Abstract
Standard vectors for high-level expression elicited undetectable levels of the gH and gL glycoproteins of rhesus monkey rhadinovirus (RRV) following transient-transfection assays under a variety of conditions. These same vectors and conditions yielded high levels of RRV gB expression. Unlike other genes of RRV, both the gH and gL genes were noted to have a highly aberrant, suboptimal codon usage. High levels of RRV gH and gL expression were achieved by two alternative means: codon optimization or coexpression of RRV ORF57. The failure of gH and gL to be expressed in the absence of ORF57 and in the absence of codon optimization could not be explained by the failure of RNA to egress from the nucleus. Rather, the defect in gH and gL expression appeared to be cytoplasmic in nature. It is not clear at the present time whether the aberrant codon usage for gH and gL of RRV is an intentional regulatory strategy used by the virus or whether it is driven by some external force, such as intrinsic immunity. In any event, our results indicate that the need of ORF57 for gH and gL expression can be circumvented by codon optimization, that RRV ORF57 acts principally to allow translation of gH and gL RNA in the cytoplasm, and that this activity of ORF57 is related in some way to the aberrant codon usage of the gH and gL RNAs.
Synthesis of herpesvirus proteins in a lytically infected cell proceeds in a distinctly ordered cascade: immediate-early, early, and late. Virally encoded control factors that are responsible for regulating the ordered synthesis act at both the transcriptional and posttranscriptional levels. All herpesviruses possess a homolog of one notable protein that acts posttranscriptionally: ORF57 for gamma-2 herpesviruses, Mta for Epstein-Barr virus, and ICP27 for herpes simplex virus. These ORF57-like proteins are made immediate-early in a lytically infected cell, are essential for viral replication, and facilitate egress of unspliced viral RNA from the nucleus while at the same time inhibiting the egress of cellular RNAs that require splicing (for reviews, see references 3, 24, 27, and 29). These last properties provide obvious advantages to a virus of limited genome size, most of whose proteins are made from unspliced RNA. In at least some cases, these ORF57-like proteins may also function in the cytoplasm to increase mRNA stability or translation (10, 16, 20, 21).
Mammalian cells show a marked preference for one codon over another for each amino acid that can be encoded by several synonymous codons (5, 11, 14, 25, 32). Codon usage in general correlates with tRNA abundance (4). Heterologous genes with poor congruence to the favored codon usage are in general poorly expressed. The levels of expression of such heterologous genes can be markedly increased by converting their sequence to a more optimal codon usage. For reasons that are not entirely clear, the increased expression of such codon-optimized gene expression cassettes, such as those for human immunodeficiency virus (HIV) gag or env (15, 22), is often due not to increased translation but to increased egress of mRNA from the nucleus. Natural expression of HIV gag and env in a lytically infected cell relies on the posttranscriptional factor Rev and a cis-acting sequence called the Rev responsive element to overcome the highly suboptimal codon usage in these HIV genes (9, 19).
Why would a virus like HIV retain such abnormal, suboptimal codon usage for its major structural proteins Gag and Env, and why would it rely on a complex Rev-dependent, Rev responsive element-dependent system for expression of its major structural genes? At least three hypotheses can be put forward to explain this: (i) the antiviral cellular protein APOBEC-3G drives G→A mutations in the virus, and this drives suboptimal codon usage (2); (ii) there is an advantage for the virus to maximally express its structural gene products as a burst late in the viral life cycle (8, 18, 28); and (iii) the virus is able to maximize utilization of the limited size of its genome by early synthesis of proteins from fully spliced RNAs followed by late synthesis of proteins from cytoplasmic mRNAs that contain one or more potential introns (9, 19).
In this report, we describe the extreme dependence of expression of gH and gL of rhesus monkey rhadinovirus (RRV) upon coexpression of ORF57, the association of poor expression of RRV gH and gL in the absence of ORF57 with poor codon usage, and the ability of codon optimization to allow high levels of gH and gL expression even in the absence of ORF57. We believe that this is the first report relating poor expression of a herpesvirus protein to suboptimal codon usage.
Codon usage of RRV gH and gL.
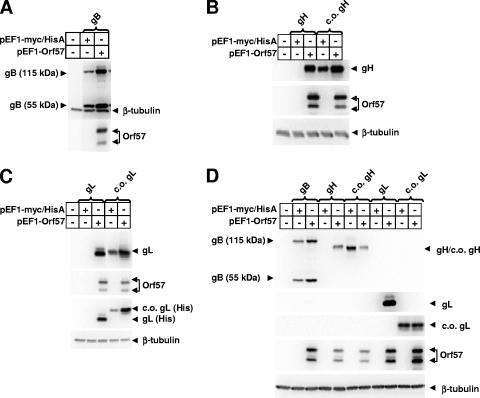
While attempting to produce gH and gL proteins of RRV for the generation of monoclonal antibodies, we encountered severe problems producing gH and gL by transient transfection of HEK293T cells. Initially, gH and gL were C-terminally tagged with glutathione S-transferase (GST); however, the production of gH-GST or gL-GST was not detectable on the basis of either anti-GST or rhesus monkey anti-RRV antibodies. We sequenced and resequenced the vector constructions to ensure that there were no inadvertent errors; there were none. We changed the C-terminal tag to Myc, His, or Myc/His without success in being able to detect gH, gL, or gH-gL together. We tried no tag at all, and this was not successful either. The promoter for these constructs was changed, and still no protein was detectable (data not shown). We encountered no such problems with the detection of gB with the same vectors and the same methods (Fig. 1A).
FIG. 1.
Achievement of gH and gL expression by codon optimization or by coexpression of ORF57. RRV gH, gL, and gB gene coding regions were cloned in-frame into the pEF1-myc/HisA expression vector (Invitrogen, Carlsbad, CA) after PCR amplification from the RRV26-95 cosmid library (1). RRV ORF57 was PCR amplified from the RRV26-95 cosmid library and inserted in-frame into the pEF1-V5/His expression vector (Invitrogen). Codon-optimized versions of RRV gH and gHt were PCR amplified from a template plasmid acquired through DNA2.0 (Menlo Park, CA) and inserted into the pEF1-myc/HisA expression vector. A codon-optimized version of RRV gL was PCR amplified from a template plasmid acquired through DNA2.0 and inserted into the pEF1-V5/HisA expression vector. Recloned products generated by PCR were sequenced to verify the absence of introduced mutations. One day postseeding, HEK293T (A to C) or Vero cells (D) (4.5 × 105 cells/well in 6-well plates) were transfected with different combinations of expression plasmids using the Transfectin reagent (Bio-Rad Laboratories, Hercules, CA) using a scaled-down procedure. At 48 h posttransfection, cultures were rinsed with phosphate-buffered saline and lysed with radioimmunoprecipitation assay buffer (Boston BioProducts, Inc., Worcester, MA). Lysates were sonicated at 20% for 10 s with a Fisher Scientific sonic dismembrator (model 500), and debris was spun down at 14,000 × g in a microcentrifuge for 1 min. Protein concentrations were determined by bicinchoninic acid assay, following the manufacturer's instructions (Pierce Biotechnology, Rockford, IL). Proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. Membranes were blocked overnight at room temperature in 5% milk in phosphate-buffered saline containing 0.1% Tween 20 (PBS-T) (Sigma, St. Louis, MO). Membranes were incubated for 1 h with mouse monoclonal antibodies (all from Santa Cruz Biotechnology, Santa Cruz, CA) diluted in 5% milk-PBS-T. The antibodies were specific for either Myc (for detection of gB, gH, c.o. gH, and gL in panels A, B, and D), six-His (for detection of gL and c.o. gL in panel C), or β-tubulin or V5 (for detection of ORF57 in all blots and c.o. gL in panel D). After successive washings in PBS-T, blots were incubated in secondary antimouse (Santa Cruz) diluted in 5% milk in PBS-T. Blots were washed in PBS-T, and antibody binding was detected using the SuperSignal West Pico chemiluminescent reagent (Pierce) and a Fuji phosphorimager. ORF57 is observed as a doublet because the transcript has multiple start sites for translation. Incubation of replicate blots or the same blot (for gB) with an anti-β-tubulin antibody was used to show equal loading. Mobility of the codon-optimized gL-V5-His protein differs slightly from that of the gL-myc-His protein because of the different epitope tag. Samples transfected with gH, c.o. gH, gL, or c.o. gL are indicated by brackets.
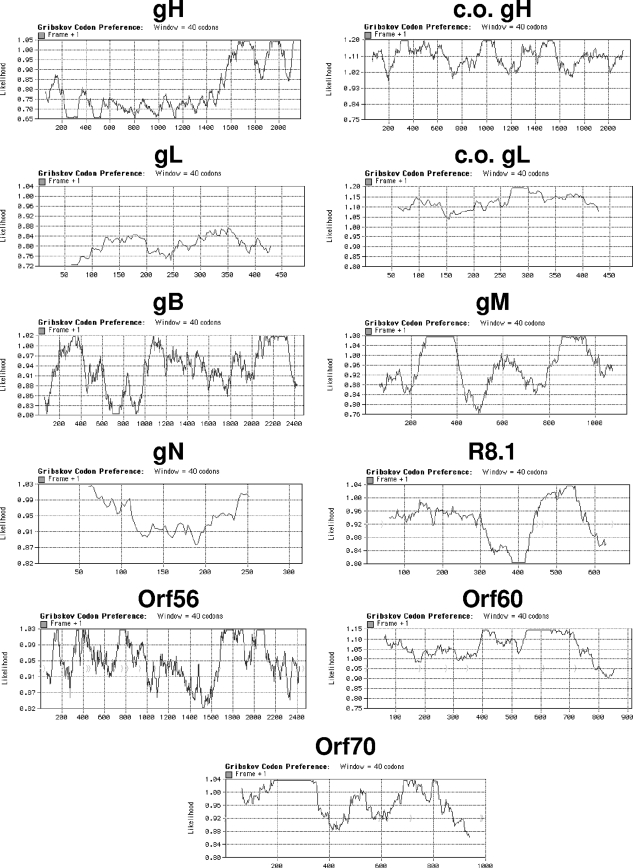
Examination of the gH and gL genomic sequence revealed an extremely abnormal codon usage. We used MacVector (MacVector, Inc., Cary, NC) software to analyze the Gribskov codon preference for all 84 RRV ORFs (data not shown) and found that gH and gL stood out for their suboptimal use of codons (Fig. 2). Orthologous glycoproteins of alpha-, beta-, and other gammaherpesviruses were also examined for their codon usage (Table 1). Several herpesvirus saimiri (HVS) glycoproteins, as well as RRV gH and gL, were found to have clearly suboptimal codon usage. Because the genomic coding sequences of HVS are so highly A:T rich (approximately 66% A+T), a high frequency of suboptimal codons is not restricted to the glycoprotein sequences of HVS as is the case for RRV gH and gL (Table 1).
FIG. 2.
Gribskov codon usage charts. The codon usage profile for each gene was determined using MacVector with codon preferences for Homo sapiens.
TABLE 1.
Herpesvirus glycoprotein codon usage
| Glycoprotein | CAI value for virusa
|
|||||
|---|---|---|---|---|---|---|
| RRV | KSHV | HVS | EBV | HSV-1 | HCMV | |
| gB | 0.388 | 0.325 | 0.104 | 0.418 | 0.628 | 0.307 |
| gH | 0.110 | 0.265 | 0.087 | 0.333 | 0.419 | 0.276 |
| gL | 0.104 | 0.208 | 0.069 | 0.294 | 0.254 | 0.399 |
| gM | 0.194 | 0.272 | 0.099 | 0.445 | 0.455 | 0.426 |
| gN | 0.255 | 0.224 | 0.078 | 0.355 | 0.378 | 0.190 |
| R8.1 | 0.256 | 0.201 | ND | ND | ND | ND |
Determined by using JCAT (Java codon adaptation tool), using the Homo sapiens algorithm (12, 32). Numbers represent codon adaptation index (CAI) values (5, 25). RRV, rhesus monkey rhadinovirus; KSHV, Kaposi's sarcoma-associated herpesvirus; HVS, herpesvirus saimiri; HSV-1, herpes simplex virus type 1; HCMV, human cytomegalovirus; ND, not determined.
Codon optimization or ORF57 coexpression leads to RRV gH and gL expression.
Clearly, the codon usage of the RRV gH and gL genes is unusual compared to the gH and gL genes of other herpesviruses and even other genes of RRV. Consequently, we synthesized codon-optimized versions of the gH and gL reading frames (c.o. gH and c.o. gL) (Fig. 2) and inserted them into standard expression vectors. We compared the levels of gH and gL expression from these vectors following transfection of HEK293T cells with the levels of expression obtained from parallel constructions in the absence of codon manipulation. Codon optimization resulted in dramatic increases in the levels of gH and gL expression (Fig. 1B and C). Coexpression of RRV ORF57 also resulted in dramatic induction of RRV gH and gL expression from the natural gH and gL cassettes in the absence of codon optimization (Fig. 1B and C). Expression levels of RRV gH, gL, and gB were not induced or altered upon coexpression of ORF50 (data not shown).
The induction by RRV ORF57 is analogous to inductions by Kaposi's sarcoma-associated herpesvirus ORF57 that have been reported previously (13, 16, 17, 31) and by Mta of Epstein-Barr virus (6, 7, 23), except that RRV gH and gL induction by ORF57 is essentially an all-or-none phenomenon. Levels of gH and gL expression go from undetectable to extremely strong with the inclusion of ORF57. Codon optimization seems to mimic the effects of ORF57 in allowing high levels of gH and gL expression (Fig. 1B and C).
We next examined whether the extreme restriction to RRV gH and gL expression was species specific. The same expression plasmids were transfected into monkey Vero cells, and the levels of expression were determined. The expression of non-codon-optimized gH and gL was also extremely restricted in Vero cells, and again this restriction could be overcome by either codon optimization or coexpression of RRV ORF57 (Fig. 1D).
Expression of other RRV genes in the presence and absence of ORF57.
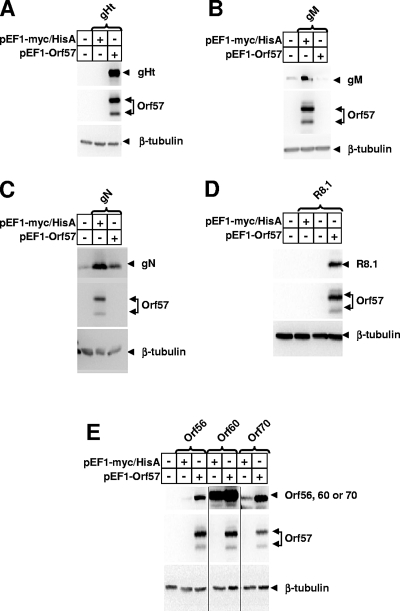
In addition to the gH, gL, and gB genes shown in Fig. 1, we also examined the expression of the RRV gM, gN, R8.1, ORF56, ORF60, and ORF70 genes with and without coexpression of RRV ORF57. These genes were selected in order to include both glyco- and nonglycoproteins and to include a range in the quality of codon usage. In all cases, the RRV genes were amplified from infectious cosmid clones that have been described previously (1) and Myc/HisA-tagged expression constructs were transfected into HEK293T cells to monitor levels of protein expression. Recloned products generated from PCR were sequenced to verify the absence of introduced mutations. The results revealed various levels of basal expression and various degrees of induction by coexpression of ORF57 (Fig. 3). Basal levels of expression of ORF60 were high, and there was minimal induction by ORF57. The RRV 8.1 glycoprotein was similar to gH and gL in yielding undetectable basal levels of expression and strong induction by ORF57 (Fig. 3). Expression of the other RRV genes and the degree of ORF57 induction fell within these extremes. As observed for RRV gH and gL, expression levels of RRV gM, gN, and R8.1 were not induced or altered upon coexpression of ORF50 (data not shown).
FIG. 3.
Expression of other RRV genes in the presence and absence of ORF57. RRV genes for gHt (A), gM (B), gN (C), R8.1 (D), ORF56, ORF60, and ORF70 (E) were cloned into the pEF1-myc/HisA vector with a Myc/His epitope at the C terminus, expressed, and detected as described in the legend to Fig. 1.
There was some, but not absolute, correlation of the basal levels of expression and the degree of ORF57 induction with the extent of optimal codon usage (Fig. 2 and 3). ORF60 had the most optimal codon usage of the genes studied, and it had the highest basal levels of expression and the least induction by ORF57. The gH and gL genes had the least optimal codon usage, and they had the lowest basal expression levels and greatest induction levels. Except for RRV R8.1, the remaining RRV genes exhibited codon usage, basal levels of expression, and levels of ORF57 induction that fell within these extremes. However, we were not able to derive a strict correlation of codon usage with basal expression levels or degree of ORF57 induction.
Expression of a truncated version of gH from which the C-terminal cytoplasmic domain was removed (gHt) was also examined to determine if trafficking or stability of full-length gH was in some way inhibited in the absence of ORF57. Again, expression of the truncated form of gH was undetectable in the absence of ORF57 and was strongly induced in the presence of ORF57 (Fig. 3A).
Export of gH and gL RNA from the nucleus.
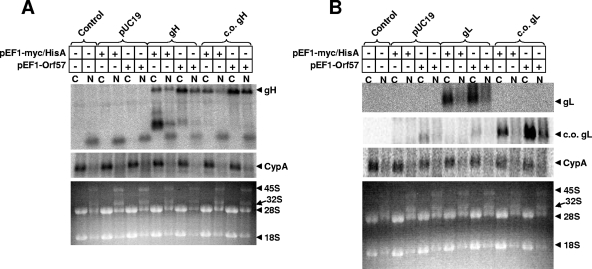
We next examined whether codon optimization or ORF57 coexpression facilitated the export of gH RNA or gL RNA from the nucleus, a known site of action of ORF57-related proteins from a number of different herpesviruses (3, 16, 20, 21, 24, 27, 29, 30). HEK293T cells were transfected with the relevant expression constructs, and cytoplasmic and nuclear RNA was isolated 48 h after transfection. RNA was electrophoresed through a 1% denaturing formaldehyde agarose gel and transferred to a membrane for hybridization. The patterns of ethidium bromide staining in the lower panels of Fig. 4A and B revealed prominent 45S and 32S pre-rRNA bands in the nuclear fraction but not the cytoplasmic fraction, a testimony to the relative purity of the fractionated components. Hybridization of the transferred RNA was performed with an antisense myc oligonucleotide corresponding to the codon-optimized tag. The labeled probe thus did not recognize cellular myc RNA. RNA corresponding to the expected full-length gH transcript (Fig. 4A) and to the expected full-length gL transcript (Fig. 4B) was efficiently exported to the cytoplasm in all cases. An overall increase in the abundance of full-length gH and gL transcripts was observed by coexpression of ORF57 (Fig. 4A and B). However, the failure to detect gH and gL protein expression in the absence of ORF57 coexpression cannot be explained by the failure of RNA to egress from the nucleus.
FIG. 4.
Cytoplasmic and nuclear RNA after transfection of HEK293T cells. (A) gH. (B) gL. At 48 h posttransfection, total cytoplasmic RNA was isolated using the RNeasy Mini kit (Qiagen, Valencia, CA), following the instructions for cytoplasmic RNA purification. Total nuclear RNA was isolated from the nuclear pellets remaining from the cytoplasmic RNA isolation procedure. The concentration of purified cytoplasmic and nuclear RNA was determined by spectrophotometry. Five micrograms of RNA was combined with formamide loading buffer (Ambion, Austin, TX), denatured at 65°C for 10 min, and loaded onto a 1% denaturing formaldehyde agarose gel containing ethidium bromide in accordance with the instructions of the NorthernMax instruction manual (Ambion). The RNA was transferred to a BrightStar-Plus membrane (Ambion) overnight and then cross-linked by UV light exposure in a commercial cross-linker (Bio-Rad). Blots were then prehybridized in UltraHyb (Ambion) for >1 h at 42°C. An end-labeled antisense oligonucleotide probe was added directly to UltraHyb and allowed to hybridize overnight at 42°C. Antisense probes were generated by 2 h of incubation of [γ-32P]ATP, antisense oligonucleotides (c-myc, 5′-CAGATCCTCTTCTGAGATGAGTTTTTGTTC-3′, or V5, 5′-ACCGAGGAGAGGGTTAGGGATAGGCTTACC-3′), and polynucleotide kinase (New England Biolabs, Beverly, MA). Following this incubation, the end-labeled probes were purified using ProbeQuant G-50 Micro columns (GE Healthcare, Pittsburgh, PA). The next day, the blots were washed in triplicate with low-stringency wash solution (Ambion) for 5 min at room temperature, followed by one wash in low-stringency wash solution for 10 min at 42°C. Blots were then exposed overnight in phosphorimager cassettes, and the screens were read using a phosphorimager (BAS 2000; Fuji Photo Film Co., Tokyo, Japan). Blots were then stripped in 0.1% sodium dodecyl sulfate, prehybridized as before, and hybridized to an antisense probe to Macaca mulatta cyclophilin A (5′-CCAAATCCTTTCTCTCCAGTGCTCAGAGC-3′) prepared as described above. Blots were rinsed, exposed to phosphorimager screens, and read as above. The lower panel for each shows the ethidium bromide-stained agarose gel. C, cytoplasmic; N, nuclear. After hybridization and exposure for detection of gH (A) or gL (B) RNA, the blot was stripped and reprobed for cyclophilin A as a control for equal RNA loading. Samples transfected with pUC19 (Invitrogen), gH, c.o. gH, gL, or c.o. gL are indicated by brackets.
In the case of gH expression, a small hybridizing RNA product of less than 500 bp in length predominated under conditions of endogenous codon usage and without ORF57 coexpression (Fig. 4A). This small product suggests extensive turnover/degradation of gH RNA in the absence of codon optimization and in the absence of ORF57 coexpression. No such small RNA was detected in the Northern blots for gL RNA. Because the gL transcript is short (∼500 bp), degradation products for this mRNA would not have been observed in these Northern blots.
There are intriguing parallels between the expression of the RRV glycoproteins gH and gL reported here and the expression of HIV glycoprotein Env (gp160) despite the fact that these viruses represent completely different virus families. Simple, standard expression cassettes for each of these three proteins produce no detectable protein expression in transfected cells in the absence of any further manipulation. All three proteins have a highly suboptimal codon usage naturally encoded in their viral genomes. Expression of all three proteins by standard expression cassettes in transfected cells can be dramatically induced simply by changing the codon usage to one that is more optimal. And each virus encodes a protein (Rev for HIV and ORF57 for RRV) that is required for dramatic induction of glycoprotein expression in the course of the viral replication cycle. But there also appear to be differences in the specifics of these activities. In the case of HIV, codon optimization and Rev induction for Env expression act at the stage of facilitating RNA egress from the nucleus. Our early results presented here for RRV suggest that ORF57 acts principally in the cytoplasm to facilitate translation of gH and gL RNA.
What may be the selective forces that have driven highly suboptimal codon usage for RRV gH and gL and, for that matter, most or all of the genes of herpesvirus saimiri, a gamma-2 herpesvirus of the more primitive New World primates? In a sense, the suboptimal codon usage of gH and gL in RRV could be viewed as a possible evolutionary remnant of what affects the entire coding region of herpesvirus saimiri. In the case of HIV, simian immunodeficiency virus, and other members of the lentivirus subfamily of retroviruses, it is generally believed that the cellular intrinsic immunity protein APOBEC-3G is responsible at least in part for driving the suboptimal codon usage (2). APOBEC-3G causes G→A hypermutation during the process of reverse transcription; lentiviruses encode a specific gene product, Vif, whose role is to counteract this activity (26). Whether an analogous intrinsic immunity protein has been responsible for driving suboptimal codon usage in some or in primitive gamma-2 herpesviruses remains to be determined. The availability of codon-optimized versions of gH and gL should facilitate investigations directed at the mechanism of ORF57 action and the consequences of codon optimization in the context of the viral genome and viral replication.
Acknowledgments
We thank Blossom Damania (University of North Carolina) for providing the RRV ORF50 expression plasmid and Jae Jung and Sun Hwa Lee for helpful insights and discussions.
This work was supported by PHS grants 1R01AI063928 and 1P01DE1438804 to R.C.D., RR00168 to NEPRC, and 5T32AI0724522 to J.P.B.
Footnotes
Published ahead of print on 14 May 2008.
REFERENCES
- 1.Bilello, J. P., J. S. Morgan, B. Damania, S. M. Lang, and R. C. Desrosiers. 2006. A genetic system for rhesus monkey rhadinovirus: use of recombinant virus to quantitate antibody-mediated neutralization. J. Virol. 801549-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, K. N., R. K. Holmes, A. M. Sheehy, and M. H. Malim. 2004. APOBEC-mediated editing of viral RNA. Science 305645. [DOI] [PubMed] [Google Scholar]
- 3.Boyne, J. R., and A. Whitehouse. 2006. Gamma-2 herpes virus post-transcriptional gene regulation. Clin. Microbiol. Infect. 12110-117. [DOI] [PubMed] [Google Scholar]
- 4.Bulmer, M. 1987. Coevolution of codon usage and transfer RNA abundance. Nature 325728-730. [DOI] [PubMed] [Google Scholar]
- 5.Carbone, A., A. Zinovyev, and F. Kepes. 2003. Codon adaptation index as a measure of dominating codon bias. Bioinformatics 192005-2015. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L., G. Liao, M. Fujimuro, O. J. Semmes, and S. D. Hayward. 2001. Properties of two EBV Mta nuclear export signal sequences. Virology 288119-128. [DOI] [PubMed] [Google Scholar]
- 7.Farjot, G., M. Buisson, M. Duc Dodon, L. Gazzolo, A. Sergeant, and I. Mikaelian. 2000. Epstein-Barr virus EB2 protein exports unspliced RNA via a Crm-1-independent pathway. J. Virol. 746068-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feinberg, M. B., R. F. Jarrett, A. Aldovini, R. C. Gallo, and F. Wong-Staal. 1986. HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell 46807-817. [DOI] [PubMed] [Google Scholar]
- 9.Felber, B. K., M. Hadzopoulou-Cladaras, C. Cladaras, T. Copeland, and G. N. Pavlakis. 1989. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 861495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontaine-Rodriguez, E. C., and D. M. Knipe. 2008. Herpes simplex virus ICP27 increases translation of a subset of viral late mRNAs. J. Virol. 823538-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouy, M., and C. Gautier. 1982. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 107055-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grote, A., K. Hiller, M. Scheer, R. Munch, B. Nortemann, D. C. Hempel, and D. Jahn. 2005. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 33W526-W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta, A. K., V. Ruvolo, C. Patterson, and S. Swaminathan. 2000. The human herpesvirus 8 homolog of Epstein-Barr virus SM protein (KS-SM) is a posttranscriptional activator of gene expression. J. Virol. 741038-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gustafsson, C., S. Govindarajan, and J. Minshull. 2004. Codon bias and heterologous protein expression. Trends Biotechnol. 22346-353. [DOI] [PubMed] [Google Scholar]
- 15.Haas, J., E. C. Park, and B. Seed. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6315-324. [DOI] [PubMed] [Google Scholar]
- 16.Kirshner, J. R., D. M. Lukac, J. Chang, and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 743586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik, P., D. J. Blackbourn, and J. B. Clements. 2004. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 27933001-33011. [DOI] [PubMed] [Google Scholar]
- 18.Malim, M. H., J. Hauber, R. Fenrick, and B. R. Cullen. 1988. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature 335181-183. [DOI] [PubMed] [Google Scholar]
- 19.Malim, M. H., J. Hauber, S. Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338254-257. [DOI] [PubMed] [Google Scholar]
- 20.Nekorchuk, M., Z. Han, T. T. Hsieh, and S. Swaminathan. 2007. Kaposi's sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export. J. Virol. 819990-9998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimura, K., K. Ueda, E. Guwanan, S. Sakakibara, E. Do, E. Osaki, K. Yada, T. Okuno, and K. Yamanishi. 2004. A posttranscriptional regulator of Kaposi's sarcoma-associated herpesvirus interacts with RNA-binding protein PCBP1 and controls gene expression through the IRES. Virology 325364-378. [DOI] [PubMed] [Google Scholar]
- 22.Qiu, J. T., R. Song, M. Dettenhofer, C. Tian, T. August, B. K. Felber, G. N. Pavlakis, and X. F. Yu. 1999. Evaluation of novel human immunodeficiency virus type 1 Gag DNA vaccines for protein expression in mammalian cells and induction of immune responses. J. Virol. 739145-9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruvolo, V., A. K. Gupta, and S. Swaminathan. 2001. Epstein-Barr virus SM protein interacts with mRNA in vivo and mediates a gene-specific increase in cytoplasmic mRNA. J. Virol. 756033-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandri-Goldin, R. M. 2004. Viral regulation of mRNA export. J. Virol. 784389-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharp, P. M., and W. H. Li. 1987. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 151281-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418646-650. [DOI] [PubMed] [Google Scholar]
- 27.Smith, R. W., P. Malik, and J. B. Clements. 2005. The herpes simplex virus ICP27 protein: a multifunctional post-transcriptional regulator of gene expression. Biochem. Soc. Trans. 33499-501. [DOI] [PubMed] [Google Scholar]
- 28.Sodroski, J., W. C. Goh, C. Rosen, A. Dayton, E. Terwilliger, and W. Haseltine. 1986. A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature 321412-417. [DOI] [PubMed] [Google Scholar]
- 29.Swaminathan, S. 2005. Post-transcriptional gene regulation by gamma herpesviruses. J. Cell Biochem. 95698-711. [DOI] [PubMed] [Google Scholar]
- 30.Whitehouse, A., M. Cooper, and D. M. Meredith. 1998. The immediate-early gene product encoded by open reading frame 57 of herpesvirus saimiri modulates gene expression at a posttranscriptional level. J. Virol. 72857-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams, B. J., J. R. Boyne, D. J. Goodwin, L. Roaden, G. M. Hautbergue, S. A. Wilson, and A. Whitehouse. 2005. The prototype gamma-2 herpesvirus nucleocytoplasmic shuttling protein, ORF 57, transports viral RNA through the cellular mRNA export pathway. Biochem. J. 387295-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeeberg, B. 2002. Shannon information theoretic computation of synonymous codon usage biases in coding regions of human and mouse genomes. Genome Res. 12944-955. [DOI] [PMC free article] [PubMed] [Google Scholar]