Abstract
APOBEC3G (A3G) is a cytidine deaminase that can inhibit a wide range of retroviruses, including the para-retrovirus hepatitis B virus (HBV). The antiviral function of A3G depends on its incorporation into assembling viral particles. However, it remains enigmatic how A3G is specifically packaged into a variety of unrelated viruses. By adopting a native agarose gel electrophoresis assay that can specifically measure the levels of A3G incorporation into HBV nucleocapsids, we found that A3G is specifically packaged into replication-competent HBV nucleocapsids in a fashion that is dependent on both the viral reverse transcriptase (RT) and viral RNA packaging signal, ɛ. In contrast, A3G is not incorporated into empty capsids formed in the absence of RT or ɛ. We demonstrated that the packaged A3G was protected from protease digestion by the nucleocapsids, thus confirming its interior localization. We also showed that A3G could bind RT specifically in an RNA-independent manner, which may be responsible for mediating the specific incorporation of A3G into replication-competent nucleocapsids. Finally, we provide evidence that the N-terminal domain of A3G is required for packaging into HBV nucleocapsids.
Hepatitis B virus (HBV) is a major cause of acute and chronic viral hepatitis worldwide (16). An estimated 350 million individuals are chronically infected with HBV, many of whom will eventually develop severe liver diseases, including cirrhosis and hepatocellular carcinoma, one of the most common forms of human cancer. HBV belongs to the family Hepadnaviridae, which also includes viruses that infect other mammalian and avian species (e.g., the duck HBV). All hepadnaviruses contain a small (ca. 3.2-kb), partially double-stranded, relaxed-circular DNA genome, replicate this DNA through reverse transcription of an RNA intermediate, the pregenomic RNA (pgRNA), and are thus classified as para-retroviruses (47, 48, 52).
Initiation of reverse transcription and nucleocapsid assembly in HBV is carried out by the virally encoded reverse transcriptase (RT) (24, 52). RT initiates DNA synthesis by using a specific tyrosine residue located in its N-terminal domain (61, 69). This protein priming reaction requires the recognition and binding of RT to a stem-loop structure located at the 5′ end of the pgRNA called epsilon (ɛ) (41, 56). In addition to protein priming, the formation of this same RT-ɛ ribonucleoprotein (RNP) complex also triggers nucleocapsid assembly, leading to the specific incorporation of both the pgRNA and RT into replication-competent nucleocapsids (4, 41).
Recently, it was reported that the human cytidine deaminase APOBEC3G (A3G) can restrict the replication of HBV, as well as a number of different retroviruses (21, 42, 55). Initially, A3G was discovered due to its ability to decrease the infectivity of Vif-deficient human immunodeficiency virus (HIV) (49). It was shown to cause dC deamination on the viral minus-strand DNA, resulting in extensive G-to-A hypermutations in the viral genome (20, 32). However, unlike HIV, the main mode of inhibition of HBV DNA synthesis is via a deaminase-independent mechanism (39, 55), although it may induce editing in a small fraction of HBV DNA (5, 43, 53, 55). Subsequently, the same editing-independent antiviral function of A3G has been extended to HIV and other retroviruses as well (13, 38). Although it is still poorly understood, the editing-independent mechanism of A3G seems to target a very early stage during viral reverse transcription to block viral DNA synthesis within the nucleocapsid (39).
The antiviral activity of A3G requires its incorporation into assembling viral particles. It has been shown that A3G is packaged into the budding virions of Vif-deficient HIV (20, 35, 49). Although the mechanism by which A3G is incorporated into viral particles is still not fully understood, it is widely accepted that the HIV Gag protein is responsible for the incorporation of A3G into virions. In particular, in vitro studies have shown that A3G binds to the nucleocapsid region of the Gag polyprotein (3, 8, 34, 44). Moreover, deletion analysis of A3G has indicated that the linker region of A3G is required and sufficient for incorporation into Gag particles (8, 19, 34). The role played by RNA, either cellular or viral, in stabilizing this interaction and/or mediating A3G incorporation is also still unclear. Some studies have reported that the packaging of A3G into viral particles requires or is enhanced by the viral RNA (30, 54); however, others have suggested that viral RNA is not needed for A3G packaging (8, 37, 57, 58, 67).
The mechanism by which A3G is incorporated into HBV nucleocapsids remains unknown. By adopting a native agarose gel electrophoresis assay, developed previously to measure viral pgRNA packaging (66), we showed that incorporation of A3G into replication-competent HBV nucleocapsids was specifically dependent on both the viral RT and ɛ. Moreover, protease digestion of the nucleocapsids revealed that A3G was located within the capsids and not just associated on its surface. Additionally, we showed that A3G interacted with RT and that the N-terminal domain of A3G was needed for incorporation into HBV nucleocapsids.
MATERIALS AND METHODS
Cell culture, transfection, and antibodies.
All cells were maintained at 37°C in complete medium supplemented with 10% fetal bovine serum, as previously described (39). HepG2 cells were seeded in 60-mm or 100-mm dishes and transfected at 30 to 40% confluence with 4 μg or 10 μg of DNA, respectively, using FuGENE 6 (Roche) (39). 293T cells were seeded in 60-mm or 100-mm dishes and transfected at 70% confluence with 10 μg or 20 μg of DNA, respectively, using a CalPhos mammalian transfection kit (Clontech) (39). Rabbit anti-HBV core antibody was from DAKO. Rabbit anti-A3G antibodies were from the NIH (29, 36, 50). Mouse anti-Flag antibody was obtained from Sigma.
Plasmids.
pCMV-HBV contains the wild-type HBV 1.1-mer over-length genomic sequence driven by the cytomegalovirus (CMV) immediate-early promoter (14). pCIdA-HBV contains wild-type HBV sequence from 1876 to 1994 in the pCI vector (Promega). It expresses all the HBV proteins, but the 5′ ɛ RNA signal is deleted. pCMV-HBVpol- has a frameshift mutation within the RT gene which eliminates RT expression (17). pcDNA4-3XFlag-HP expresses an HBV RT protein that is tagged with a triple Flag epitope at its N terminus. pA3G-Flag is an SRα-driven expression vector encoding the human A3G gene fused to the Flag epitope tag at its amino terminus (39). pcDNA3.1-A3G plasmid (38) was kindly provided by Michael Malim (King's College London, London, England). A3G 1-104, A3G 1-180, A3G 105-384, and A3G 182-384 were derived from pA3G-Flag with appropriate restriction digestion and subcloning. They all retained the N-terminal Flag tag. A3G 104-384 and A3G 1-156 were constructed by using PCR to amplify the appropriate segment of A3G and cloned into either the same vector as the full-length tagged A3G (for A3G 1-156, retaining the N-terminal Flag tag) or into pcDNA3 without any tag (for A3G 104-384). All constructs were sequenced to confirm the deletions.
Native agarose gel electrophoresis assay.
Cell lysate was prepared from transfected cells on day 3 posttransfection by lysing cells in NP-40 lysis buffer (50 mM Tris, pH 8.0, 1 mM EDTA, pH 8.0, 1% NP-40) supplemented with complete protease inhibitors (Roche) for 10 min at 37°C. The cellular lysate was treated with 60 U of micrococcal nuclease and 5 mM CaCl2 at 37°C for 30 min. Subsequently, an additional 60 U of micrococcal nuclease was added and incubated for another 30 min. HBV RNA and DNA in cytoplasmic core particles were detected by resolving the nucleocapsid particles on a 0.8% agarose gel followed by Southern blot analysis (25, 39, 60). In principle, both the pgRNA and plus-strand DNA in the nucleocapsids could be detected using the plus-strand-specific riboprobe. However, the nucleic acid signals detected under conditions for assaying pgRNA packaging were predominantly derived from pgRNA packaged into the nucleocapsid, with very little contribution by the plus-strand DNA (25). For detection of viral DNA associated with nucleocapsids, the agarose gel was treated with 0.5 M NaOH to remove RNA before transfer onto a nitrocellulose membrane. The amount of A3G incorporated into the cytoplasmic core particles and the levels of HBV capsids were determined subsequently by Western blot analysis. Following the nucleic acid analyses, the nitrocellulose membrane was rewet in distilled H2O, blocked, and probed with the anti-A3G antibodies (1:2,000) overnight at 4°C. The next day, the membrane was washed and probed for 1 hour at room temperature with a peroxidase-labeled polyclonal anti-rabbit immunoglobulin G (IgG; diluted 1:20,000; Southern Biotechnology Associates), developed with the Western Lightning chemiluminescent reagent plus (Perkin-Elmer), and exposed to light-sensitive films. The same membrane was subsequently reprobed for assembled HBV capsids by stripping for 30 min at 50°C in Western stripping solution (62.5 mM Tris, pH 6.8, 2% sodium dodecyl sulfate [SDS], 100 mM β-mercaptoethanol [β-ME]), blocked, and then probed for 1 hour at room temperature with the anti-HBV core antibody (1:2,000). The membrane was washed and then probed for 30 min at room temperature with the polyclonal anti-rabbit IgG (1:20,000) and developed as described above.
Immunoprecipitation assay.
Forty-eight hours after transfection in 100-mm dishes, transfected cells were washed twice with phosphate-buffered saline (PBS) and then lysed in immunoprecipitation (IP) lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40) containing complete protease inhibitors (Roche) for 30 min on ice. Cell lysates were either left untreated or treated with 200 μg/ml RNase A, precleared, and then incubated with anti-Flag antibodies overnight at 4°C. Immune complexes were pulled down with immobilized protein A/G (Pierce) for 3 h at 4°C and washed with IP lysis buffer before boiling in 2× SDS electrophoresis sample buffer. Samples were loaded onto an SDS-polyacrylamide gel electrophoresis (PAGE) gel and subjected to Western blot analysis.
Sucrose gradient ultracentrifugation.
Three days posttransfection, transfected HepG2 cells were lysed in 50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% NP-40, and 0.05% β-ME supplemented with complete protease inhibitors (Roche) for 30 min on ice. Cell lysates were treated with 200 μg/ml RNase A for 60 min at 37°C, followed by an additional treatment by adding 200 μg/ml RNase A and 20 mM EDTA, pH 8.0, and incubating for 60 min at 37°C. Lysates were subjected to ultracentrifugation over a 10% to 20% sucrose gradient in a Beckman SW-41 rotor at 26,000 rpm for 16 h to pellet the viral nucleocapsids (40). The capsid pellets were resuspended in 20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM EDTA, 0.01% Triton X-100, 0.1% NP-40, and 0.05% β-ME, incubated at 4°C overnight, and sonicated briefly the next day. Resuspended capsids were subjected to native agarose gel analysis to determine the incorporation of A3G into the capsids, as well as SDS-PAGE and Western blot analysis using anti-A3G and anti-HBV core antibodies for the detection of A3G and HBV core protein, respectively.
Protease protection assay.
Transfected cells were lysed in NP-40 lysis buffer containing complete protease inhibitors (Roche), and the lysate was treated with the indicated concentrations of proteinase K (Roche) for 1 h at 37°C. Proteinase K-treated lysates were then loaded onto a 0.8% agarose gel to resolve the nucleocapsid particles, followed by transfer onto a nitrocellulose membrane. Western blot analysis was performed using anti-A3G to determine the amount of incorporated A3G, and the amount of assembled capsid particles was determined by subsequent reprobing of the same membrane using the anti-HBV core antibody.
A3G and RT titration.
For A3G titration, HepG2 cells were transfected with 2 μg pCMV-HBV along with increasing concentrations of pcDNA3.1-A3G plasmid (up to 2 μg DNA) using FuGENE 6 (Roche). For RT titration, HepG2 cells were transfected with 2 μg pCMV-HBVpol-, 2 μg pcDNA3.1-A3G, along with increasing concentrations of pcDNA4-3XFlag-HP (up to 1 μg DNA) using FuGENE 6. Twenty-four hours posttransfection, cells were treated with 20 μM lamivudine (3TC) for up to 3 days to block viral DNA synthesis. Subsequently, transfected cells were lysed, and lysates were treated with 60 U micrococcal nuclease (Roche) for 1 h at 37°C and loaded onto an SDS-PAGE gel or an agarose gel for Western blot or Southern blot analysis.
RESULTS
Native agarose gel electrophoresis assay as an approach to measuring specific A3G association with HBV nucleocapsids.
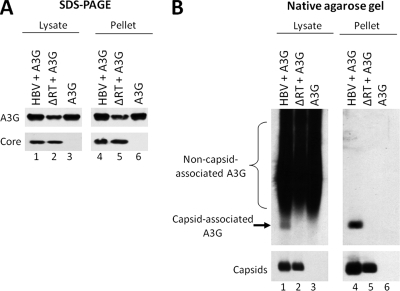
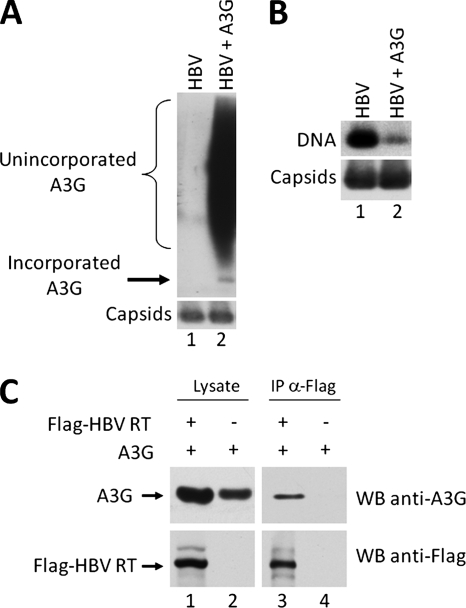
To determine the most reliable approach for measuring specific A3G association with HBV nucleocapsids, we compared the widely used method of viral nucleocapsid isolation by sucrose gradient ultracentrifugation to a native agarose gel electrophoresis assay, which was initially developed to measure RNA packaging into HBV nucleocapsids (60, 66). A plasmid expressing either the wild-type HBV or an HBV polymerase-deficient mutant (ΔRT) was cotransfected together with an A3G expression plasmid into HepG2 cells. The cells were lysed and HBV nucleocapsids were partially purified by pelleting through a sucrose cushion. The resuspended pellets, along with transfected cell lysates, were resolved by either SDS-PAGE or native agarose gel electrophoresis to determine the association of A3G with HBV capsids. As seen in Fig. 1A, the apparent association of A3G with HBV nucleocapsids was unaffected by the presence or absence of the viral RT protein, when measured by ultracentrifugation. Moreover, in the absence of any HBV proteins, A3G was also found in the gradient pellet. However, based on the agarose gel assay, A3G association with capsids was dependent on RT and it was not associated with empty viral nucleocapsids in the absence of RT (Fig. 1B), even though the levels of A3G in the cellular lysates that was not associated with the nucleocapsids (evident as smears above the viral capsid band) were similar (Fig. 1B, lanes 1 to 3). Furthermore, after sucrose gradient ultracentrifugation, A3G that was not associated with the nucleocapsids was removed, whereas the A3G signal comigrating with the nucleocapsids was not affected (Fig. 1B, lanes 4 to 6). These results indicated that pelleting through a sucrose cushion may not be a dependable assay for measuring A3G incorporation into HBV nucleocapsids. This may be due to the nonspecific association of A3G with the outer surface of the nucleocapsids and/or the known property of A3G to form high-molecular-weight (HMW) complexes that may be pelleted by ultracentrifugation (10, 42). In contrast, the native agarose gel electrophoresis assay could clearly resolve A3G that was associated with nucleocapsids from the nonassociated A3G, and thus was a reliable method for measuring specific A3G association with (and indeed, incorporation into [see below]) HBV nucleocapsids.
FIG. 1.
Native agarose gel electrophoresis assay as a reliable method for determining A3G incorporation into HBV nucleocapsids. HepG2 cells were cotransfected with plasmids expressing either wild-type HBV or a polymerase-deficient HBV mutant (ΔRT) along with A3G. Cell lysates were prepared 72 h after transfection and subjected to ultracentrifugation over a sucrose gradient to separate HBV capsids from most cellular proteins. The unfractionated lysates (lanes 1 to 3) and the resuspended pellets (lanes 4 to 6) were loaded onto an SDS-PAGE gel (A) and a native agarose gel (B). Western blot analysis was used to detect the levels of HBV core protein (A, bottom panel), capsid (B, bottom panel), or A3G protein (A and B, upper panels). For native agarose gel analysis (B), A3G was detected by immunoblotting with the anti-A3G antibodies (B, upper panel), followed by subsequent reprobing of the same membrane for HBV capsids using the anti-HBV core protein antibody (B, bottom panel).
A3G incorporated into HBV nucleocapsids was protected from protease digestion.
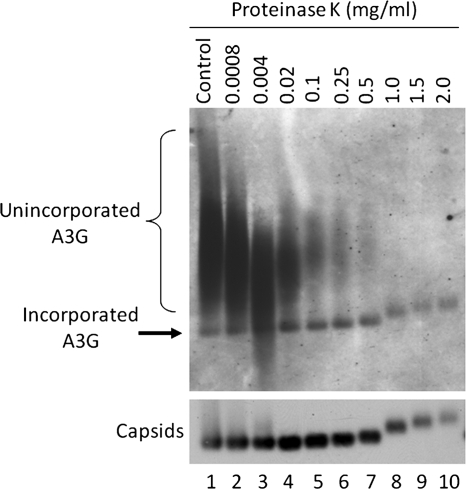
To confirm the interior localization of A3G in the HBV nucleocapsids, we employed a protease protection assay. Cell lysates from transfected HepG2 cells were treated with various concentrations of proteinase K prior to resolution on an agarose gel. If A3G were associated with the exterior of HBV nucleocapsids, the proteinase K would digest the exposed proteins. However, if A3G were located inside the nucleocapsids, it would be protected from the protease. As seen in Fig. 2, increasing the concentration of proteinase K led to the progressive removal of unincorporated A3G (Fig. 2, smears), whereas the A3G signal associated with the nucleocapsid remained constant, indicating the interior localization of A3G. Moreover, at higher concentrations of proteinase K, the capsid signal shifted upward, which was accompanied by the same upward shift of the A3G signal (Fig. 2, lanes 8 to 10), further demonstrating that A3G was indeed incorporated into HBV nucleocapsids.
FIG. 2.
Protection of nucleocapsid-associated A3G from protease digestion. HepG2 cells were transfected with plasmids expressing HBV and A3G. Three days later, NP-40 cell lysates were prepared and treated with the indicated concentration of proteinase K. Lysates were then resolved on a native agarose gel and transferred onto a nitrocellulose membrane. Western blot analysis was used to detect A3G (upper panel) followed by detection of assembled capsids (bottom panel) on the same membrane using the anti-A3G and anti-HBV core antibodies, respectively.
Encapsidation of A3G into HBV nucleocapsids was dependent on the viral RT and RNA packaging signal, ɛ.
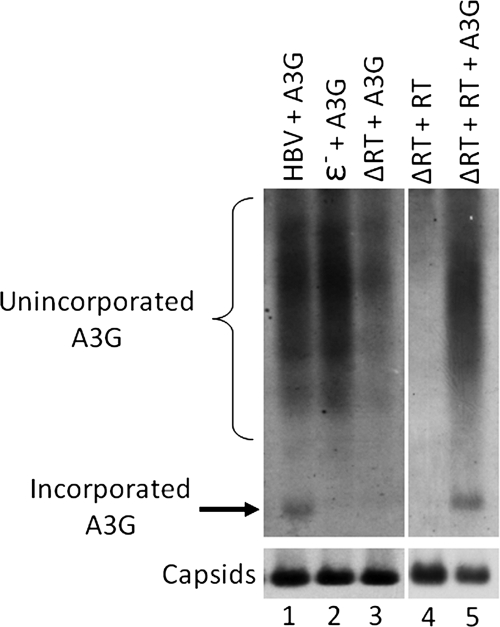
Since assembly of authentic HBV nucleocapsids is well known to require the binding of the viral RT to ɛ, we hypothesized that A3G required one or both of these viral components for its encapsidation into HBV nucleocapsids. Having established that A3G was not incorporated into empty capsids (Fig. 1), we wanted to know whether RT or ɛ was required for A3G packaging. HepG2 cells were transfected with plasmids expressing either a wild-type, an RT-deleted (ΔRT), or an ɛ-deleted (ɛ−) HBV genome, together with an A3G-expressing plasmid. Three days after transfection, cell lysates were prepared and the nucleocapsids were resolved on an agarose gel. As seen in Fig. 3, A3G was packaged into authentic nucleocapsids containing RT and pgRNA. However, it was not packaged in the absence of either RT or ɛ (Fig. 3, lanes 2 and 3). Furthermore, transcomplementation of the HBV polymerase-deficient mutant with a wild-type HBV RT resulted in the reincorporation of A3G into HBV nucleocapsids (Fig. 3, lane 5). Taken together, these results showed that both RT and ɛ are necessary to facilitate A3G incorporation into HBV nucleocapsids. However, since HBV RT and ɛ incorporation is mutually dependent (4, 41, 48), these results were consistent with a requirement for either RT or ɛ or both in A3G incorporation.
FIG. 3.
A3G incorporation was dependent on the viral RT protein and ɛ RNA. HepG2 cells were transfected with plasmids expressing HBV, a polymerase-deficient HBV mutant (ΔRT), or a ɛ− HBV mutant together with A3G. For transcomplementation of HBV RT, HepG2 cells were transfected with plasmids expressing ΔRT and RT in the presence or absence of A3G. Cells were lysed, and A3G packaging into HBV nucleocapsids was measured by native agarose gel analysis. A3G levels (upper panel) and assembled HBV capsids (bottom panel) were detected on the same membrane, as described for Fig. 1B.
Interaction of A3G with HBV RT.
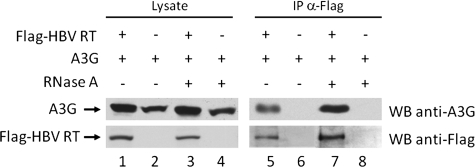
To directly examine the potential interaction between A3G and HBV RT, we performed a coimmunoprecipitation (co-IP) assay. Flag-tagged HBV RT and A3G were expressed in HepG2 cells, and 48 h after transfection, the interaction between A3G and HBV RT was determined. HBV RT was immunoprecipitated from the cell lysate using the anti-Flag antibody, and the presence of coprecipitated A3G in the immunoprecipitate was detected by immunoblotting with anti-A3G. As seen in Fig. 4, A3G indeed coimmunoprecipitated with HBV RT, indicating an interaction between A3G and the HBV RT.
FIG. 4.
HBV RT interacts with A3G in HepG2 cells. Plasmids expressing Flag-tagged HBV RT and A3G were cotransfected into HepG2 cells. Forty-eight hours after transfection the interaction between A3G and HBV RT was determined. HBV RT was immunoprecipitated from the cell lysate using the anti-Flag antibody, and the presence of A3G in the immunoprecipitate was detected by immunoblotting with anti-A3G. To determine if the interaction between A3G and HBV RT was dependent on RNA, the cell lysate was treated with 200 μg/ml RNase A prior to IP (lanes 3, 4, 7, and 8).
Studies using HIV have shown that the interaction between A3G and the viral Gag polyprotein, as well as numerous cellular RNA-binding proteins present within the A3G HMW complexes, is dependent on the presence of cellular or viral RNA (11, 15). To determine if the A3G interaction with HBV RT was reliant upon RNA, we treated the cell lysate with RNase A prior to immunoprecipitation. The RNase treatment did not affect A3G coimmunoprecipitation with RT (Fig. 4), indicating that the interaction between A3G and HBV RT was independent of RNA.
We also examined the interaction between A3G and HBV RT in the 293T human embryonic kidney cell line. As in HepG2 cells, A3G was packaged into HBV nucleocapsids and was able to inhibit HBV DNA synthesis (Fig. 5A and B). Similarly, co-IP between A3G and HBV RT indicated an interaction between the two proteins in 293T cells as well (Fig. 5C).
FIG. 5.
A3G interaction with HBV RT in 293T cells. (A) Transfected 293T cells were lysed, and A3G packaging into HBV nucleocapsids was measured by native agarose gel analysis. A3G levels (upper panel) were detected by A3G-specific antibodies, followed by subsequent reprobing of the same membrane for HBV capsids (bottom panel) using the anti-HBV core protein antibody. (B) 293T cells were transfected with plasmids expressing HBV and A3G, cytoplasmic nucleocapsids were extracted from transfected cells, and capsid-associated DNA levels were determined by native agarose gel analysis. HBV DNA was detected by using a minus-strand-specific riboprobe (top). Capsids were detected by probing the same membrane with anti-HBV core protein antibody (bottom). (C) Co-IP of Flag-tagged HBV RT and A3G. Transfected 293T cell lysate was incubated with the anti-Flag antibody, and the immunoprecipitated proteins were analyzed by Western blotting (WB) with anti-A3G (upper panel) or anti-Flag antibodies (lower panel).
Correlation of A3G incorporation levels with both pgRNA incorporation and A3G expression.
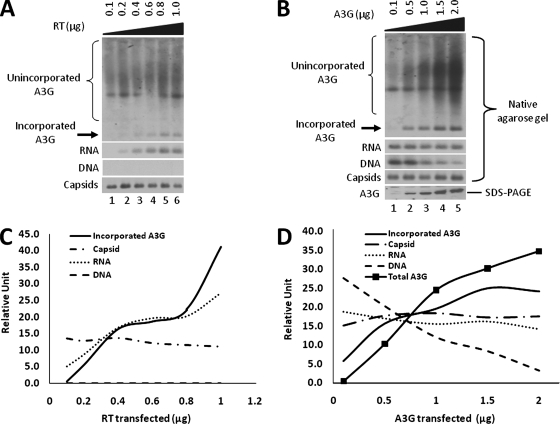
To further demonstrate that A3G interaction with RT mediated its incorporation into HBV nucleocapsids, we reasoned that the incorporation levels of A3G should correspond to the expression level of RT, but not with that of the capsid protein. To this end, we transfected HepG2 cells with equal amounts of plasmids expressing the RT-defective HBV mutant and A3G, together with increasing amounts of the RT expression plasmid. Due to the inability to detect the RT protein incorporated into nucleocapsids directly by Western blotting, we instead quantified the levels of pgRNA as an indirect measurement of RT incorporation. To measure specifically the pgRNA level and not viral DNA, 24 h posttransfection, the cells were treated with an RT inhibitor, 3TC, to inhibit DNA synthesis (25). Cytoplasmic nucleocapsids were then isolated from transfected cells and core-associated DNA, RNA, A3G, and capsid levels were determined by native agarose gel electrophoresis. Southern blot analysis using a riboprobe specific for HBV plus-strand DNA showed that HBV DNA synthesis was completely inhibited by the 3TC treatment, as expected, and that the level of pgRNA increased with increasing amounts of transfected RT DNA (Fig. 6A and C). Furthermore, the increase in packaged pgRNA levels correlated well with an increase in the amount of A3G packaged into HBV nucleocapsids. On the other hand, there was no correlation between the amount of A3G packaged and capsid protein levels (Fig. 6A and C). Taken together, these results further demonstrated that A3G encapsidation required RT and that the capsid protein was not sufficient for packaging A3G.
FIG. 6.
Correlation between levels of A3G incorporation and levels of pgRNA incorporation and A3G expression. (A) RT titration. HepG2 cells were transfected with constant amounts of RT-defective HBV and A3G plasmid, along with increasing amounts of the HBV RT plasmid. Twenty-four hours after transfection, the cells were treated with 20 μM 3TC to inhibit DNA synthesis. Cytoplasmic nucleocapsids were then isolated from transfected cells and loaded onto an agarose gel. For DNA analysis, the agarose gel was treated with NaOH to remove RNA. A3G (first panel) and capsid levels (fourth panel) were analyzed by Western blotting of the same membrane as described for Fig. 1B, while core-associated RNA (second panel) and DNA (third panel) levels were determined by Southern blotting using a plus-strand-specific riboprobe for HBV plus-strand DNA. (B) A3G titration. HepG2 cells were transfected with plasmids expressing wild-type HBV and increasing amounts of A3G. Cytoplasmic nucleocapsids were extracted from transfected cells and analyzed by native agarose gel assay (panels 1 to 4) as in panel A. Total A3G expression was analyzed by SDS-PAGE and Western blotting (fifth panel). (C and D) Quantitative correlation between incorporated A3G levels, HBV RNA and DNA, capsid protein, and total A3G expression levels. Nucleic acid signals and protein bands were quantified with a phosphorimager and densitometer, respectively.
In addition to examining the relationship between A3G and RT levels, we wanted to determine whether A3G incorporation levels were also dependent on the total A3G expression level. Therefore, we transfected HepG2 cells with the HBV expressing plasmid along with increasing amounts of the A3G plasmid. As the A3G expression level increased, HBV DNA replication was inhibited in a dose-dependent manner (Fig. 6B and D), as reported before (39). Moreover, higher A3G expression led to a proportionate increase in A3G incorporation (Fig. 6B and D). In agreement with previous studies (39, 42), increasing A3G expression and incorporation had little to no effect on RNA packaging.
The A3G N-terminal domain was required for packaging into HBV nucleocapsids.
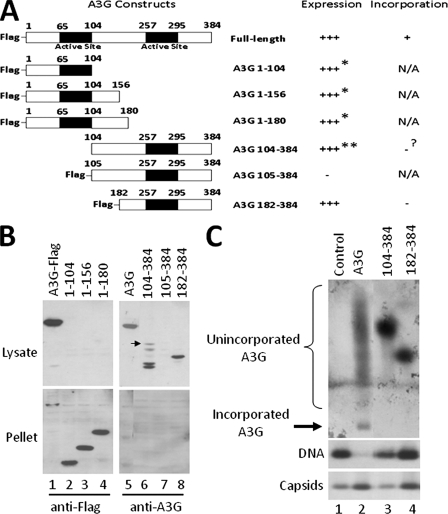
To define the minimal domain of A3G capable of incorporation into HBV nucleocapsids, we constructed a series of Flag-tagged A3G deletion mutants (Fig. 7A). The expression of the mutants was determined by SDS-PAGE and Western blot analysis, and their incorporation into HBV nucleocapsids was measured in an agarose gel assay. With the exception of A3G 105-384, all the deletion mutants were stably expressed in HepG2 cells. However, only the C-terminal fragments were soluble, while the N-terminal fragments were present in the insoluble pellet (Fig. 7B). Interestingly, addition of the Flag tag appeared to destabilize the C-terminal fragment A3G 105-384 (Fig. 7B, lanes 6 and 7).
FIG. 7.
Requirement for the A3G N-terminal domain in encapsidation. (A) Schematic representation of the A3G deletion mutants and their expression and incorporation. All deletion constructs contain a Flag tag at the amino terminus of the protein, with the exception of A3G 104-384. (B and C) HepG2 cells were transfected with a plasmid expressing either the full-length A3G or the A3G deletion mutants together with the HBV plasmid. Seventy-two hours posttransfection, cells were lysed in NP-40 and total A3G levels in the lysate (soluble) and pellet (insoluble) (B) and the amount of packaged A3G (C) were determined by SDS-PAGE and native agarose gel assay, respectively. Expression of C-terminal A3G fragments and incorporated A3G levels were detected using anti-A3G antibody, while N-terminal A3G fragments were detected by using anti-Flag antibody. For the native agarose gel analysis (C), A3G was detected by immunoblotting with anti-A3G antibodies (B, upper panel), followed by reprobing of the same membrane for HBV capsids using the anti-HBV core antibody (B, bottom panel). *, protein expressed but insoluble; **, protein expressed but partially degraded; ?, incorporation status uncertain due to protein degradation. The arrow in panel B indicates the intact A3G 104-384 protein.
A previous study examining the incorporation of A3G into HIV particles mapped the region required for A3G encapsidation to amino acids 104 to 156, which is part of the linker region of A3G (8). Based on these results, we tested the incorporation of two C-terminal A3G fragments into HBV nucleocapsids using our agarose gel assay. Although the expression level of A3G 182-384 was comparable to full-length A3G (Fig. 7B and C), it was, however, not incorporated into the HBV nucleocapsid. On the other hand, A3G 104-384, although expressed, was degraded extensively, with only low levels of the intact protein (Fig. 7B). Nevertheless, based on our A3G titration experiment (Fig. 6B), this amount of intact A3G 104-384 should have led to detectable levels of A3G incorporation, if it were packaging competent. However, we were unable to detect any incorporation of A3G 104-384 into the HBV nucleocapsids (Fig. 7C). Examination of the core-associated DNA levels revealed that incorporated full-length A3G inhibited DNA synthesis, while A3G 104-384 and A3G 182-384 had little to no effect (Fig. 7C). Taken together, these results suggested that the N-terminal region of A3G from 1-103 was required for its packaging into HBV nucleocapsids. Unfortunately, the N-terminal fragments of A3G were insoluble (Fig. 7B, lanes 2 to 4) and as a result, we were unable to identify the specific region of A3G sufficient for incorporation.
DISCUSSION
In the present study, we sought to identify factors needed for the incorporation of the antiviral deaminase protein A3G into HBV nucleocapsids. By adapting the native agarose gel electrophoresis assay, which was originally developed to measure RNA packaging into HBV nucleocapsids, we were able to measure the levels of incorporated A3G specifically and reliably. Surprisingly, when we compared the agarose gel assay to the widely used method of copelleting the virus and A3G by sucrose gradient ultracentrifugation, we found that A3G could be copelleted with authentic viral nucleocapsids or empty capsids, but on the agarose gel, the pelleted A3G only comigrated with authentic nucleocapsids and not with empty capsids. These results indicated that pelleting through a sucrose cushion may not be a reliable assay for measuring A3G incorporation. Such a discrepancy between the two assays could be due to A3G association with the exterior of the nucleocapsids and/or the fact that A3G exists in various complexes (10) that may have similar sedimentation profiles as the viral capsids (42), but nevertheless have a different mobility on the agarose gel. Furthermore, the limited proteinase K digestion assay firmly established that A3G resides within the nucleocapsid, and is not just associated with its exterior. Unexpectedly, we found that at higher concentrations of proteinase K, the capsid signal displayed an upshift pattern. The cause of the shift could be due to the removal of associated proteins from the exterior of the capsid or the cleavage of the C-terminal tail of the core protein, which is believed to extend out from the capsid, at least sometimes (7, 45, 46). Interestingly, a similar pattern was observed when HBV capsids were treated with SDS. In this case, the formation of a disulfide bridge by the carboxyl-terminal cysteine of the core protein was necessary for the SDS-induced upshift (38). These results suggest that the upward shift in the agarose gel electrophoresis may be induced by some conformational change of the capsids.
Our findings showed that the mechanism of A3G incorporation into HBV nucleocapsids is drastically different from that for HIV. In HIV, encapsidation of A3G is facilitated by the nucleocapsid region of the Gag polyprotein and is thought to require either viral or cellular RNA (30, 37, 54, 57, 58, 67). However, in HBV, we found that A3G is specifically incorporated into replication-competent HBV nucleocapsids in a manner that depends on the viral RT and ɛ RNA packaging signal. Moreover, we found that A3G specifically interacted with the viral RT in an RNA-independent manner, which suggests that A3G is incorporated into the HBV nucleocapsid via its interaction with RT. We also found a direct correlation between A3G packaging levels to the levels of RT, but not with that of the capsid protein, indicating that A3G packaging was quantitatively dependent on RT. These results are consistent with a recent study that demonstrated that the binding between A3G and HBV core protein occurred only indirectly with the coexpression of RT and pgRNA, and not with core protein alone (5). The difference in how A3G is packaged into HBV versus HIV particles reflects the different mechanisms of nucleocapsid assembly in these viruses. Whereas assembly of HIV particles is mediated mainly by the Gag polyprotein (including the Gag-Pol fusion) interacting with the genomic RNA (33), the formation of replication-competent HBV nucleocapsids requires the specific interaction between RT and ɛ RNA, which then nucleates the formation of the nucleocapsids (4, 41).
Whether viral or cellular RNA is involved in the incorporation of A3G into HIV virions is still controversial. However, in HBV, it is clear that encapsidation of A3G into the nucleocapsid is dependent on the viral ɛ RNA. Since the packaging of RT and ɛ RNA is mutually dependent, we are still uncertain whether A3G actually binds to the RNA or the requirement for ɛ in A3G encapsidation simply reflects the essential role of ɛ in RT packaging. Although it has been demonstrated that A3G can bind RNA nonspecifically in vitro (26), preliminary efforts to detect sequence specific binding of A3G to the HBV ɛ RNA have been unsuccessful so far (D. Flores and J. Hu, unpublished data).
It is possible that low levels of A3G were packaged into the empty HBV capsids by nonspecific interaction with cellular RNA, as HBV capsids formed in bacteria are known to package cellular RNA nonspecifically even without the RT protein (6, 12), but our agarose gel assay might not be sensitive enough to detect such low levels of incorporation. Alternatively, cellular RNAs that are in complex with A3G may be somehow excluded from incorporation into empty capsids. However, studies examining the structure of the HBV nucleocapsids using recombinant core proteins, expressed in insect cells, have found that the assembled cores are devoid of any RNA (23, 31). It is thus possible that HBV capsid formed in human hepatoma cells may also lack cellular RNA, which may explain the lack of A3G detection in these empty capsids. This would be intriguing, since the HBV capsid protein is known to be phosphorylated in insect cells and mammalian cells, which may downregulate its nonspecific RNA packaging activity (22, 28) while facilitating specific pgRNA-RT packaging (18, 65).
As mentioned earlier, previous work examining the A3G region needed for incorporation in HIV particles found that the linker region of A3G, amino acids 104 to 156, was necessary for incorporation (8, 34), but the N-terminal residues from 1 to 103 were dispensable. In our attempt to determine the A3G domain needed for HBV nucleocapsid incorporation, we found that residues 1 to 103 of A3G appeared to be critical for its incorporation into HBV nucleocapsids, since A3G C-terminal fragments lacking these sequences were not packaged. Such a difference may be due to the fact that A3G must interact with two very different viral proteins for its incorporation into HIV versus HBV particles: HIV Gag versus HBV RT. Nevertheless, since the A3G 104-384 deletion mutant was unstable, as has been reported for a number of other A3G deletion mutants (8, 34, 37), we cannot completely rule out the possibility that residues 1 to 103 are also dispensable for A3G packaging into HBV, as in HIV.
Unfortunately, due to the insolubility of the N-terminal fragments, we have not been able to determine the exact region of A3G that mediates nucleocapsid incorporation. Moreover, the fact that the N-terminal fragments were insoluble may indicate that the C-terminal domain of A3G is needed for protein solubility in HepG2 cells. In contrast to this idea, an A3G C-terminal fragment (residues 198 to 384), expressed in Escherichia coli, was found to be insoluble (9). However, we found that a similar C-terminal fragment, A3G 182-384, was soluble in HepG2 cells. These results suggest the solubility of A3G may differ in bacterial versus mammalian cells. As for the C-terminal fragments, since they appeared as distinct bands on the native agarose gel (compared to the smear seen from full-length A3G), the N-terminal domain may be required for the formation of some A3G complexes, consistent with the recent finding demonstrating that the N-terminal cytidine deaminase motif of A3G was necessary for HMW complex formation (59). Interestingly, we noted that addition of the Flag epitope to the N terminus of A3G 105-384 destabilized the protein, similar to a previous report showing that the terminal tag can interfere with wild-type or mutant A3G expression (38). We attempted to examine the subcellular localization of the full-length and the truncated A3G proteins by immunofluorescence staining. As reported by others (15), the full-length protein was localized mainly in the cytoplasm in a diffuse distribution with a fine granular pattern, as was the C-terminal fragment 104-384. The other C-terminal fragment, 182-384, was also localized diffusely in the cytoplasm but showed a diffuse nuclear staining as well. These staining patterns, together with their detergent solubilities, suggest that the inability of these C-terminal A3G fragments to be incorporated into HBV nucleocapsids was unlikely due to their altered subcellular distribution. On the other hand, the N-terminal fragments were localized in both the cytoplasm and nucleus with a large granular pattern, which may be related to their detergent insolubility (Nguyen and Hu, unpublished).
The observation that A3G can bind to HBV RT independent of RNA, as well as to RNA nonspecifically, suggests possible mechanisms for the editing-independent inhibition of HBV reverse transcription by A3G (39). One possibility is that A3G could bind RT and directly affect RT function in DNA synthesis. Alternatively, A3G can randomly bind to pgRNA and thus could act as a physical barrier during DNA elongation. Additionally, it was reported that long-range base pairing between cis-acting elements found on the HBV pgRNA is necessary for the synthesis of HBV minus-strand DNA (1, 2). It would be interesting to see whether A3G inhibits reverse transcription by disrupting these interactions.
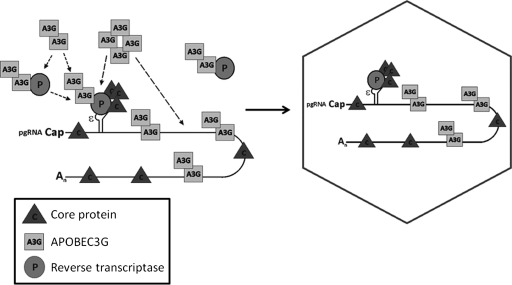
It is still unknown exactly how many molecules of A3G are packaged into the HBV nucleocapsid. In principle, if one molecule of RT is packaged per HBV nucleocapsid (4, 68), it should be reasoned that one molecule of A3G would also be packaged, assuming a 1:1 binding of RT to A3G. However, preliminary experiments examining the stoichiometry of A3G in the HBV nucleocapsid suggest that the level of A3G packaged per nucleocapsid may significantly exceed that of RT (Nguyen and Hu, unpublished). It has also been reported that more than one molecule of A3G can be packaged per Vif-deficient HIV virion (64). Consistent with these results, data from our A3G titration experiment indicated that the amount of A3G packaged into the nucleocapsid was directly related to the overall expression level of A3G in transfected cells, even when RT levels were constant. The data suggest that either more than one A3G molecule can bind to the viral RT or that additional A3G molecules can be incorporated via binding nonspecifically to pgRNA due to its known nonspecific RNA binding activity (26, 37). Based on these results, we propose a model (Fig. 8) whereby A3G binds to the viral RT for its selective incorporation into authentic, RT-containing nucleocapsid. A3G can bind RT either before or after formation of the RT-ɛ RNP complex, but only A3G molecules that are part of this viral RNP complex are packaged, while the A3G-RT complex, in the absence of ɛ RNA, remains unincorporated. Although the oligomeric state of A3G remains to be fully characterized, recent reports suggest that the full-length A3G may form dimers (62, 63) and that it can further exist as HMW complexes (10, 51, 59, 62). Thus, to account for the excess amount (relative to that of RT) of incorporated A3G in the HBV nucleocapsids, A3G complexes, containing multiple copies of A3G, may be incorporated into the nucleocapsids via its interaction with RT. Alternatively, the viral RNP complex may also bring along additional A3G molecules that are nonspecifically associated with the pgRNA, thus increasing the amount of A3G incorporated. Even if the in vivo significance of the A3G antiviral pathway in defending against HBV remains to be more thoroughly investigated (27), elucidating its mechanism of action against HBV DNA synthesis, including factors responsible for its incorporation into HBV particles, will likely provide important insights into HBV reverse transcription and help develop effective antivirals by harnessing the power of innate antiviral defense.
FIG. 8.
Proposed model of A3G incorporation into HBV nucleocapsids. Incorporation of A3G into the HBV nucleocapsid is dependent on its interaction with viral RT. During HBV nucleocapsid assembly, A3G binds to RT and/or to the RT-ɛ complex, leading to its specific packaging into the assembling nucleocapsid. Additional A3G proteins can also be packaged by their nonspecific binding to the pgRNA that is packaged via RT interaction or, alternatively, multiple A3G molecules (e.g., as HMW complexes) can be bound by one RT molecule and incorporated into the nucleocapsids. The pgRNA and ɛ RNA packaging signal are indicated.
Acknowledgments
We thank Michael Malim for providing the pcDNA3.1-A3G plasmid. The anti-A3G antibodies were obtained from Warner C. Greene, Jaisri Lingappa, Klaus Strebel, and Sandra Kao, through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health.
This work was supported by a Public Health Service grant, R01 AI43453, from the National Institutes of Health.
Footnotes
Published ahead of print on 14 May 2008.
REFERENCES
- 1.Abraham, T. M., and D. D. Loeb. 2006. Base pairing between the 5′ half of epsilon and a cis-acting sequence, Φ, makes a contribution to the synthesis of minus-strand DNA for human hepatitis B virus. J. Virol. 804380-4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham, T. M., and D. D. Loeb. 2007. The topology of hepatitis B virus pregenomic RNA promotes its replication. J. Virol. 8111577-11584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 27934083-34086. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., and H. Schaller. 1992. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 113413-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumert, T. F., C. Rosler, M. H. Malim, and F. von Weizsacker. 2007. Hepatitis B virus DNA is subject to extensive editing by the human deaminase APOBEC3C. Hepatology 46682-689. [DOI] [PubMed] [Google Scholar]
- 6.Birnbaum, F., and M. Nassal. 1990. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J. Virol. 643319-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borisova, G. P., I. Berzins, P. M. Pushko, P. Pumpen, E. J. Gren, V. V. Tsibinogin, V. Loseva, V. Ose, R. Ulrich, H. Siakkou, et al. 1989. Recombinant core particles of hepatitis B virus exposing foreign antigenic determinants on their surface. FEBS Lett. 259121-124. [DOI] [PubMed] [Google Scholar]
- 8.Cen, S., F. Guo, M. Niu, J. Saadatmand, J. Deflassieux, and L. Kleiman. 2004. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 27933177-33184. [DOI] [PubMed] [Google Scholar]
- 9.Chen, K. M., N. Martemyanova, Y. Lu, K. Shindo, H. Matsuo, and R. S. Harris. 2007. Extensive mutagenesis experiments corroborate a structural model for the DNA deaminase domain of APOBEC3G. FEBS Lett. 5814761-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435108-114. [DOI] [PubMed] [Google Scholar]
- 11.Chiu, Y. L., H. E. Witkowska, S. C. Hall, M. Santiago, V. B. Soros, C. Esnault, T. Heidmann, and W. C. Greene. 2006. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. USA 10315588-15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crowther, R. A., N. A. Kiselev, B. Bottcher, J. A. Berriman, G. P. Borisova, V. Ose, and P. Pumpens. 1994. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell 77943-950. [DOI] [PubMed] [Google Scholar]
- 13.Cullen, B. R. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 801067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fallows, D. A., and S. P. Goff. 1995. Mutations in the epsilon sequences of human hepatitis B virus affect both RNA encapsidation and reverse transcription. J. Virol. 693067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallois-Montbrun, S., B. Kramer, C. M. Swanson, H. Byers, S. Lynham, M. Ward, and M. H. Malim. 2007. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 812165-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganem, D., and A. M. Prince. 2004. Hepatitis B virus infection: natural history and clinical consequences. N. Engl. J. Med. 3501118-1129. [DOI] [PubMed] [Google Scholar]
- 17.Gao, W., and J. Hu. 2007. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J. Virol. 816164-6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gazina, E. V., J. E. Fielding, B. Lin, and D. A. Anderson. 2000. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J. Virol. 744721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, F., S. Cen, M. Niu, J. Saadatmand, and L. Kleiman. 2006. Inhibition of tRNA3Lys-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 8011710-11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113803-809. [DOI] [PubMed] [Google Scholar]
- 21.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4868-877. [DOI] [PubMed] [Google Scholar]
- 22.Hatton, T., S. Zhou, and D. N. Standring. 1992. RNA- and DNA-binding activities in hepatitis B virus capsid protein: a model for their roles in viral replication. J. Virol. 665232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hilditch, C. M., L. J. Rogers, and D. H. Bishop. 1990. Physicochemical analysis of the hepatitis B virus core antigen produced by a baculovirus expression vector. J. Gen. Virol. 712755-2759. [DOI] [PubMed] [Google Scholar]
- 24.Hirsch, R. C., J. E. Lavine, L. J. Chang, H. E. Varmus, and D. Ganem. 1990. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature 344552-555. [DOI] [PubMed] [Google Scholar]
- 25.Hu, J., D. Flores, D. Toft, X. Wang, and D. Nguyen. 2004. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J. Virol. 7813122-13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79285-296. [DOI] [PubMed] [Google Scholar]
- 27.Jost, S., P. Turelli, B. Mangeat, U. Protzer, and D. Trono. 2007. Induction of antiviral cytidine deaminases does not explain the inhibition of hepatitis B virus replication by interferons. J. Virol. 8110588-10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kann, M., and W. H. Gerlich. 1994. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J. Virol. 687993-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 7711398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan, M. A., S. Kao, E. Miyagi, H. Takeuchi, R. Goila-Gaur, S. Opi, C. L. Gipson, T. G. Parslow, H. Ly, and K. Strebel. 2005. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J. Virol. 795870-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanford, R. E., and L. Notvall. 1990. Expression of hepatitis B virus core and precore antigens in insect cells and characterization of a core-associated kinase activity. Virology 176222-233. [DOI] [PubMed] [Google Scholar]
- 32.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 3001112. [DOI] [PubMed] [Google Scholar]
- 33.Luciw, P. A. 2001. Human immunodeficiency viruses and their replication, p. 1881-1952. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 34.Luo, K., B. Liu, Z. Xiao, Y. Yu, X. Yu, R. Gorelick, and X. F. Yu. 2004. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 7811841-11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 42499-103. [DOI] [PubMed] [Google Scholar]
- 36.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 11421-31. [DOI] [PubMed] [Google Scholar]
- 37.Navarro, F., B. Bollman, H. Chen, R. Konig, Q. Yu, K. Chiles, and N. R. Landau. 2005. Complementary function of the two catalytic domains of APOBEC3G. Virology 333374-386. [DOI] [PubMed] [Google Scholar]
- 38.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15166-170. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen, D. H., S. Gummuluru, and J. Hu. 2007. Deamination-independent inhibition of hepatitis B virus reverse transcription by APOBEC3G. J. Virol. 814465-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perlman, D. H., E. A. Berg, P. B. O'Connor, C. E. Costello, and J. Hu. 2005. Reverse transcription-associated dephosphorylation of hepadnavirus nucleocapsids. Proc. Natl. Acad. Sci. USA 1029020-9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollack, J. R., and D. Ganem. 1994. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J. Virol. 685579-5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosler, C., J. Kock, M. Kann, M. H. Malim, H. E. Blum, T. F. Baumert, and F. von Weizsacker. 2005. APOBEC-mediated interference with hepadnavirus production. Hepatology 42301-309. [DOI] [PubMed] [Google Scholar]
- 43.Rosler, C., J. Kock, M. H. Malim, H. E. Blum, and F. von Weizsacker. 2004. Comment on inhibition of hepatitis B virus replication by APOBEC3G. Science 3051403. [DOI] [PubMed] [Google Scholar]
- 44.Schafer, A., H. P. Bogerd, and B. R. Cullen. 2004. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 328163-168. [DOI] [PubMed] [Google Scholar]
- 45.Schlicht, H. J., R. Bartenschlager, and H. Schaller. 1989. The duck hepatitis B virus core protein contains a highly phosphorylated C terminus that is essential for replication but not for RNA packaging. J. Virol. 632995-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schodel, F., A. M. Moriarty, D. L. Peterson, J. A. Zheng, J. L. Hughes, H. Will, D. J. Leturcq, J. S. McGee, and D. R. Milich. 1992. The position of heterologous epitopes inserted in hepatitis B virus core particles determines their immunogenicity. J. Virol. 66106-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 6451-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seeger, C., W. S. Mason, and F. Zoulim. 2007. Hepadnaviruses, p. 2977-3029. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 49.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418646-650. [DOI] [PubMed] [Google Scholar]
- 50.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12591-601. [DOI] [PubMed] [Google Scholar]
- 51.Stopak, K. S., Y. L. Chiu, J. Kropp, R. M. Grant, and W. C. Greene. 2007. Distinct patterns of cytokine regulation of APOBEC3G expression and activity in primary lymphocytes, macrophages, and dendritic cells. J. Biol. Chem. 2823539-3546. [DOI] [PubMed] [Google Scholar]
- 52.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29403-415. [DOI] [PubMed] [Google Scholar]
- 53.Suspene, R., D. Guetard, M. Henry, P. Sommer, S. Wain-Hobson, and J. P. Vartanian. 2005. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. USA 1028321-8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Svarovskaia, E. S., H. Xu, J. L. Mbisa, R. Barr, R. J. Gorelick, A. Ono, E. O. Freed, W. S. Hu, and V. K. Pathak. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 27935822-35828. [DOI] [PubMed] [Google Scholar]
- 55.Turelli, P., B. Mangeat, S. Jost, S. Vianin, and D. Trono. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 3031829. [DOI] [PubMed] [Google Scholar]
- 56.Wang, G. H., F. Zoulim, E. H. Leber, J. Kitson, and C. Seeger. 1994. Role of RNA in enzymatic activity of the reverse transcriptase of hepatitis B viruses. J. Virol. 688437-8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, T., C. Tian, W. Zhang, K. Luo, P. T. Sarkis, L. Yu, B. Liu, Y. Yu, and X. F. Yu. 2007. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J. Virol. 8113112-13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, T., C. Tian, W. Zhang, P. T. Sarkis, and X. F. Yu. 2008. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. J. Mol. Biol. 3751098-1112. [DOI] [PubMed] [Google Scholar]
- 59.Wang, X., P. T. Dolan, Y. Dang, and Y. H. Zheng. 2007. Biochemical differentiation of APOBEC3F and APOBEC3G proteins associated with HIV-1 life cycle. J. Biol. Chem. 2821585-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, X., N. Grammatikakis, and J. Hu. 2002. Role of p50/CDC37 in hepadnavirus assembly and replication. J. Biol. Chem. 27724361-24367. [DOI] [PubMed] [Google Scholar]
- 61.Weber, M., V. Bronsema, H. Bartos, A. Bosserhoff, R. Bartenschlager, and H. Schaller. 1994. Hepadnavirus P protein utilizes a tyrosine residue in the TP domain to prime reverse transcription. J. Virol. 682994-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wedekind, J. E., R. Gillilan, A. Janda, J. Krucinska, J. D. Salter, R. P. Bennett, J. Raina, and H. C. Smith. 2006. Nanostructures of APOBEC3G support a hierarchical assembly model of high molecular mass ribonucleoprotein particles from dimeric subunits. J. Biol. Chem. 28138122-38126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 232451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu, H., E. Chertova, J. Chen, D. E. Ott, J. D. Roser, W. S. Hu, and V. K. Pathak. 2007. Stoichiometry of the antiviral protein APOBEC3G in HIV-1 virions. Virology 360247-256. [DOI] [PubMed] [Google Scholar]
- 65.Yeh, C. T., and J. H. Ou. 1991. Phosphorylation of hepatitis B virus precore and core proteins. J. Virol. 652327-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu, M., and J. Summers. 1994. Multiple functions of capsid protein phosphorylation in duck hepatitis B virus replication. J. Virol. 684341-4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zennou, V., D. Perez-Caballero, H. Gottlinger, and P. D. Bieniasz. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 7812058-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, Z., and J. E. Tavis. 2006. The duck hepatitis B virus reverse transcriptase functions as a full-length monomer. J. Biol. Chem. 28135794-35801. [DOI] [PubMed] [Google Scholar]
- 69.Zoulim, F., and C. Seeger. 1994. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J. Virol. 686-13. [DOI] [PMC free article] [PubMed] [Google Scholar]