins bound only to p92C (Table 2) and not to p33. The list includes an RNA helicase (Dpb3p), a methylase (Dot1p), an aminopeptidase (Map1p), an RNA-binding protein (Npl3p), and a translation factor (Tef2p) (Table 2). Thus, the protoarray approach allowed the identification of host proteins that selectively bound to p33 or the nonoverlapping portion of p92pol in vitro.
TABLE 2.
The names and functions of yeast proteins bound to TBSV p92C
| Genea | Binding of corresponding protein to p92Cb | Protein functionc |
|---|---|---|
| DBP3 | + | Putative ATP-dependent RNA helicase of the DEAD-box family |
| DOT1 | ++ | Nucleosomal histone H3-Lys79 methylase |
| MAP1 | + | Methionine aminopeptidase |
| NOB1 | ++++ | Essential nuclear protein involved in proteasome maturation |
| NPL3 | ++++ | RNA-binding protein that carries poly(A)+ mRNA from the nucleus into the cytoplasm; phosphorylation by Sky1p in the cytoplasm may promote release of mRNAs |
| RDS2 | ++ | Involved in response to xenobiotic stimulus |
| TEF2 | ++ | Translational elongation factor EF-1 alpha |
| UTP7 | ++++ | Component of the SSU processome |
| VPS66 | ++ | Cytoplasmic protein involved in vacuolar protein sorting |
| YKL023W | ++++ | Cytoplasmic protein of unknown function |
| YOR309C | ++++ | Hypothetical protein |
Genes corresponding to proteins binding specifically to p92C but not to p33.
Binding of the yeast protein to p92C was calculated according to the following formula: (signal − background signal/amount of the selected protein on the array) × 10,000. Relative binding values are indicated as follows: ++++, above 1,000; ++, between 350 and 1000; +, between 150 and 350.
Documented functions of the yeast proteins are based on the Saccharomyces Genome Database at http://www.yeastgenome.org/. SSU, small ribosomal subunit.
Interaction of recombinant p33 with selected yeast proteins in vitro.
Since the protoarray approach might be prone to identify false positives (i.e., proteins that interact with the viral p33 only on the chip under the conditions applied), we used additional approaches to confirm that the recombinant p33 could bind to the identified host proteins as found in the above-described protoarray experiments. To this end, we performed protein pulldown experiments with purified recombinant p33C and 16 host proteins chosen based on their intriguing functions in yeast. For the protein pulldown experiments, yeast lysates containing soluble proteins prepared from yeast overexpressing one of the 16 GST/His6-tagged host proteins were applied to columns containing immobilized MBP-p33C fusion protein. After elution of the bound proteins from the column, we analyzed whether the particular host protein was present in the eluted fraction by using Western blotting (Fig. 2). Similarly prepared recombinant MBP bound to beads was the negative control to exclude nonspecific binders. These experiments are thus complementary to the protoarray experiments, which had the yeast proteins fixed to a solid surface, while the p33C was the probe protein present in solution.
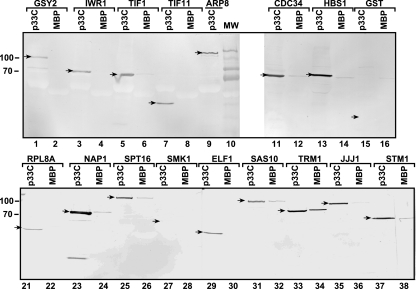
FIG. 2.
Binding of selected host proteins to p33 replication protein in vitro. MBP-tagged p33C and MBP (1 μg each) were separately immobilized on amylose beads, followed by incubation with cytosolic extracts prepared from yeast individually expressing the given GST-His6-tagged host proteins. The bound host proteins were eluted from the beads and were analyzed by 10% SDS-PAGE and detected via anti-His6 antibody. We tested a total of 17 host proteins out of the 58 host proteins identified via the protoarray for their abilities to bind to p33C. Protein bands of the expected sized are depicted with arrows. Molecular mass markers are shown on the left. Note that Hbs1p was expressed as a His6-tagged protein from plasmid pYES.
As expected, GST control bound to neither MBP nor p33C (Fig. 2, lanes 15 and 16). In contrast, 14 host proteins, including Arp8p, Iwr1p, Tif1p, Tif11p, Gsy2p (not shown), Cdc34p, Hbs1p, Rpl8Ap, Nap1p, Spt16p, Jjj1p, Sas10p, Elf1p, and Stm1p bound to p33C-MBP much more efficiently than to MBP (Fig. 2), confirming that these host proteins can interact with the C-terminal domain of p33 in vitro. The binding of Trm1p to p33C-MBP was only about twice as efficient as that to MBP (Fig. 2, lanes 33 and 34). Smk1p mid-sporulation-specific mitogen-activated protein kinase, which bound weakly to p33 on the protoarray (not shown), did not interact with p33C in vitro (Fig. 2, lanes 27 and 28). Overall, data on 16 yeast proteins from the protein pulldown experiments are mostly in agreement with the protoarray experiments, demonstrating efficient interactions between p33 and the selected yeast proteins under in vitro conditions. The major difference in the results from the protoarray and protein pulldown experiments is that Tif1p did not bind to p33C (only to the full-length p33) in the protoarray (Table 1), while it did bind efficiently to p33C based on protein pulldown experiments (Fig. 2, lanes 5 and 6).
In vivo interaction between identified host proteins and the tombusvirus p33 based on the split-Ub two-hybrid assay.
To confirm that the identified p33-host protein interactions can also take place on subcellular membrane surfaces within yeast cells, where p33 is localized (22, 33), we used the split-ubiquitin yeast two-hybrid assay. This assay is based on the ability of the N-terminal (NubG) and C-terminal (Cub) halves of Ub to reconstitute a functional protein (9, 10). When NubG and Cub, both fused separately to interacting proteins, are brought into close proximity and reconstitute a functional Ub protein, cleavage by endogenous Ub-specific proteases (UBPs) leads to the release of an artificial transcription factor, LexA-VP16, fused to Cub. This allows the activation of LexA-driven HIS3 and ADE2 expression in the nucleus. In summary, the split-Ub system, unlike the original yeast two-hybrid system, does not require interacting proteins to be localized to the nucleus, allowing an analysis of protein interactions on the cytosolic surfaces of membranes, which are the natural subcellular locations of the membrane-bound p33 protein (22, 33).
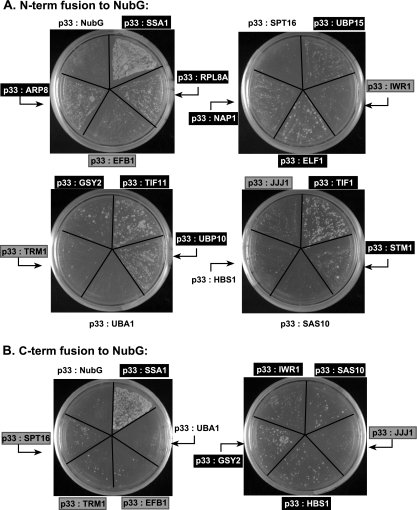
The split-Ub assay with 19 of the identified host proteins revealed that Rpl8Ap, Arp8p, Ubp15p, Elf1p, Nap1p, Tif11p, Ubp10p, Gsy2p, Tif1p, and Stm1p interacted with the membrane-bound p33 when used as N-terminal fusion with NubG (Fig. 3A). Additional host proteins, such as Sas10p, Hbs1p, and Iwr1p, interacted with the membrane-bound p33 when used as C-terminal fusion with NubG (Fig. 3B). The interaction of Efb1p, Trm1p, Spt16p, and Jjj1p with p33 was detectable, but weak, in this assay. Only Uba1p, which is a Ub-activating E1 protein, and Cdc34p (not shown), which is an E2 Ub-conjugating enzyme, have not been found to interact with p33 in the split-Ub assay (Fig. 3B). Since Uba1p and Cdc34p are involved in ubiquitination, they might interfere with Ub cleavage in this assay (putative false negatives). Altogether, the split-ubiquitination assay confirmed that 18 of the identified host proteins interact with membrane-bound p33 in yeast.
FIG. 3.
Confirmation of host protein-p33 interactions via the split-Ub two-hybrid assay. (A) The full-length sequences of 19 host proteins were fused to NubG as N-terminal (N-term) fusions. Heat shock protein 70 (SSA1) was used as a positive control because it is known to interact with p33 (49). The number of colonies formed reflects the strength of protein-protein interaction. Those host factors that had positive interactions with p33 are boxed in black, whereas those with weak interactions are shown in gray boxes. Negative interactors are in boldface on a plain background. Note that the split-Ub two-hybrid assay is based on the ability of N-terminal (Nub) and C-terminal (Cub) halves of Ub to reconstitute a functional protein. A single-amino-acid mutation in Nub (NubG) reduces its affinity for Cub. However, when NubG and Cub are fused to interacting proteins they are brought to close proximity and reconstitute a functional Ub protein, which is cleaved by endogenous UBPs. This cleavage releases an artificial transcription factor, LexA-VP16, which is fused to Cub, allowing the activation of LexA-driven HIS3 and ADE2 genes in the nucleus. Unlike the yeast two-hybrid system, the split-Ub system does not require interacting proteins to be localized to the nucleus, allowing the analysis of protein interactions involving membrane-bound proteins in their natural cell location. (B) Sequences of those host proteins from panel A that interacted weakly or did not interact with p33 were fused to the C terminus (C-term) of NubG and tested for their interactions with p33 as described in the legend to panel A.
Effect of overexpression of selected host proteins on TBSV repRNA replication in yeast.
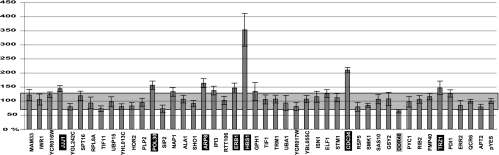
To test if the host proteins that interacted with p33 could affect tombusvirus RNA replication, we took advantage of the previously developed efficient tombusvirus replication system in yeast (34, 35) with some modifications. First, we separately overexpressed 44 of the identified host proteins from the galactose-inducible GAL1 promoter (12) in yeast cells for 20 h. Subsequently, we launched TBSV repRNA replication from the CUP1 promoter in the same cells. Comparable amounts of yeast cells were harvested 24 h later, followed by Northern blotting to measure the level of TBSV repRNA produced. We used rRNA as a loading control for the normalization of data on repRNA accumulation in yeast. The accumulation level of repRNA in yeast carrying pYES plasmid, which expresses only a short peptide, was taken as 100% (Fig. 4). As an additional control, we overexpressed a pseudogene (APT2) which has no enzymatic activity when expressed (2) and failed to interact with p33 based on the protoarray experiments (not shown). Overexpression of Apt2p led to 79% ± 9% repRNA accumulation compared with that seen for yeast carrying the pYES control (Fig. 4). This suggests that protein overexpression in general might reduce the ability of yeast cells to support repRNA accumulation under the protein overexpression condition. Based on the pYES and APT2 controls, we considered the overexpression of a protein to be inhibitory if it significantly reduced repRNA accumulation below 70% and stimulatory if it increased repRNA accumulation significantly, i.e., above 130% (Fig. 4).
FIG. 4.
Effect of overexpression of selected host proteins interacting with p33 replication proteins on TBSV repRNA accumulation in yeast. A total of 44 zz domain-tagged yeast proteins (Open Biosystems) were expressed separately from the galactose-inducible GAL1 promoter 20 h prior to launching TBSV repRNA replication. The accumulation of repRNA was measured with Northern blotting using total RNA extracts from yeast 24 h after the addition of 50 μM copper sulfate, which induced TBSV repRNA replication. The accumulation levels of repRNA in yeast strains overexpressing the APT2 pseudogene and a short peptide from pYES were used as controls to calculate what values were not significantly different (shaded areas). Host factors whose overexpression significantly enhanced repRNA accumulation are boxed in black, whereas one host factor that decreased repRNA accumulation is boxed in gray. Each value represents data from 6 to 12 independent samples. Note that Cdc34p was expressed without the zz tag from a pYES vector.
These protein overexpression experiments revealed that two of the host proteins tested affected repRNA accumulation dramatically (Fig. 4). These were Hbs1p translation factor and Cdc34p E2 Ub-conjugating enzyme, which increased repRNA levels by ∼3.5- and ∼2.2-fold, respectively. An additional five host proteins, namely, the Jjj1p cochaperone, the Pol30p proliferating cell nuclear antigen transcription factor, the Arp8p actin-related protein, the Erb1p rRNA maturation protein, and the Trz1p tRNA-processing protein, increased repRNA accumulation by 40 to 60% compared to what was seen for yeast carrying the pYES expression vector (Fig. 4). In contrast, overexpression of Ddr48p, a DNA damage-responsive protein, led to a 63% ± 5% reduction in repRNA accumulation in yeast (Fig. 4), which is only slightly (but significantly) less than the inhibitory effect of the Apt2p control. Overexpression of the remaining 36 host proteins affected repRNA accumulation by less than 40% (Fig. 4). Overall, the above-described experiments demonstrated that ∼18% of the 44 host proteins tested could affect tombusvirus repRNA accumulation when overexpressed in yeast. We could not exclude the possibility that several of the overexpressed tagged proteins are not fully functional under the expression conditions.
Effect of overexpression of selected host proteins on TBSV RNA recombination in yeast.
The effect on tombusvirus RNA recombination was tested for 44 of the identified host proteins that interacted with p33 by taking advantage of the previously developed tombusvirus recombination system in yeast (51) with some modifications. The recombination assay was similar to the above-described replication assay, except that it was based on a highly recombinogenic repRNA termed DI-AU-FP, which contains an AU-rich recombination hot spot (51, 54). In this assay, recombination takes place between two molecules of DI-AU-FP RNAs (50, 51). The accumulation of the recombinant RNAs (recRNAs) was estimated using Northern blotting (Fig. 5). Importantly, we calculated the ratio of recRNA to repRNA (DI-AU-FP), which is more informative about recombination than recRNA levels alone (50, 51). Among the 44 host proteins tested, 4 affected TBSV RNA recombination dramatically by enhancing the ratio of recRNA by ∼8- to 12-fold (Fig. 5). Overexpression of other host proteins tested had lesser effects on TBSV recRNA accumulation (not shown).
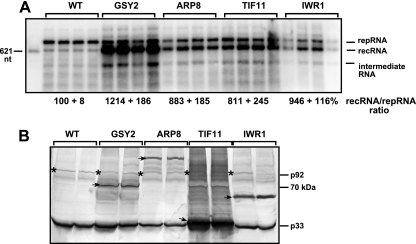
FIG. 5.
Effect of overexpression of selected host proteins interacting with p33 replication proteins on recombination by a TBSV repRNA in yeast. (A) Northern blotting of total RNA obtained from yeast overexpressing zz-tagged yeast proteins as shown. The host proteins were overexpressed separately from the galactose-inducible GAL1 promoter 20 h prior to launching the replication of DI-AU TBSV repRNA, a highly recombinogenic repRNA (7). Then, the yeast culture was grown in the presence of 50 μM copper sulfate at 29°C for 24 h, followed by a 1:10 dilution into the same medium. The recombinant repRNAs were measured in samples obtained from yeast cultures 48 h after the dilution step. Note that the ratio of recRNA to repRNA was calculated, since that reflects the frequency of recombination more accurately than does the amount of recRNA, which is dependent on both the frequency of recombination and the amount of the template (original parental repRNA). The ratio of recRNA to repRNA in the control yeast expressing a short peptide was set to 100%. Each experiment was performed six to eight times. (B) Western blotting showing the overexpressed zz-tagged host proteins (marked with arrows) and the tombusvirus p33 and p92pol (marked with asterisks) replication proteins.
Among the identified proteins, Gsy2p glycogen synthase enhanced the recRNA ratio by 12-fold, the most among the 44 host proteins tested (Fig. 5A). Overexpression of the Arp8p actin-related protein, the Tif11p translation initiation factor eIF1A, and Iwr1p (unknown function) increased the ratio of recRNA to repRNA by 8- to 9.5-fold (Fig. 5A). Because a high amount of p92pol has been shown to increase TBSV recombination (16), we also tested p33 and p92pol levels in yeast overexpressing host proteins. The Western analysis showed no significant increase in p92pol levels in comparison with p33 in the Gsy2p, Arp8p, Tif11p, and Iwr1p overexpression strains (Fig. 5B), suggesting that these host proteins do not affect RNA recombination through changing the p33: p92pol ratio but via a different, yet-uncharacterized mechanism(s).
Cdc34p is present within the purified tombusvirus replicase complex.
To further test the functional relevance of the identified host proteins interacting with p33, we selected Cdc34p based on its intriguing function as an E2 Ub-conjugating enzyme. Moreover, Cdc34p might be present within the tombusvirus replicase complex, since Cdc34p matches well with an unidentified yeast protein (termed X factor) with a pI value of ∼4 and a molecular mass of ∼35 kDa that has been detected in the highly purified tombusvirus replicase complex via two-dimensional gel electrophoresis (49). On the other hand, the other intriguing host protein, Hbs1p, is far less characterized and its function will be studied in the future.
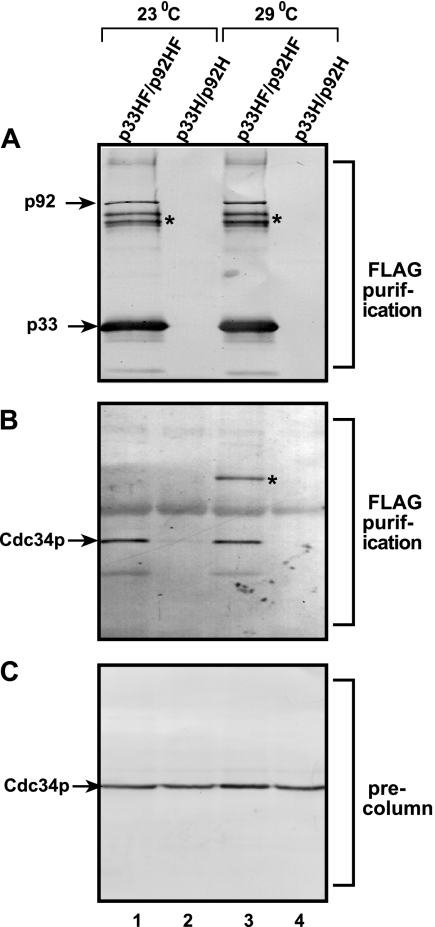
To test if Cdc34p is present within the highly purified tombusvirus replicase complex, we performed Western blotting with Cdc34p-specific antibody by use of a purified preparation of the detergent-solubilized tombusvirus replicase obtained from yeast grown at 23°C (optimal for TBSV replication) or 29°C (optimal for protein expression) (Fig. 6). The Flag affinity-purified tombusvirus replicase containing Flag-tagged p33 and p92pol replication proteins (Fig. 6A, lanes 1 and 3) contained the native Cdc34p expressed from its natural chromosomal position (Fig. 6B, lanes 1 and 3). Copurification of Cdc34p with the Flag-tagged replication proteins indicated that Cdc34p is a component of the membrane-bound replicase complex. The preparation was RNase treated prior to loading to the affinity column, suggesting that Cdc34p was likely retained on the Flag column via its binding to the replication proteins and not via the viral repRNA bound to the replication proteins. The control Flag-purified preparations from yeast expressing His6-tagged p33 and p92pol (Fig. 6A, lanes 2 and 4) lacked these replication proteins as well as Cdc34p (Fig. 6B, lanes 2 and 4), excluding the possibility that Cdc34p is a contaminating protein bound nonspecifically to the beads. Therefore, we propose that the previously detected X factor in the tombusvirus replicase complex (49) is Cdc34p, based on its copurification with the viral replicase (Fig. 6) and its physical properties (pI of 3.96 and molecular mass of 34 kDa).
FIG. 6.
Copurification of Cdc34p with the tombusvirus replicase complex. (A) FLAG- and His6-tagged p33 (p33HF) and p92pol (p92HF) replication proteins were purified after detergent-based solubilization of a membrane-enriched fraction of yeast on a FLAG affinity column. Western blotting with anti-His6 antibody detected the presence of p33 and p92pol in the purified replicase complex active in an in vitro replication assay (not shown), as demonstrated by Serva and Nagy (49). The control samples were from yeast expressing His6-tagged p33 (p33H) and p92pol (p92H) prepared as described above for p33HF and p92HF. Asterisks show p33 dimers that are partly resistant to denaturing conditions. (B) Western blotting of the same samples as in panel A with anti-Cdc34p antibody. Note that native Cdc34p was expressed from its original promoter and its original location on the chromosome. Occasionally, the Cdc34p formed a dimer when the yeast was grown at 29°C (marked with an asterisk in lane 3). (C) Western blotting analysis with anti-Cdc34p antibody showing similar amounts of Cdc34p present in the membrane-enriched fractions prior to affinity purification.
Downregulation of the Cdc34p level decreases TBSV repRNA accumulation in yeast.
To test if downregulation of the Cdc34p level affects TBSV repRNA accumulation, we used doxycycline-regulatable expression of Cdc34p from its original chromosomal location (25). Because our preliminary experiments suggested that Cdc34p has a long half-life (not shown), we downregulated Cdc34p expression 6 h prior to launching repRNA replication. Indeed, the level of Cdc34p expression was ∼10 times lower in yeast grown in the presence of doxycycline than it was in its absence (Fig. 7B, bottom, lanes 3 and 4 versus 1 and 2). The accumulation of repRNA decreased ∼3-fold in yeast grown in the presence of doxycycline (Fig. 7B, top, lanes 4 to 6), whereas the amount of p33 replication protein did not change (Fig. 7B, bottom, lanes 1 to 4), demonstrating that Cdc34p is important for tombusvirus replication.
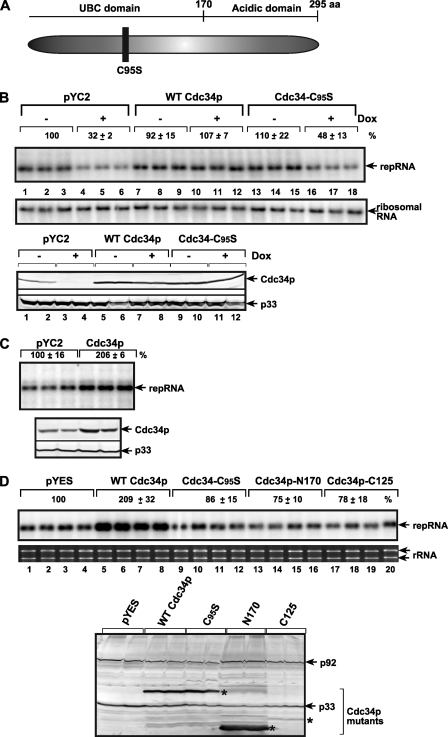
FIG. 7.
The effect of downregulation and overexpression of WT and mutant Cdc34p on TBSV repRNA accumulation in yeast. (A) Schematic representation of the known domains in Cdc34p. The active-site mutation that abolishes the E2 Ub conjugation activity of Cdc34p is shown with a black box. (B) Northern blotting of repRNA accumulation in yeast with downregulated expression of Cdc34p. The native Cdc34p was expressed from its original location but via the doxycycline (Dox)-regulatable Tet expression promoter. The yeast also expressed a short peptide from pYC2 (as a control), the WT Cdc34p, and the C95S active-site mutant of Cdc34p from low-copy-number plasmid pYC2. Yeast was grown in the absence or presence of 10 μg/ml doxycycline as indicated. Doxycycline was added 6 h prior to launching TBSV repRNA replication. The accumulation level of repRNA was measured 22 h after induction using ImageQuant software. rRNA was used as a loading control (see bottom panel). Western blotting results show the levels of Cdc34p and p33 for the above-described yeast samples. (C) Overexpression of Cdc34p increases repRNA accumulation in yeast. (Top) Northern blotting shows the accumulation of TBSV repRNA in yeast overexpressing of a short peptide from pYC2 or WT Cdc34p in BY4741; (bottom) Western blotting results show the levels of Cdc34p and p33 in the above-described yeast samples. Further details are as described for panel B. (D) Effect of overexpression of WT or mutated His6-tagged Cdc34p from high-copy-number plasmid pYES on repRNA accumulation in BY4741 yeast. The shown Cdc34p mutants are the E2 site mutant (C95S) and the C-terminally (N170; deletion of aa 171 to 295) and the N-terminally (C125, deletion of aa 1 to 170) truncated versions (Fig. 7A). (Bottom) Western blotting to show the overexpressed His6-tagged Cdc34p mutants. Further details are as described for panel B.
Downregulation of Cdc34p level decreases the activity of the tombusvirus replicase.
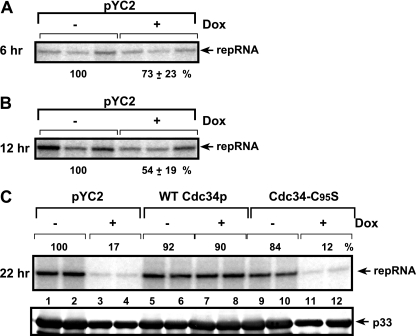
To test if a decreased amount of Cdc34p could directly affect the activity of the tombusvirus replicase, we performed in vitro replicase assays with a membrane-enriched fraction derived from yeast. The membrane-enriched fraction contains the tombusvirus replicase and is capable of performing TBSV repRNA synthesis in vitro using the copurified repRNA as a template. To compare similar amounts of replicase complexes, we adjusted the p33 content in the membrane-enriched fractions obtained from yeast cultured with or without doxycycline (17, 38, 39). These in vitro experiments have shown that the tombusvirus replicase obtained from yeast with downregulated Cdc34p 22 h after launching TBSV replication was only 17% as active as the control preparation from yeast grown in the absence of doxycycline (Fig. 8C, lanes 3 and 4 versus 1 and 2). Similarly prepared replicase preparations at earlier time points after launching TBSV replication, such as 6 and 12 h, showed only 73% and 54% decreases (Fig. 8A and B), suggesting that Cdc34p was still more readily available during the formation of the replicase complex at the early time points than at the late time point. Altogether, the in vitro assay demonstrated that Cdc34p is critical for efficient tombusvirus replicase activity.
FIG. 8.
Decreased replicase activity in the presence of a low Cdc34p level. A replicase activity assay with membrane-enriched preparations obtained from yeast expressing high or low levels of Cdc34p, based on the addition of 10 μg/ml doxycycline (Dox) to the growth medium. Doxycycline was added 6 h prior to launching TBSV replication. The yeast samples were taken 6 h (A), 12 h (B), or 22 h (C) after the induction of TBSV replication. The membrane-enriched fraction contained the endogenous repRNA template that was used during the in vitro replicase assay in the presence of 32P-UTP and the other unlabeled ribonucleotide triphosphates. Panel C also shows the replicase assay with WT and C95S Cdc34p proteins. Note that the in vitro activities of the tombusvirus replicase were normalized based on p33 levels (to adjust for some differences in the p33 levels, as shown in the bottom panel).
The Ub conjugation function of Cdc34p is important for TBSV repRNA replication.
Cdc34p has two functional domains: the 170-aa N-terminal UBC Ub conjugation domain conserved in E2s and the unique C-terminal acidic domain involved in protein/substrate binding (18). To test which function/domain of Cdc34p is important for tombusvirus replication, we used Cdc34p mutants in a complementation assay. We found that overexpression of Cdc34-C95S with inactive Ub-conjugating function from a plasmid could not complement TBSV repRNA accumulation in yeast with downregulated Cdc34p expression from the chromosome, whereas the full-length wild-type (WT) Cdc34p could complement repRNA accumulation (Fig. 7B, lanes 16 to 18 versus 10 to 12). Moreover, the N-terminal and C-terminal portions of Cdc34p rendered Cdc34p nonfunctional in a complementation assay (Fig. 7D, lanes 13 to 20), suggesting that both the UBC and the acidic domains of Cdc34p are important during TBSV replication.
In addition, we found a normal level of replicase activity obtained from yeast expressing the WT Cdc34p from a centromeric, low-copy-number plasmid, while Cdc34p was downregulated from the chromosomal location (Fig. 8C, lanes 7 and 8). Thus, the plasmid-borne WT Cdc34p can function in the viral replicase. On the other hand, the mutated Cdc34-C95S protein with inactive Ub-conjugating function expressed from a plasmid could not complement the downregulated Cdc34p from the chromosomal location in a replicase assay (Fig. 8C, lanes 11 and 12). These experiments suggest that the Ub conjugation function of Cdc34p is important for the function of the tombusvirus replicase.
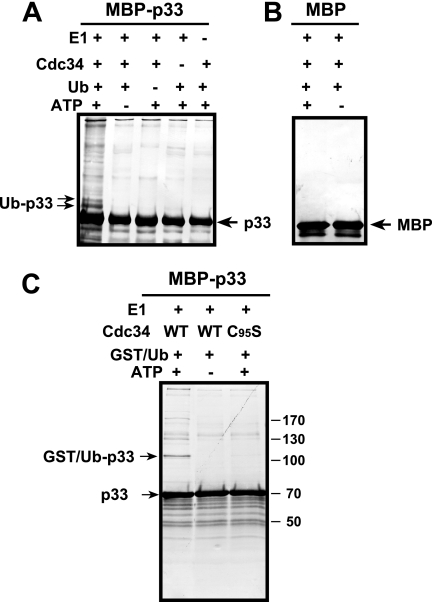
Ubiquitination of p33 by Cdc34p in vitro.
Since Cdc34p is part of the tombusvirus replicase complex, it also binds to p33 directly and the ubiquitination function of Cdc34p is important for the replicase function (Fig. 8); therefore, we wanted to test if Cdc34p could directly ubiquitinate p33 replication protein in the absence of Skp1p/cullin/F-box (SCF) E3 ligase complex, which is involved in substrate selection during normal cellular functions (48). To this end, we performed an in vitro ubiquitination assay with purified recombinant proteins. The in vitro assay demonstrated that Cdc34p could ubiquitinate a fraction of p33 replication protein, causing ∼9-and ∼18-kDa shifts in the mobility of p33 (Fig. 9A, lane 1). This shift in mobility is expected if a single Ub or two Ubs are added to p33 (mono- and biubiquitination). The higher-molecular-mass products detected on the gels could represent p33 proteins with multi- and/or polyubiquitination (Fig. 9A, lane 1). The ubiquitination of p33 by Cdc34p required Uba1p E1 protein, Ub, and ATP, and it did not take place if one of these components was missing in the assay (Fig. 9A, lanes 2 to 5). A similar assay with the purified MBP did not yield ubiquitination of MBP at a detectable level (Fig. 9B, lanes 6 and 7), suggesting that p33 is specifically ubiquitinated by Cdc34p. Moreover, the increase in the molecular mass of p33 was ∼35 kDa when we used GST-tagged Ub during a standard in vitro ubiquitination assay (Fig. 9C), confirming that Ub was added to p33 in the in vitro ubiquitination assay. A mutated Cdc34-C95S protein with inactive Ub-conjugating function (Fig. 9C), as well as the N- or C-terminally truncated Cdc34p variants (not shown), could not ubiquitinate p33, supporting the model that the ubiquitination of p33 was a specific feature of Cdc34p.
FIG. 9.
Ubiquitination of p33 by Cdc34p in vitro is independent of SCF E3 ligase complex. The in vitro ubiquitination was carried out in reaction mixtures containing 50 nM GST-Uba1p E1-activating enzyme (purified from yeast), 500 nM purified recombinant GST-Cdc34p (purified from E. coli), 100 μM His-Ub, 2 mM ATP, and purified recombinant MBP-p33 (A) or MBP as the negative control (B). The ubiquitination was analyzed by Western blotting using an anti-MBP antibody. The mono- and biubiquitinated p33s are marked with arrows. Note that monoubiquitination introduces an increase of ∼9 kDa to the molecular mass of p33. (C) The in vitro ubiquitination of p33 by Cdc34p was carried out as described above except that GST-tagged Ub (GST/Ub) was used, leading to a 35-kDa increase in the size of MBP-p33 (shown as p33) as well as higher-molecular-mass products indicating mono-, bi-, and multiubiquitination of p33. Note that we also tested an ubiquitination-deficient mutant of Cdc34p (C95S).
Ubiquitination of the p33 replication protein in yeast.
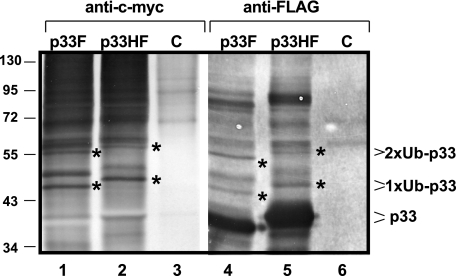
To demonstrate the possible ubiquitination of p33, we FLAG affinity purified His6/FLAG-tagged p33HF from yeast coexpressing Ub tagged with c-Myc from a plasmid. In these experiments, p33HF was solubilized from the membrane fraction, followed by purification and Western blotting to detect the addition of c-Myc-Ub to p33HF via anti-c-Myc antibody. Interestingly, we found mono- and biubiquitinated p33HF based on detection by anti-c-Myc antibody as well as a shift in the molecular mass of p33HF (mono- and biubiquitination cause ∼8- and 16-kDa increases, respectively) (Fig. 10, lane 2). The identified bands likely represent ubiquitinated p33HF because similar experiments with p33 tagged with FLAG only (termed p33F, which is about 2 kDa smaller than p33HF) resulted in slightly faster-migrating mono- and biubiquitinated p33F (Fig. 10, lane 1). The change in the migration pattern of the purified p33HF versus p33F proteins supports the idea that these bands represent various ubiquitinated p33 and excludes the possibility that they represent ubiquitinated contaminating host proteins in our FLAG affinity-purified samples. Moreover, the control sample containing His6/FLAG-tagged peptide (expressed from pESC-His plus YEp105 plus pYC-HF) lacked similar ubiquitinated proteins (Fig. 10, lane 3). We also detected putative higher-molecular-mass p33 derivatives with multiubiquitination. However, these bands could also represent high-molecular-mass ubiquitinated host proteins which could have been copurified with p33. These host proteins could be part of complexes containing p33, since similar proteins were absent from the control samples (Fig. 10, lanes 3 and 6).
FIG. 10.
Ubiquitination of p33 replication protein in yeast. The membrane-bound p33HF (tagged with FLAG and His6) or p33F (tagged with FLAG only) was purified via FLAG affinity chromatography after solubilization from yeast coexpressing c-Myc-tagged Ub from a plasmid. The ubiquitinated p33 was detected by Western blotting using anti-c-Myc antibody, as were the expected 8- and 16-kDa increases in molecular mass for mono- and biubiquitination (1xUb and 2xUb, respectively). The p33-specific bands can be recognized due to the 2-kDa difference between p33HF (lane 2) and p33F (lane 1). Note that the identity of the p33-specific band between the mono- and biubiquitinated bands is currently unknown. The origin of high-molecular-mass ubiquitinated proteins is also unknown; they are probably copurified ubiquitinated host proteins. These products, however, are missing from the control sample (a His6-FLAG peptide expressed from pYC-HF). Lanes 4 to 6 show a Western blot of the same samples as shown in lanes 1 to 3, except using anti-FLAG antibody. The ubiquitinated p33 was identified based on the expected 8- and 16-kDa increases in molecular mass for mono- and biubiquitination, respectively (bands marked with asterisks), and the 2-kDa difference between p33HF (lane 5) and p33F (lane 4).
To further demonstrate that the identified bands represent ubiquitinated p33HF, we also analyzed the same FLAG-purified p33HF or p33F samples by SDS-PAGE/Western blotting with anti-FLAG antibody (49). Accordingly, we detected p33-specific bands with ∼8- and 16-kDa molecular mass increases (Fig. 10, lanes 4 and 5) that are consistent with mono- and biubiquitinated p33HF or p33F. The mono- and biubiquitinated p33 proteins represented a small fraction (less than 5%) of the total p33, suggesting that not all p33 is ubiquitinated or that the deubiquitination process is rapid in yeast cells.
DISCUSSION
To exploit host cells, tombusvirus-coded replication proteins p33 and p92pol likely interact with a currently unknown number of host proteins. The interacting host proteins could be recruited and be part of the replicase complex to aid replication, and/or they might facilitate the transport of viral proteins in the cells and/or the assembly of the viral replicase or provide regulatory functions. To catalogue the interacting host proteins, we have taken a proteome-wide approach with a yeast protoarray carrying 4,088 purified proteins that covers ∼70% of yeast proteins. This led to the identification of 58 proteins binding to p33 (Table 1), whereas 11 additional host proteins bound only to p92C and not to p33 (Table 2). Similar to other genome-wide approaches, protoarray might lead to false positives and false negatives as well. The false negatives could be due to many factors, including among others (i) the use of general binding conditions, which are not optimized for individual protein-protein interactions; (ii) the absence of cofactors or membrane surfaces under the in vitro conditions; and (iii) inactive or denatured proteins present on the surface of the chip. Indeed, we did not detect significant binding between the purified p33 and Ssa1p, a heat shock 70 protein which has been shown to be part of the replicase complex (49) and bind to p33 in the split-Ub assay (Fig. 3). This suggests that multiple complementary approaches are needed to identify all host proteins interacting with the replication proteins.
The identification of many host proteins binding to p33 makes the characterization of their interactions with p33 protein challenging. A fraction of interactions could be due to false positives, which interact with p33 only under in vitro conditions. However, we performed additional in vitro binding and split-Ub two-hybrid assays for ∼20 host proteins, which further demonstrated interaction between the given host protein and p33. Overall, these data support the model that p33 might interact with a relatively large number of host proteins in infected cells.
Comparison of the set of host proteins interacting with p33 with the host genes identified during genome-wide screens of deletion/downregulatable gene libraries (17, 35, 50, 51) revealed only two common hits: BUD21 and GPH1. This could be due to redundant functions provided by many host proteins during TBSV replication, as demonstrated for the heat shock protein 70 chaperone family and the TDH2 and TDH3 genes coding for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), which are required for TBSV replication (49, 62). Therefore, it is likely that multiple, noncomplementary genomics- and proteomics-based screens will be needed to identify all the host genes affecting TBSV replication.
Many of the identified host proteins binding to p33 were translation factors, ribosomal proteins, and RNA-binding proteins. These host proteins could affect/regulate the activity of p33, which is important for the recruitment of the TBSV RNA into replication and the assembly of the viral replicase complex (33, 39, 41, 45) and possibly for the rescue of the viral RNA from translation to allow the viral RNA to enter replication. Similar host proteins, including translation factors, ribosomal proteins, and RNA-binding proteins, have also been identified during genome-wide screens (17, 35, 50, 51), suggesting that these factors play critical, though somewhat redundant, roles in TBSV replication.
Interaction of p33 with Ub-specific proteins.
A major group of host proteins that bound to p33 are involved in the Ub pathway of protein modification/degradation. These proteins include E2 Ub-conjugating enzyme (CDC34), a Ub protein ligase (RSP5), a Ub-activating enzyme (UBA1), and two UBPs (UBP10 and UBP15). In addition, Ub-associated factors, such as the RAD6 E2 Ub-conjugating enzyme, the BRE1 Ub protein ligase, protein monoubiquitination-involved LGE1, the DOA4 deubiquitination enzyme, UBP3, and the BRE5 Ub protease cofactor, have been identified during genome-wide screens (17, 35, 50, 51). The identification of many host proteins involved in the Ub pathway that affected TBSV replication/recombination suggests that ubiquitination plays a critical role in TBSV replication.
The binding of p33 with Ub-specific proteins suggests that p33 might be modified posttranslationally by ubiquitination. Posttranslational modification of host proteins by the highly conserved 76-aa Ub regulates many cellular processes, including immune response, signal transduction, transcription, protein trafficking, and autophagy. Four types of enzymes are involved in protein ubiquitination: E1 is involved in Ub activation, E2s (Ub-conjugating enzymes) function in Ub transfer to the proteins, and E3s are involved in substrate selection, while de-ubiquitination enzymes, which remove the Ub from the proteins, are the fourth type of enzyme. Viruses are known to take advantage of the ubiquitination pathway (6, 52, 53, 60). Accordingly, ubiquitination and deubiquitination (3, 21, 59) of viral proteins are major mechanisms regulating protein stability and function in infections with DNA and RNA viruses (13, 23, 24, 30, 32, 42, 61, 65). For plus-stranded RNA viruses, ubiquitination has been documented in the cases of hepatitis C virus, coxsackievirus, and coronaviruses (5, 47, 59, 64), and a host Ub gene sequence was found in bovine viral diarrhea virus genomic RNA via RNA recombination (4). Interaction with a Ub-like protein was found to enhance the ubiquitination and degradation of hepatitis C virus RNA-dependent RNA polymerase (11). In spite of intensive efforts, the roles of ubiquitination in plus-stranded RNA replication and infections are currently poorly characterized.
Cdc34p Ub-conjugating enzyme affects replicase activity and is a component of the tombusvirus replicase complex.
An important finding from the protoarray approach is the identification of Cdc34p-p33 interaction, which was also confirmed in in vitro binding experiments (Fig. 2). Also, Cdc34p has been shown to be part of the tombusvirus replicase complex based on copurification experiments (Fig. 6). Moreover, Cdc34p matches well with a previously detected protein in a two-dimensional gel of the double-affinity-purified replicase complex based on its rare features (pI, ∼4; molecular mass, 34 kDa), but its amount was too little for mass spectrometry-based identification (49). Overall, the identification of Cdc34p within the tombusvirus replicase complex validates the usefulness of the protoarray approach.
Cdc34p, also called Ubc3p, is an essential protein involved in many cellular processes and is present in both the cytoplasm and the nucleus. Its known activities include cell cycle regulation, maintenance of cell wall integrity, and roles in mitotic spindle function and in the degradation of cyclin-dependent kinase (40, 48). The best-characterized interaction of Cdc34p is with the SCF(Cdc4) E3 Ub ligase, which ubiquitinates several host proteins in vivo (40, 48). However, ubiquitination of p33 did not require SCF(Cdc4) E3 Ub ligase in vitro, and recombinant Cdc34p was able to perform this reaction in the absence of an E3 ligase (Fig. 9). Interestingly, RAD6 (UBC2) E2 Ub-conjugating enzyme, which is similar to the CDC34 enzyme, has also been shown to affect TBSV replication in yeast (35). Thus, it is possible that more than one E2 Ub-conjugating enzyme are involved in TBSV replication.
The relevance of Cdc34p-p33 interaction is supported by the findings that the downregulation of Cdc34p decreased TBSV repRNA accumulation by 3-fold, whereas the overexpression of Cdc34p led to an ∼2-fold increase in repRNA levels. Therefore, Cdc34p might have a regulatory function in the replicase complex. This function could be due at least in part to the ability of Cdc34p to ubiquitinate p33, since a mutant of Cdc34p inactive in ubiquitination could not complement the downregulated Cdc34p to support TBSV accumulation and increase the activity of the tombusvirus replicase (Fig. 7 and 8). It is also possible that Cdc34p might affect TBSV replication through modulating the levels/activities of other host factors.
We found that p33 is mono-, bi-, and polyubiquitinated in vitro, which could modify the function of p33. Importantly, we have also detected mono- and biubiquitinated p33 in yeast cells (Fig. 10), suggesting that the ubiquitination of p33 also takes place in intact cells. It is possible that p33 ubiquitination plays a role in the recruitment of host factors such as Vps23p (D. Barajas and P. D. Nagy, unpublished data), which binds to ubiquitinated membrane proteins and it also affects TBSV repRNA replication (35).
Usefulness of the protoarray approach in the identification of host proteins.
In addition to the above-described data on Cdc34p, future works will likely establish the roles of other host proteins interacting with p33 replication proteins in TBSV replication. It is worth noting that the presented data are likely an underestimation of the number of host proteins binding to p33 and p92pol. This is due to the presence of some possibly inactive purified proteins on the array, the lack of optimization of binding conditions for each protein, or the lack of host protein complexes on the array. In addition, the protoarray contains only 70% of the known/predicted number of proteins expressed in yeast. Regardless of these limitations, the protoarray approach could provide a rapid tool for the identification or confirmation of p33 binding host proteins.
Acknowledgments
We thank Judit Pogany and David Smith for valuable comments. We are grateful to have obtained the Cdc34 antibody from Mark Goebl (Indiana University), the GST-tagged yeast ORF library from Brenda Andrew (University of Toronto), and YEp105 from Ralf Erdmann.
This work was supported by the National Science Foundation (IOB-0517218), NIH-NIAID, and the Kentucky Tobacco Research and Development Center. D. Barajas was supported by a fellowship from the Spanish Ministry of Education and Science.
Footnotes
Published ahead of print on 7 May 2008.
REFERENCES
- 1.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 778181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfonzo, J. D., T. R. Crother, M. L. Guetsova, B. Daignan-Fornier, and M. W. Taylor. 1999. APT1, but not APT2, codes for a functional adenine phosphoribosyltransferase in Saccharomyces cerevisiae. J. Bacteriol. 181347-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balakirev, M. Y., M. Jaquinod, A. L. Haas, and J. Chroboczek. 2002. Deubiquitinating function of adenovirus proteinase. J. Virol. 766323-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baroth, M., M. Orlich, H. J. Thiel, and P. Becher. 2000. Insertion of cellular NEDD8 coding sequences in a pestivirus. Virology 278456-466. [DOI] [PubMed] [Google Scholar]
- 5.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 7915189-15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barry, M., and K. Fruh. 2006. Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci. STKE 2006pe21. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, C. P., E. Serviene, and P. D. Nagy. 2006. Suppression of viral RNA recombination by a host exoribonuclease. J. Virol. 802631-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellison, M. J., and M. Hochstrasser. 1991. Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J. Biol. Chem. 26621150-21157. [PubMed] [Google Scholar]
- 9.Fetchko, M., D. Auerbach, and I. Stagljar. 2003. Yeast genetic methods for the detection of membrane protein interactions: potential use in drug discovery. BioDrugs 17413-424. [DOI] [PubMed] [Google Scholar]
- 10.Fetchko, M., and I. Stagljar. 2004. Application of the split-ubiquitin membrane yeast two-hybrid system to investigate membrane protein interactions. Methods 32349-362. [DOI] [PubMed] [Google Scholar]
- 11.Gao, L., H. Tu, S. T. Shi, K. J. Lee, M. Asanaka, S. B. Hwang, and M. M. Lai. 2003. Interaction with a ubiquitin-like protein enhances the ubiquitination and degradation of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 774149-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gelperin, D. M., M. A. White, M. L. Wilkinson, Y. Kon, L. A. Kung, K. J. Wise, N. Lopez-Hoyo, L. Jiang, S. Piccirillo, H. Yu, M. Gerstein, M. E. Dumont, E. M. Phizicky, M. Snyder, and E. J. Grayhack. 2005. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 192816-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geoffroy, M. C., G. Chadeuf, A. Orr, A. Salvetti, and R. D. Everett. 2006. Impact of the interaction between herpes simplex virus type 1 regulatory protein ICP0 and ubiquitin-specific protease USP7 on activation of adeno-associated virus type 2 rep gene expression. J. Virol. 803650-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, D. A., H. Zhu, X. Zhu, T. Royce, M. Gerstein, and M. Snyder. 2004. Regulation of gene expression by a metabolic enzyme. Science 306482-484. [DOI] [PubMed] [Google Scholar]
- 15.Huang, J., H. Zhu, S. J. Haggarty, D. R. Spring, H. Hwang, F. Jin, M. Snyder, and S. L. Schreiber. 2004. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc. Natl. Acad. Sci. USA 10116594-16599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaag, H. M., J. Stork, and P. D. Nagy. 2007. Host transcription factor Rpb11p affects tombusvirus replication and recombination via regulating the accumulation of viral replication proteins. Virology 368388-404. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, Y., E. Serviene, J. Gal, T. Panavas, and P. D. Nagy. 2006. Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J. Virol. 807394-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolman, C. J., J. Toth, and D. K. Gonda. 1992. Identification of a portable determinant of cell cycle function within the carboxyl-terminal domain of the yeast CDC34 (UBC3) ubiquitin conjugating (E2) enzyme. EMBO J. 113081-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kushner, D. B., B. D. Lindenbach, V. Z. Grdzelishvili, A. O. Noueiry, S. M. Paul, and P. Ahlquist. 2003. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. USA 10015764-15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, F., and S. W. Ding. 2006. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60503-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindner, H. A. 2007. Deubiquitination in virus infection. Virology 362245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCartney, A. W., J. S. Greenwood, M. R. Fabian, K. A. White, and R. T. Mullen. 2005. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 173513-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mechali, F., C. Y. Hsu, A. Castro, T. Lorca, and C. Bonne-Andrea. 2004. Bovine papillomavirus replicative helicase E1 is a target of the ubiquitin ligase APC. J. Virol. 782615-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, C. L., J. S. Parker, J. B. Dinoso, C. D. Piggott, M. J. Perron, and M. L. Nibert. 2004. Increased ubiquitination and other covariant phenotypes attributed to a strain- and temperature-dependent defect of reovirus core protein μ2. J. Virol. 7810291-10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mnaimneh, S., A. P. Davierwala, J. Haynes, J. Moffat, W. T. Peng, W. Zhang, X. Yang, J. Pootoolal, G. Chua, A. Lopez, M. Trochesset, D. Morse, N. J. Krogan, S. L. Hiley, Z. Li, Q. Morris, J. Grigull, N. Mitsakakis, C. J. Roberts, J. F. Greenblatt, C. Boone, C. A. Kaiser, B. J. Andrews, and T. R. Hughes. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 11831-44. [DOI] [PubMed] [Google Scholar]
- 26.Monkewich, S., H. X. Lin, M. R. Fabian, W. Xu, H. Na, D. Ray, O. A. Chernysheva, P. D. Nagy, and K. A. White. 2005. The p92 polymerase coding region contains an internal RNA element required at an early step in tombusvirus genome replication. J. Virol. 794848-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy, P. D. 2008. Yeast as a model host to explore plant virus-host interactions. Annu. Rev. Phytopathol. doi: 10.1146/annurev.phyto.121407.093958. [DOI] [PubMed]
- 28.Nagy, P. D., and J. Pogany. 2008. Multiple roles of viral replication proteins in plant RNA virus replication. Methods Mol. Biol. 45155-68. [DOI] [PubMed] [Google Scholar]
- 29.Nagy, P. D., and J. Pogany. 2006. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology 344211-220. [DOI] [PubMed] [Google Scholar]
- 30.Nerenberg, B. T., J. Taylor, E. Bartee, K. Gouveia, M. Barry, and K. Fruh. 2005. The poxviral RING protein p28 is a ubiquitin ligase that targets ubiquitin to viral replication factories. J. Virol. 79597-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oster, S. K., B. Wu, and K. A. White. 1998. Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J. Virol. 725845-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278111-121. [DOI] [PubMed] [Google Scholar]
- 33.Panavas, T., C. M. Hawkins, Z. Panaviene, and P. D. Nagy. 2005. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology 33881-95. [DOI] [PubMed] [Google Scholar]
- 34.Panavas, T., and P. D. Nagy. 2003. Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology 314315-325. [DOI] [PubMed] [Google Scholar]
- 35.Panavas, T., E. Serviene, J. Brasher, and P. D. Nagy. 2005. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc. Natl. Acad. Sci. USA 1027326-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panavas, T., E. Serviene, J. Pogany, and P. D. Nagy. 2008. Genome-wide screens for identification of host factors in viral replication. Methods Mol. Biol. 451615-624. [DOI] [PubMed] [Google Scholar]
- 37.Panaviene, Z., J. M. Baker, and P. D. Nagy. 2003. The overlapping RNA-binding domains of p33 and p92 replicase proteins are essential for tombusvirus replication. Virology 308191-205. [DOI] [PubMed] [Google Scholar]
- 38.Panaviene, Z., T. Panavas, and P. D. Nagy. 2005. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J. Virol. 7910608-10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panaviene, Z., T. Panavas, S. Serva, and P. D. Nagy. 2004. Purification of the cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J. Virol. 788254-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petroski, M. D., and R. J. Deshaies. 2005. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 1231107-1120. [DOI] [PubMed] [Google Scholar]
- 41.Pogany, J., K. A. White, and P. D. Nagy. 2005. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 794859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poon, A. P., H. Gu, and B. Roizman. 2006. ICP0 and the US3 protein kinase of herpes simplex virus 1 independently block histone deacetylation to enable gene expression. Proc. Natl. Acad. Sci. USA 1039993-9998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajendran, K. S., and P. D. Nagy. 2003. Characterization of the RNA-binding domains in the replicase proteins of tomato bushy stunt virus. J. Virol. 779244-9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajendran, K. S., and P. D. Nagy. 2004. Interaction between the replicase proteins of Tomato bushy stunt virus in vitro and in vivo. Virology 326250-261. [DOI] [PubMed] [Google Scholar]
- 45.Rajendran, K. S., and P. D. Nagy. 2006. Kinetics and functional studies on interaction between the replicase proteins of Tomato bushy stunt virus: requirement of p33:p92 interaction for replicase assembly. Virology 345270-279. [DOI] [PubMed] [Google Scholar]
- 46.Rajendran, K. S., and P. D. Nagy. 2008. Surface plasmon resonance analysis of interactions between replicase proteins of tomato bushy stunt virus. Methods Mol. Biol. 451267-277. [DOI] [PubMed] [Google Scholar]
- 47.Ratia, K., K. S. Saikatendu, B. D. Santarsiero, N. Barretto, S. C. Baker, R. C. Stevens, and A. D. Mesecar. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA 1035717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadowski, M., A. Mawson, R. Baker, and B. Sarcevic. 2007. Cdc34 C-terminal tail phosphorylation regulates Skp1/cullin/F-box (SCF)-mediated ubiquitination and cell cycle progression. Biochem. J. 405569-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serva, S., and P. D. Nagy. 2006. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J. Virol. 802162-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serviene, E., Y. Jiang, C. P. Cheng, J. Baker, and P. D. Nagy. 2006. Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J. Virol. 801231-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serviene, E., N. Shapka, C. P. Cheng, T. Panavas, B. Phuangrat, J. Baker, and P. D. Nagy. 2005. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc. Natl. Acad. Sci. USA 10210545-10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shackelford, J., and J. S. Pagano. 2005. Targeting of host-cell ubiquitin pathways by viruses. Essays Biochem. 41139-156. [DOI] [PubMed] [Google Scholar]
- 53.Shackelford, J., and J. S. Pagano. 2004. Tumor viruses and cell signaling pathways: deubiquitination versus ubiquitination. Mol. Cell. Biol. 245089-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shapka, N., and P. D. Nagy. 2004. The AU-rich RNA recombination hot spot sequence of Brome mosaic virus is functional in tombusviruses: implications for the mechanism of RNA recombination. J. Virol. 782288-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shapka, N., J. Stork, and P. D. Nagy. 2005. Phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus adjacent to the RNA binding site affects viral RNA replication. Virology 34365-78. [DOI] [PubMed] [Google Scholar]
- 56.Smith, M. G., G. Jona, J. Ptacek, G. Devgan, H. Zhu, X. Zhu, and M. Snyder. 2005. Global analysis of protein function using protein microarrays. Mech. Ageing Dev. 126171-175. [DOI] [PubMed] [Google Scholar]
- 57.Sopko, R., B. Papp, S. G. Oliver, and B. J. Andrews. 2006. Phenotypic activation to discover biological pathways and kinase substrates. Cell Cycle 51397-1402. [DOI] [PubMed] [Google Scholar]
- 58.Stork, J., Z. Panaviene, and P. D. Nagy. 2005. Inhibition of in vitro RNA binding and replicase activity by phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus. Virology 34379-92. [DOI] [PubMed] [Google Scholar]
- 59.Sulea, T., H. A. Lindner, E. O. Purisima, and R. Menard. 2005. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J. Virol. 794550-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor, J. M., and M. Barry. 2006. Near death experiences: poxvirus regulation of apoptotic death. Virology 344139-150. [DOI] [PubMed] [Google Scholar]
- 61.Wang, J., A. N. Loveland, L. M. Kattenhorn, H. L. Ploegh, and W. Gibson. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 806003-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, R. Y., and P. D. Nagy. 2008. Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe 3178-187. [DOI] [PubMed] [Google Scholar]
- 63.White, K. A., and P. D. Nagy. 2004. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acid Res. Mol. Biol. 78187-226. [DOI] [PubMed] [Google Scholar]
- 64.Wong, J., J. Zhang, X. Si, G. Gao, and H. Luo. 2007. Inhibition of the extracellular signal-regulated kinase signaling pathway is correlated with proteasome inhibitor suppression of coxsackievirus replication. Biochem. Biophys. Res. Commun. 358903-907. [DOI] [PubMed] [Google Scholar]
- 65.Woo, J. L., and A. J. Berk. 2007. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 81575-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu, H., M. Bilgin, R. Bangham, D. Hall, A. Casamayor, P. Bertone, N. Lan, R. Jansen, S. Bidlingmaier, T. Houfek, T. Mitchell, P. Miller, R. A. Dean, M. Gerstein, and M. Snyder. 2001. Global analysis of protein activities using proteome chips. Science 2932101-2105. [DOI] [PubMed] [Google Scholar]
- 67.Zhu, H., M. Bilgin, and M. Snyder. 2003. Proteomics. Annu. Rev. Biochem. 72783-812. [DOI] [PubMed] [Google Scholar]
- 68.Zhu, J., K. Gopinath, A. Murali, G. Yi, S. D. Hayward, H. Zhu, and C. Kao. 2007. RNA-binding proteins that inhibit RNA virus infection. Proc. Natl. Acad. Sci. USA 1043129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]