Abstract
Recently, complete replication of hepatitis C virus (HCV) in tissue culture was established using the JFH1 isolate. To analyze determinants of HCV genome packaging and virion assembly, we developed a system that supports particle production based on trans-packaging of subgenomic viral RNAs. Using JFH1 helper viruses, we show that subgenomic JFH1 replicons lacking the entire core to NS2 coding region are efficiently encapsidated into infectious virus-like particles. Similarly, chimeric helper viruses with heterologous structural proteins trans-package subgenomic JFH1 replicons. Like authentic cell culture-produced HCV (HCVcc) particles, these trans-complemented HCV particles (HCVTCP) penetrate target cells in a CD81 receptor-dependent fashion. Since HCVTCP production was limited by competition between the helper and subgenomic RNA and to avoid contamination of HCVTCP stocks with helper viruses, we created HCV packaging cells. These cells encapsidate various HCV replicons with high efficiency, reaching infectivity titers up to 106 tissue culture infectious doses 50 per milliliter. The produced particles display a buoyant density comparable to HCVcc particles and can be propagated in the packaging cell line but support only a single-round infection in naïve cells. Together, this work demonstrates that subgenomic HCV replicons are assembly competent, thus excluding cis-acting RNA elements in the core-to-NS2 genomic region essential for RNA packaging. The experimental system described here should be helpful to decipher the mechanisms of HCV assembly and to identify RNA elements and viral proteins involved in particle formation. Similar to other vector systems of plus-strand RNA viruses, HCVTCP may prove valuable for gene delivery or vaccination approaches.
Hepatitis C virus (HCV) is an enveloped plus-strand RNA virus of the genus Hepacivirus within the family Flaviviridae (34). The HCV genome is approximately 9.6 kb in length and consists of a single open reading frame encoding a polyprotein of ca. 3,000 amino acids and nontranslated regions (NTRs) located at the 5′ and 3′ termini. These NTRs are highly structured RNA segments encompassing critical cis-active RNA elements essential for genome replication and RNA translation (31). Viral proteins are expressed in a cap-independent manner by means of an internal ribosome entry site (IRES) located in the 5′ NTR. Co- and posttranslational cleavages liberate 10 viral proteins: core; envelope protein 1 (E1) and E2, representing the structural proteins that constitute the virion; a small membrane-associated ion channel protein designated p7 that is essential for virus assembly (16, 22, 43, 57); and six nonstructural (NS) proteins (NSs 2, 3, 4A, 4B, 5A, and 5B). HCV proteins NS3 to NS5B are both necessary and sufficient to establish membrane-bound replication complexes catalyzing RNA replication (13, 36). More recent data indicate that the NS2 protease that catalyzes cleavage at the NS2-NS3 site in addition participates in assembly and release of infectious viruses (22). Finally, ribosomal frame-shifting and internal translation initiation yield a group of additional proteins designated ARFP (alternative reading frame protein) or core+1 proteins. However, their function for the HCV replication cycle is currently not known.
One hallmark of HCV is its high propensity to establish a persistent infection, which frequently causes progressive morbidity ranging from hepatic fibrosis to cirrhosis and hepatocellular carcinoma (20). Despite considerable progress in the treatment of HCV infection, the currently available therapy (a combination of pegylated interferon alpha with ribavirin) is not well tolerated and is efficacious in only ca. 50% of patients infected with the most prevalent genotype 1 (38). Therapeutic or prophylactic vaccines are not available. For these reasons and with currently ca. 170 million persistently infected individuals, HCV infection represents a considerable global health problem necessitating pertinent basic and applied research efforts.
In recent years three major advances enabled analysis of the HCV replication cycle in tissue culture. First, Lohmann and colleagues developed subgenomic HCV replicons (36). These autonomously replicating RNA molecules carry all the genetic elements necessary for self-replication (the NTRs and NS3 to NS5B), including a selectable marker or a reporter gene in place of the viral structural proteins, and an internal IRES for expression of the HCV replicase genes (reviewed in reference 45). Second, HCV pseudotype particles, i.e., retroviral particles surrounded by an envelope carrying HCV E1-E2 complexes in place of their cognate envelope proteins, were established (3, 21). As these particles carry functional HCV glycoprotein complexes on their surface, HCV pseudotype particles have been instrumental for the analysis of E1-E2 receptor interactions and the early events of HCV infection (reviewed in reference 2). Finally, in 2005 fully permissive cell culture systems which are based on the JFH1 clone were described (33, 66, 72). This isolate replicates with unprecedented efficiency in transfected Huh7 human hepatoma cells and produces particles infectious both in vitro and in vivo, thus providing a model system reproducing the complete HCV replication cycle.
Use of these novel models has considerably expanded our knowledge of viral and host cell factors involved in HCV replication (for a recent review, see reference 59). It is now known that similar to virtually all other plus-strand RNA viruses, HCV induces intracellular membrane alterations and replicates its genome in conjunction with vesicular membrane structures, the so-called “membranous web” (10, 13). Presumably as a consequence of this specific, rather secluded architecture of the membrane-associated replication machinery, all viral proteins involved in RNA replication, with the exception of NS5A function in cis, cannot be complemented in trans (1). Restricted trans-complementation of viral replicase proteins has been observed for other plus-strand RNA viruses as well, thus indicating that a rather “closed” replication machinery is a shared feature of these viruses (15, 27, 60). In contrast, for a number of plus-strand RNA viruses from diverse virus families like Picornaviridae (poliovirus), Alphaviridae (Sindbis virus, Semliki Forest virus, and Venezuelan equine encephalitis virus), Coronaviridae (human coronavirus E229), and Flaviviridae (tick-borne encephalitis virus, Kunjin virus, West Nile virus, and yellow fever virus), assembly of progeny viruses can be achieved when structural proteins are expressed in trans and independent from the RNA molecule that encodes the replicase proteins. Similarly, Miyanari recently reported that HCV genomes with lethal mutations in core protein can be rescued by ectopic expression of functional core protein (39). This flexibility has been extensively used to create viral vectors for gene delivery as well as viral vector-based immunization approaches (32, 48, 49, 61, 68) (for a recent review on alphaviral vectors, the most frequently used among plus strand RNA vectors, see reference 37). In these systems the viral genome region encoding the structural proteins is replaced by a transgene. The resulting defective vector genomes are capable of RNA replication but due to the lack of structural proteins are unable to produce progeny virus particles. This defect is rescued by expression of the structural proteins in trans via helper viruses (28, 55) or, in some cases, by DNA constructs stably expressed in packaging cell lines (17). The resulting virus-like particles are infectious but support only single-round infection and are unable to spread, thus improving the safety of the viral transduction system.
Given the success of plus-strand RNA vector technology for basic and applied clinical research, in this study we developed a trans-complementation system for HCV that provided new insights into the basic principles of HCV particle assembly.
MATERIALS AND METHODS
Cell culture, cell lines, and antibodies.
All cell lines were grown in Dulbecco's modified minimal essential medium (DMEM; Invitrogen, Karlsruhe, Germany) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 10% fetal calf serum (DMEM complete). Huh7.5 and Huh7-Lunet cells (subclones of the Huh7 hepatoma cell line [4, 12]) that are highly permissive for HCV RNA replication and, in the case of the latter, also for HCV infection served as parental cell lines for construction of HCV packaging cells and control cells transduced with the empty lentiviral vector pWPI. Huh7-Lunet[CE1][E2p7NS2] cells were subcloned by limiting dilution. Briefly, cells were seeded into 96-well plates at a density of ca. 0.8 cells per well and cultivated in DMEM complete medium supplemented with blasticidin at a concentration of 5 μg/ml. Individual cell clones were expanded and analyzed as described in the text.
Plasmid construction.
The plasmids pFK-JFH1, pFK-Luc-JFH1/ΔE1E2 (where Luc is luciferase) (66), pFK-Jc1 (46), pFK-Luc-Jc1 (30), and pFKJc1/Δp7 (57) have been described recently. The plasmid pFKi389Luc-EI/NS3-3′_JFH1_dg encodes a bicistronic subgenomic JFH1 replicon (Luc-NS3-5B) consisting of the JFH1-derived nucleotides 1 through 389 encoding the 5′ NTR and the 16 amino-terminal residues of core fused in frame via an AscI restriction site to the gene encoding firefly luciferase, followed by the encephalomyocarditis virus (EMCV) IRES, the JFH1-derived coding sequence of NS3 to NS5B, and the 3′ NTR of JFH1. Plasmid pFKi389Venus-EI/NS3-3′_JFH1_dg encodes the Venus-NS3-5B subgenomic JFH1 replicon with analogous composition to Luc-NS3-5B but comprising the Venus variant of green fluorescent protein (GFP) (41) in place of luciferase. To permit generation of in vitro transcripts, the HCV replicon is flanked at the 5′ end by the promoter sequence of T7 polymerase and at the 3′ end by the hepatitis delta virus genomic ribozyme (dg), followed by the T7 terminator sequence (V. Lohmann, unpublished data). The plasmid pFK-SG-Luc-JFH1/5AGFP, a derivative of pFKI389Luc-EI/NS3-3′_dg_JFH1, was described recently (50) and was generated by insertion of GFP into domain 3 of the NS5A coding sequence (amino acid position 383). For construction of HCV packaging cell lines, we created derivatives of the lentiviral self-inactivating vector pWPI (44). In this vector the gene of interest is transcribed from an internal human elongation factor 1α (EF1-α) promoter. The transcribed unit also contains an IRES element from EMCV that allows internal initiation of translation of GFP. First, we replaced GFP with the gene encoding the blasticidin S deaminase (BSD) from Aspergillus terreus, which confers resistance to blasticidin to create pWPI-BSD. Then, we transferred the core and E1 coding region of the HCV isolate J6CF (70) by using a PCR-based strategy and employing the following oligonucleotides: S-Core_BamHI (5′-GCGCCGGATCCATGAGCACAAATCCTAAACCTCAAAG-3′) and A-E1-SpeI (5′-CCGGACTAGTTTATTACGCGTCCACCCCGGCGGCCAAC-3′). After restriction digestion with BamHI and SpeI, the amplicon was inserted into the digested pWPI-BSD vector, creating pWPI-CE1-BSD. For pWPI-SpE2p7NS2-BSD which encodes the 14 most-C-terminal residues of E1 (signal peptide), E2, p7, and NS2, a similar cloning strategy was employed using the oligonucleotides S-E2-BamHI (5′-GGCCAGATCTAAAGTCGTTGTCATCCTTCTG-3′) and A-NS2-SpeI (5′-GGCCACTAGTTTATTAAAGGAGCTTCCACCCCTTGGAGG-3′). Note that PCR amplification was performed using pFK-Jc1 as a template, which encodes a chimeric HCV polyprotein consisting of J6CF- and JFH1-derived sequences fused with each other at a junction within the NS2 coding region (46). As a consequence, the lentiviral vector transduces a chimeric polyprotein encompassing codons 370 to 846 of J6CF and codons 847 to 1030 of JFH1. Inserts generated by a PCR-based strategy were verified by automated nucleotide sequencing using an ABI 310 sequencer (Applied Biosystems, Darmstadt, Germany). Further details regarding the cloning strategies and exact nucleotide sequences can be obtained upon request.
In vitro transcription, electroporation, and transient HCV replication assays using luciferase reporter genomes.
In vitro transcripts of the individual constructs were generated by linearizing 5 to 10 μg of the respective plasmid by digestion for one hour with MluI. Plasmid DNA was extracted with phenol and chloroform and, after precipitation with ethanol, dissolved in RNase-free water. In vitro transcription reaction mixtures contained 80 mM HEPES (pH 7.5), 12 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol (DTT), a 3.125 mM concentration of each ribonucleoside triphosphate, 1 U of RNasin (Promega, Mannheim, Germany) per μl, 0.1 μg of plasmid DNA/μl, and 0.6 U of T7 RNA polymerase (Promega) per μl. After incubation for 2 h at 37°C, an additional 0.3 U of T7 RNA polymerase/μl of reaction mixture was added, followed by another 2 h at 37°C. Transcription was terminated by the addition of 1.2 U of RNase-free DNase (Promega) per μg of plasmid DNA and a 30-min incubation at 37°C. The RNA was extracted with acidic phenol and chloroform, precipitated with isopropanol, and dissolved in RNase-free water. The concentration was determined by measurement of the optical density at 260 nm. Denaturing agarose gel electrophoresis was used to check RNA integrity.
For electroporation of HCV RNA into Huh7-Lunet cells, single-cell suspensions were prepared by trypsinization of monolayers and subsequent resuspension with DMEM complete. Huh7-Lunet cells were washed with phosphate-buffered saline (PBS), counted, and resuspended at 1 × 107 cells per ml in Cytomix (65) containing 2 mM ATP and 5 mM glutathione whereas Huh7.5 cells were resuspended at 1.5 × 107 cells per ml. Five to 10 μg of in vitro transcribed RNA was mixed with 400 μl of cell suspension by pipetting and then electroporated with a Gene Pulser system (Bio-Rad, Munich, Germany) in a cuvette with a gap width of 0.4 cm (Bio-Rad) at 975 μF and 270 V. Cells were immediately transferred to 16 to 20 ml of DMEM complete, and 2 ml of the cell suspension was seeded per well of a six-well plate.
Quantification of luciferase reporter activity was used to determine transient HCV RNA replication as described previously (30). For assaying the luciferase activity, cells were washed once with PBS, lysed directly on the plate with 1 ml of ice-cold lysis buffer (0.1% Triton X-100, 25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, and 1 mM DTT, pH 7.8), and frozen. After being thawed, lysates were resuspended by pipetting. For each well, 100 μl of lysate was mixed with 360 μl of assay buffer (25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT, 2 mM ATP, and 15 mM K2PO4, pH 7.8) and, after addition of 200 μl of a luciferin solution (200 μM luciferin, 25 mM glycylglycine, pH 8.0), measured for 20 s in a luminometer (Lumat LB9507; Berthold, Freiburg, Germany). The kinetics of replication was determined by normalizing the relative light units of the different time points to the respective 4-h value. All luciferase assays were done at least in duplicate measurements.
Western blot analysis.
Cells were washed once with PBS, detached from the plate by trypsinization, and subsequently resuspended in DMEM complete. Cells contained in a small aliquot of the suspension were counted and centrifuged, and the cell pellet was harvested in 150 μl of 2× sample buffer (400 mM Tris, pH 8.8, 10 mM EDTA, 0.2% bromophenol blue, 20% sucrose, 3% sodium dodecyl sulfate [SDS], 2% β-mercaptoethanol, 1 × 10−4 U/ml aprotinin, 4 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride. After incubation at 37°C for 30 min with 50 to 75 U of benzonase (Merck, Darmstadt, Germany), samples were heated for 5 min at 95°C, and lysate equivalent to ca. 400,000 cells was loaded onto an 15% polyacrylamide-SDS gel. After electrophoresis, proteins were transferred to a polyvinylidene difluoride membrane using a semidry blotter (Bio-Rad, Munich, Germany) according to the manufacturer's instructions. The membrane was blocked in PBS supplemented with 0.5% Tween (PBS-T) and 5% milk for at least 1 h. Upon blocking, the membrane was incubated with primary antibodies diluted in PBS-T supplemented with 5% milk and, after extensive washing with PBS-T, incubated with horseradish peroxidase-conjugated secondary antibodies diluted in PBS-T supplemented with 5% milk. As primary antibodies a core-specific mouse monoclonal antibody (C7/50), kindly provided by D. Moradpour, or a polyclonal rabbit antibody raised against recombinant E2 protein derived from the J6CF strain was used at a dilution of 1:1,000 or 1:500, respectively. Bound antibodies were detected after a washing step with the ECL Plus Western Blotting Detection System (GE Healthcare Europe, Freiburg, Germany).
Lentiviral gene transfer.
Human immunodeficiency virus-based pseudotypes bearing vesicular stomatitis virus glycoprotein were generated by Lipofectamine 2000-mediated cotransfection of 293T cells (8). Briefly, 2.5 × 105 293T cells were seeded in 6-cm-diameter plates 1 day before transfection with 1 μg of envelope protein expression construct (pczVSV-G [23]), 3 μg of human immunodeficiency virus Gag-Pol expression construct (pCMVΔR8.74 [9]), and 3 μg of the lentiviral vector pWPI (44). The medium was replaced 6 h after transfection. Supernatants containing the pseudo-particles were harvested 48 h later. Target cells were transduced with filtered supernatants and selected 72 h later the by addition of 5 μg/ml blasticidin.
Luciferase infection assay.
For standard infection assays, Huh7.5 cells were seeded at a density of 6 × 104 cells per well of a 12-well plate 24 h prior to inoculation with 350 μl of virus preparation. Cells were inoculated for 4 h, washed, and lysed in 350 μl of lysis buffer 72 h later. Luciferase activity was determined as described above. For neutralization experiments a CD81-specific antibody (JS-81) or isotope-matched control antibodies (WM15) directed against CD13 were used as described in the text.
Indirect immunofluorescence.
Huh7.5 cells were seeded onto glass coverslips in 24-well plates at a density of 4 × 104 to 6 × 104 cells per well. Infection was performed 24 h after seeding by inoculation with 250 μl of filtered cell culture supernatant. Cells were fixed 72 h postinfection with 500 μl of PBS supplemented with 3% (wt/vol) paraformaldehyde for 10 min at room temperature. Subsequently, cells were washed three times, permeabilized with 0.5% Triton X-100 in PBS, and washed three times with PBS prior to incubation with the first antibody. Staining of NS5A was performed by using the 9E10 hybridoma supernatant (33) at a dilution of 1:2,000. Immunostaining of NS3 was performed using a rabbit polyclonal serum raised against the helicase domain of the Con1 strain at a dilution of 1:1,000 and the E2-specific antibody AP33 (42) at a concentration of 20 ng/μl in PBS supplemented with 5% normal goat serum. After three washes with PBS, bound primary antibodies were detected by using goat antibodies specific to rabbit immunoglobulin G (IgG) conjugated to Alexa-Fluor 546 or goat antibodies to murine IgG conjugated with Alexa-Fluor 488 (Invitrogen, Karlsruhe, Germany) at a dilution of 1:1,000 in PBS containing 5% normal goat serum for 30 min in the dark. DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole dihydrochloride) (Invitrogen) for 1 min. Finally, cells were washed three times with PBS and once with water and mounted on glass slides with Fluoromount G (Southern Biotechnology Associates, Birmingham, AL).
Immunohistochemical staining and virus titration.
Virus titers were determined as described elsewhere with slight modifications (33). In brief, Huh7.5 cells were seeded in 96-well plates at a density of 1 × 104 cells per well 24 h prior to inoculation with dilutions of filtered cell culture supernatant (at least six wells were used per dilution). After 2 to 3 days, cells were washed with PBS, fixed for 20 min with ice-cold methanol at −20°C, washed three times with PBS, and then permeabilized and blocked for 1 h with PBS containing 0.5% saponin, 1% bovine serum albumin, 0.2% dried skim milk, and 0.02% sodium acid. Endogenous peroxidases were blocked by incubating cells for 5 min with PBS containing 0.3% hydrogen peroxide. After three washes with PBS and one wash with PBS containing 0.5% saponin (PBS-saponin), NS5A was detected with a 1:1,000 dilution of hybridoma supernatant 9E10 (33) in PBS-saponin for 1 h at room temperature or overnight at 4°C. Cells were washed as described above, and bound 9E10 was detected by incubation with peroxidase-conjugated antibodies specific to murine IgG (Sigma-Aldrich, Steinheim, Germany) diluted 1:200 in PBS-saponin. After a 1-h incubation at room temperature, cells were washed as specified above. Finally, peroxidase activity was detected by using a Vector NovaRED substrate kit (Linaris Biologische Produkte GmbH, Wertheim, Germany). Virus titers (50% tissue culture infective dose [TCID50/ml]) were calculated based on the method of Spearman and Kärber (24, 56).
Density gradient.
Cell-cultured HCV ([HCVcc] Jc1) particles or trans-complemented HCV particles ([HCVTCP] Luc-NS3-5B) containing culture fluid were harvested 24 h postelectroporation of appropriate HCV RNA into Huh7.5 cells or Huh7.5[CE1][E2p7NS2] cells and passed though 0.45-μm-pore-size filters. One milliliter of the preparation was layered under a 0, 10, 20, 30, and 40% iodixanol step gradient (Optiprep; Axis-Shield, Oslo, Norway) prepared in a cell suspension medium containing 0.85% (wt/vol) NaCl and 10 mM Tricine-NaOH, pH 7.4. Gradients were centrifuged for 16 h at 154,000 × g in a TH-641 swing-out rotor at 4°C using a Sorvall Ultra WX80 centrifuge. Fractions (10 of 1 ml each) were collected from the bottom, and virus infectivity and the quantity of core protein were determined using a limiting dilution assay and a core-specific enzyme-linked immunosorbent assay, respectively. The density of the fractions was quantified by refractometry.
Quantitative detection of HCV core protein.
HCV core protein was measured using an HCV core antigen kit (Wako Chemicals, Neuss, Germany) according to the instructions of the manufacturer. Cell culture medium was filtered through 0.45-μm-pore-size filters and either directly used for enzyme-linked immunosorbent assay or diluted with PBS prior to measurement.
RNA quantification by RT-PCR.
Viral RNA was isolated from infected cells using a Nucleo Spin RNAII Kit (Macherey-Nagel, Düren, Germany), as recommended by the manufacturer. Two microliters of the RNA sample was used for quantitative reverse transcription-PCR (RT-PCR) analysis using a Light Cycler 480 (Roche, Mannheim, Germany). HCV-specific RT-PCRs were conducted in duplicates utilizing a one-step RT-PCR LightCycler 480 RNA Master Hydrolysis Probes Kit (Roche, Mannheim, Germany) and the following JFH1-specific probe (TIB Molbiol, Berlin, Germany) and primers (MWG-Biotech, Martinsried, Germany): A-195, 5′-6-carboxy-fluorescein-AAA GGA CCC AGT CTT CCC GGC AAT T-tetra-chloro-6-carboxy-fluorescein-3′; S-146, 5′-TCT GCG GAA CCG GTG AGT A-3′; and A-219, 5′-GGG CAT AGA GTG GGT TTA TCC A-3′. Reactions were performed in three stages by using the following conditions: stage 1, 3 min at 63°C (reverse transcription); stage 2, 30 s at 95°C (initial denaturation); and stage 3, 35 cycles of 15 s at 95°C and 30 s at 60°C (amplification). The amount of HCV RNA was calculated by comparison to serially diluted in vitro transcripts.
RESULTS
Helper virus-dependent trans-complementation.
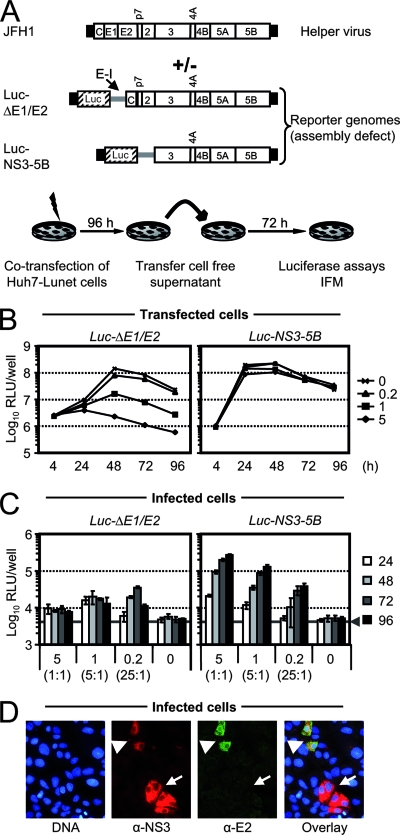
To explore if assembly-deficient JFH1 genomes with deletions of structural and additional genes can be rescued by trans-complementation, we cotransfected HCV luciferase reporter constructs with deletions of viral glycoprotein genes (Luc-ΔE1/E2 [66]) or with a deletion of the region from core to NS2 (Luc-NS3-5B) together with the full-length JFH1 genome as a helper virus into Huh7-Lunet cells (Fig. 1). Since HCV RNA genomes coreplicating in the same cell compete for limiting host factors crucial for replication (11, 35), we transfected 5 μg of reporter genome and varied the amount of cotransfected helper RNA from 0.2 to 5 μg in order to find a ratio between mutant and helper RNA optimal for efficient coreplication and trans-packaging. Nonspecific transduction of luciferase activity into inoculated target cells was controlled by transfection of the reporter RNAs alone. For both reporter replicons, replication in transfected Huh7-Lunet cells was most efficient in the absence of helper virus, reaching peak reporter activity by 24 h posttransfection in the case of Luc-NS3-5B and at 48 h for the Luc-ΔE1/E2 construct (Fig. 1B). The delayed replication kinetics of the latter is likely a consequence of the greater length of the replicon due to the insertion of the luciferase gene and the additional IRES, which reduces the efficiency of RNA replication (30). As a consequence of this diminished replication fitness, cotransfection of increasing quantities of the helper virus RNA dramatically reduced replication of Luc-ΔE1/E2 (Fig. 1B, left panel). In contrast, replication of Luc-NS3-5B was only slightly affected by the helper virus, probably because this construct displays replication fitness comparable to the wild-type JFH1 genome.
FIG. 1.
JFH1 helper virus-dependent trans-complementation. (A) Schematic representation of the JFH1 helper virus genome, the luciferase reporter construct, and the experimental setup. The JFH1 genome is depicted at the top, and the HCV polyprotein is indicated by a large box. HCV 5′ and 3′ NTRs are denoted as black bars, and the internal IRES of the EMCV (E-I) is given as a gray bar. The firefly luciferase gene is depicted as a hatched box (Luc). To analyze trans-complementation of the assembly-defective reporter genomes, 5 μg of the reporter RNA was transfected alone or cotransfected with increasing quantities of helper RNA. Transfected Huh7-Lunet cells were seeded into parallel wells and harvested at given time points to determine luciferase reporter activity. In addition, cell-free culture fluids were collected and used to inoculate naïve Huh7.5 target cells. Luciferase activity in the inoculated cells was quantified 72 h later. C, core protein. (B) Transient replication of reporter constructs in the presence or absence of helper virus RNA. In each transfection, 5 μg of the respective reporter construct was applied either alone or together with 0.2, 1, or 5 μg of helper RNA. Mean values of duplicate measurements are given. (C) Luciferase reporter activity in Huh7.5 cells inoculated with culture fluids of cells transfected as described in panel B. The quantity of cotransfected helper virus RNA (μg) is indicates below the graph, and the ratio between defective and helper RNA is given in brackets. The gray bar represents the background level of luciferase activity measured in uninfected Huh7.5 cells. Mean values of quadruplicate measurements and the standard errors of the means are given. (D) Indirect immunofluorescence analysis of HCV protein expression in Huh7.5 cells inoculated with culture fluid from Luc-NS3-5B and JFH1 cotransfected cells. HCV NS3 and E2 were detected by simultaneous staining with a rabbit polyclonal serum directed against NS3 (red staining) and the E2-specific mouse monoclonal antibody AP33 (green staining) and appropriate secondary antibodies. Nuclei were counterstained with DAPI (blue). White arrowheads denote cells infected with JFH1, whereas arrows point to cells infected with a virus particle carrying the subgenomic reporter genome. α, anti; RLU, relative light units; IFM, immunofluorescence microscopy.
To assess if the assembly-defective subgenomes are trans-packaged into infectious virus-like particles, we collected cell culture fluids of transfected cells and inoculated naïve Huh7.5 cells. After 72 h reporter activity in these cells was determined as a quantitative measure for productive infection with virus-like particles carrying the genome of the reporter constructs. As can be seen in Fig. 1C, both assembly-defective reporter RNAs were encapsidated into infectious particles, as evidenced by easily detectable reporter activity in the inoculated cells. However, trans-complementation was much more efficient when JFH1 was cotransfected with Luc-NS3-5B than with Luc-ΔE1/E2. In the case of the former, increasing doses of the helper RNA substantially enhanced trans-complementation of Luc-NS3-5B, reaching peak efficiency at a 1:1 ratio of replicon and helper RNA. Further augmentation of helper RNA, however, did not stimulate trans-packaging, indicating that availability of the helper RNA is no longer limiting once a 1:1 ratio is reached (data not shown).
To directly show that virus-like particles carrying the subgenomic HCV replicon RNA had productively infected the target cells, we assessed infection events at the single-cell level by using indirect immunofluorescence. Employing antibodies directed against HCV E2 and NS3, we detected foci of cells expressing both proteins simultaneously as well as cell nests expressing NS3 only (Fig. 1D). These data provide strong evidence that, besides viruses with the full-length HCV genome, particles containing the subgenomic HCV RNA had successfully infected the cells, resulting in either simultaneous E2 and NS3 production or in NS3 expression only. Although this experiment does not fully exclude the possibility that recombination between the helper virus and the replicon may also occur, based on these results, we conclude that transduction of the reporter activity was accomplished by packaging of the subgenomic reporter replicon rather than by a recombinant full-length HCV genome that had acquired the luciferase reporter gene. Moreover, these data indicate that two types of particles are generated: on one hand, replication-competent infectious HCV particles carrying the complete HCV genome and, on the other hand, virus-like particles carrying the subgenomic replicon RNA and thus supporting only single-round infection. These particles arising by trans-complementation were designated HCVTCP.
CD81-dependent infection of target cells by HCVTCP.
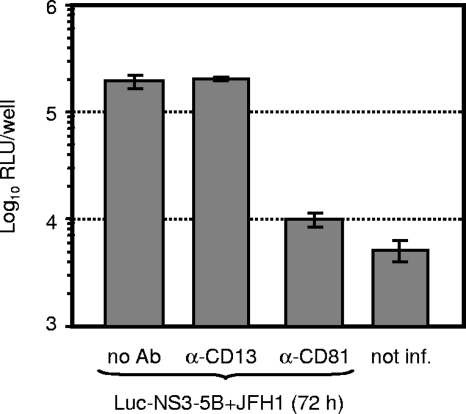
About 10 years ago the tetraspanin CD81 was identified as a molecule that binds E2 protein, and it was therefore implicated as a candidate receptor for HCV (47). In the meantime, a large body of evidence has accumulated that CD81, among other essential host factors, is a key component of the HCV receptor complex (for a recent review, see references 2 and 18). Thus, to firmly establish that HCVTCP enter Huh7.5 target cells in the same way as HCVcc particles and that luciferase expression in the inoculated cells resulted from authentic infection rather than nonspecific transfer of reporter gene activity, we inoculated cells with HCVTCP in the presence of CD81-specific antibodies or control antibodies. As is depicted in Fig. 2, anti-CD81 antibodies reduced transduction of luciferase reporter activity by more than 90% to almost background levels, whereas antibodies directed against CD13, another protein expressed on the surface of Huh7 cells, did not affect infection. In summary, these data show that a JFH1 helper virus efficiently trans-packages subgenomic JFH1 replicons and that HCVTCP, like HCVcc particles, infect target cells in a CD81-dependent fashion.
FIG. 2.
CD81-dependent infection of Huh7.5 cells with HCVTCP. Huh7.5 cells were inoculated with HCVTCP harvested 72 h postcotransfection of JFH1 and Luc-NS3-5B in the absence of antibodies (no Ab) or in the presence of 5 μg/ml CD81-specific (JS-81; Becton Dickinson) or CD13-specific (WM15; Becton Dickinson) antibodies. Infection efficiency was determined 72 h later by measuring luciferase activity in the inoculated cells. Background luciferase activity was determined in uninfected cells (not inf.). Mean values of quadruplicate measurements with the standard errors of the means are given. RLU, relative light units; α, anti.
Genotype dependence of trans-packaging of JFH1 subgenomes.
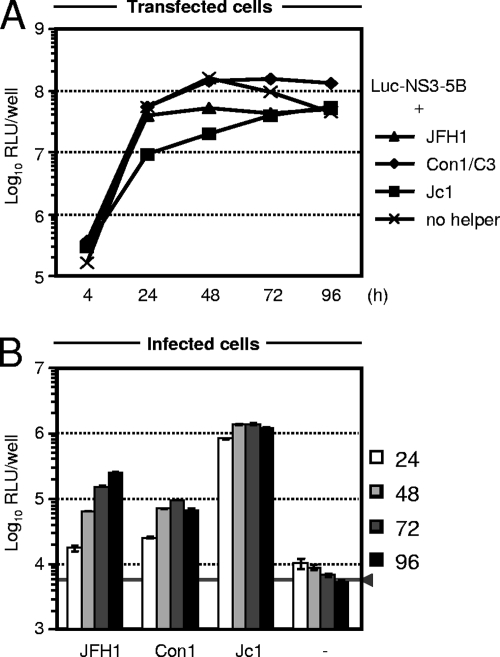
The scope of the JFH1-based infection system was recently expanded by the construction of chimeric HCV genomes (14, 33, 46, 71). These chimeras encode JFH1 NS3 to NS5B and the JFH1 NTRs and core protein to NS2 from alternative HCV isolates of various genotypes. Due to the highly efficient JFH1 replicase, these constructs amplify to high levels in transfected Huh7 cells and produce various quantities of infectious particles, arguing for genotype-specific determinants of assembly (52, 57). To determine whether genotype-specific determinants also affect trans-packaging efficiency and to identify the most efficient helper virus for trans-packaging, we cotransfected the JFH1 Luc-NS3-5B replicon with transcripts of either the JFH1 genome or the virus chimeras Con1/C3 or Jc1 (46). The latter constructs encode Con1-derived (genotype 1b) or J6CF-derived (genotype 2a) genome segments, respectively, fused with JFH1 at a junction within the NS2 coding region. As presented in Fig. 3, the subgenomic JFH1 luciferase replicon amplified efficiently, irrespective of which helper virus was cotransfected, with luciferase expression levels increasing more than 100-fold between 4 and 72 h posttransfection. Notably, however, replication was somewhat less efficient when the subgenomic replicon was cotransfected with JFH1 and, in particular, with Jc1 helper RNA. While all helper viruses complemented the subgenomic replicon, the efficacy of trans-packaging was quite variable (Fig. 3B). Production of infectious HCVTCP was most efficient with the Jc1 helper virus and least effective in the case of Con1/C3. These data show that JFH1 subgenomic replicons can be trans-packaged into HCVTCP by not only an autologous helper virus (JFH1) but also chimeric viruses that provide heterologous structural proteins from different viral subtypes or genotypes, as in case of Jc1 or Con1, respectively. The efficiency of trans-packaging attained with these helper viruses roughly correlates with their capacity to assemble infectious HCVcc particles, and the process is most efficient for Jc1 and least efficacious for Con1/C3 (46).
FIG. 3.
Complementation of JFH1 replicon by chimeric HCV helper viruses. (A) Five micrograms of the reporter RNA was transfected alone or cotransfected with 5 μg of the indicated helper virus RNA into Huh7-Lunet cells. Replication of the reporter replicon was assessed by luciferase assays of cell lysates harvested at the given time points. Mean values of duplicate measurements are given. (B) Release of infectious HCVTCP was quantified by inoculation of Huh7.5 cells with cell-free culture fluid harvested from transfected cells at 24, 48, 72, and 96 h posttransfection. Reporter gene activity was determined 72 h later and is given as mean values of quadruplicate measurements together with the standard errors. RLU, relative light units.
Construction and characterization of HCV packaging cell lines and HCVTCP.
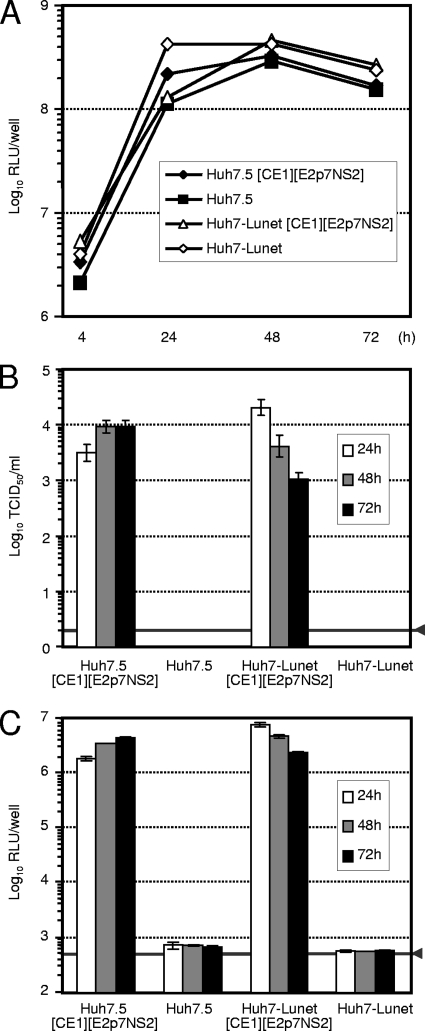
Recently, DNA-based expression systems have been described for initiation of HCV replication and virus production in tissue culture (19, 25). These reports demonstrate that functional full-length HCV RNA can be generated and that productive replication can be initiated by RNA polymerase II promoter-dependent nuclear expression of HCV RNA. Beyond this, Harvey and colleagues recently developed tetracycline-inducible packaging cell lines for production of flavivirus-like particles transducing replicon RNA (17). These reports encouraged us to construct stable packaging cell lines for production of high-titer HCVTCP preparations. To improve the biosafety of the packaging cell lines and to permit trans-packaging of subgenomic replicons lacking the complete core-to-NS2 coding region, we separated this part of the HCV genome into two distinct genetic modules that were stably coexpressed by independent lentiviral vectors (Fig. 4A). Since the Jc1 helper virus proved to be most efficient in trans-complementing JFH1 replicons, we chose the Jc1 genome for construction of these vectors. By transducing Huh7.5 and Huh7-Lunet cells, two cell clones known to support HCV RNA replication and particle production with high efficiency (4, 12, 29), with both lentiviral vectors simultaneously and by subsequent selection with blasticidin, we derived two stable cell lines designated Huh7.5[CE1][E2p7NS2] and Huh7-Lunet[CE1][E2p7NS2], respectively (Fig. 4B). Expression of both transgene cassettes was validated by Western blot analysis using an E2-specific polyclonal rabbit serum and a core-specific mouse monoclonal antibody (Fig. 4C and D). To assess the efficiency of trans-packaging of JFH1 replicons, we transfected both the parental cell lines and the novel packaging cell lines with the Luc-NS3-5B replicon and monitored RNA replication by luciferase assays and HCVTCP production by inoculation of naïve Huh7.5 cells (Fig. 5). The subgenomic replicon amplified to comparable levels in all cell lines tested. As expected, infectious HCVTCP were produced only in Huh7.5[CE1][E2p7NS2] and Huh7-Lunet[CE1][E2p7NS2] packaging cell lines and reached peak titers in the range of 1 × 104 TCID50/ml, with transduction of reporter activity into target cells more than four orders of magnitude above the assay background.
FIG. 4.
Construction of stable HCV packaging cell lines. (A) Lentiviral vectors employed for generation of Huh7.5 and Huh7-Lunet packaging cell lines. The Jc1 chimera (46) consisting of J6CF (gray bars) and JFH1-derived (open bars) genome segments and depicted at the top was utilized to create (1) pWPI-Core-E1 and (2) pWPI-E2-p7-NS2 vectors encoding the core-to-E1 region and the E2-p7-NS2 genome segment of Jc1, respectively. Note that the latter vector also encodes the 14 most-C-terminal residues of E1 which serve as a signal peptide (Sp) for E2 and that this construct encodes a chimeric NS2 protein consisting of J6CF-derived and JFH1-derived residues. For further details of pWPI vector features, see the Materials and Methods section and Pham et al. (44). The JFH1 replicon (3) Luc-NS3-5B is given at the bottom. LTR, long terminal repeat; SIN, self-inactivating; C, core. (B) Experimental setup to create HCVTCP in stable HCV packaging cell lines. (C and D) Analysis of transgene expression by Western blotting. Cell lysates of the cell lines indicated above the lanes were separated by denaturing SDS-polyacrylamide gel electrophoresis, blotted, and analyzed with actin-, HCV core-, and HCV E2-specific antibodies.
FIG. 5.
Efficient complementation of a subgenomic JFH1 replicon in Huh7.5 and Huh7-Lunet packaging cells. (A) Transient replication of Luc-NS3-5B RNA in transfected Huh7.5 and Lunet packaging cells as well as the respective parental cell lines. Luc-NS3-5B RNA was transfected into the indicated cell lines; cells were harvested at the time points given to determine reporter gene activity. Mean values of duplicate measurements are shown. (B and C) Infection of Huh7.5 target cells by HCVTCP generated upon transient transfection of packaging cells. Infectivity was determined by a limiting dilution assay (B) or reporter gene assays (C). RLU, relative light units.
To further enhance the efficiency of the trans-packaging system, we explored two strategies. First, we analyzed whether various adaptive mutations that had recently been identified by passaging intergenotypic JFH1 chimeras in Huh7.5 cells can boost HCVTCP production. Neither the V2440L mutation within the C-terminal portion of NS5A that elevated HCVcc production of JFH1 as well as genotype 1a, 1b, and 3a chimeras (26) nor the Q1251L and K1404Q mutations that had been identified in genotype 1a/2a and GT 3a/2a chimeras, respectively (14, 71), improved HCVTCP production in the packaging cell lines (data not shown).
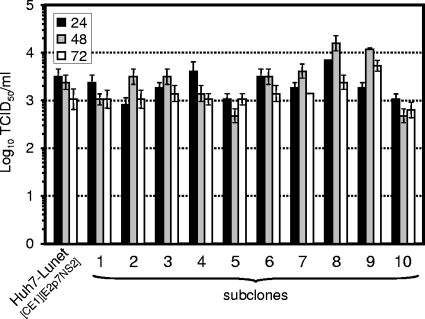
As a second approach to improve the efficacy of HCVTCP production, we subcloned the Huh7-Lunet[CE1][E2p7NS2] packaging cell line by limiting dilution and derived 10 independent subclones of the initial population of cells. Subsequently, we assessed the efficiency of HCVTCP production in the subclones (Fig. 6). Although most subclones permitted comparable HCV RNA replication (data not shown) and displayed comparable levels of HCVTCP production, we observed approximately threefold higher peak luciferase transduction in cell clones Lunet#8 and Lunet#9 than in the bulk culture of Huh7-Lunet[CE1][E2p7NS2] cells. Given that HCV RNA replication in these cell lines is comparable (data not shown), these data indicate that HCV transgene expression or host factor(s) for virus assembly in the former cells is somewhat more favorable for virus production, thus permitting generation of HCVTCP preparations with higher infectious titers.
FIG. 6.
HCVTCP production in Huh7-Lunet[CE1][E2p7NS2] subclones. Huh7-Lunet[CE1][E2p7NS2] cells were subcloned by limiting dilution as detailed in the Materials and Methods section. Subsequently, individual cell clones and the parental packaging cell line were transfected with Luc-NS3-5B RNA, and production of HCVTCP was monitored by using a limiting dilution assay.
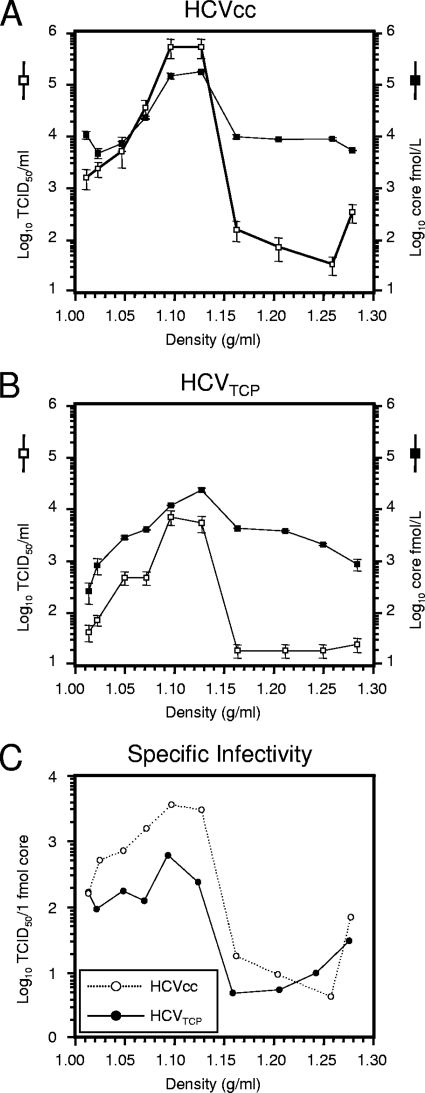
Next, we compared the biophysical properties of HCVTCP with those of HCVcc particles. To this end, we compared the distribution of infectivity and core protein between HCVcc and HCVTCP upon equilibrium centrifugation through iodixanol gradients (Fig. 7). Based on the quantification of core protein, the majority of HCVcc particles floated to a low density between 1.07 to 1.12 g/ml, with those particles between 1.10 and 1.12 g/ml displaying highest specific infectivity (Fig. 7C). HCVTCP showed a comparable distribution, and both core protein quantity and infectivity peaked between 1.10 g/ml and 1.12 g/ml. Interestingly, in total the HCVcc preparation contained approximately sevenfold higher numbers of core protein (as judged by the total amount of core in the gradient) than the HCVTCP, but ca. 76-fold higher infectivity, suggesting that HCVcc particles have an approximately 10-fold higher specific infectivity. In agreement with this notion, the highly infectious fractions of HCVcc and HCVTCP in the range between 1.02 and 1.12 g/ml differed in terms of specific infectivity by a factor of approximately 5 to 10 (Fig. 7C). Relating the infectivity measurements of HCVcc and HCVTCP to the amount of core protein in the fraction with highest specific infectivity (1.10 g/ml) and assuming that one HCV particle may harbor 180 copies of core, then one TCID50 unit corresponds to approximately 900 and 5,600 particles, respectively.
FIG. 7.
Buoyant density and specific infectivity of HCVcc and HCVTCP. Jc1 HCVcc particles (A) and Luc-NS3-5B HCVTCP (B) harvested 24 h postelectroporation of Huh7.5 or Huh7.5[CE1][E2p7NS2] cells were resolved by using an iodixanol step gradient. Ten fractions were harvested from the bottom, and infectivity and the amount of core protein were determined. (C) Specific infectivity of HCVcc particles and HCVTCP expressed as TCID50 units per 1 fmol of core protein.
In conclusion, these data indicate that HCVcc and HCVTCP have comparable buoyant density; however, HCVcc preparations display an approximately 5 to 10 times higher ratio of infectious to noninfectious particles.
During virus production in the packaging cell lines, the HCV genome is separated into three distinct genetic units, substantially lowering the likelihood for reconstitution of fully replication-competent viruses by RNA recombination. However, given the existence of some homologous coding regions on the distinct RNA molecules (e.g., EMCV IRES) that may initiate recombination, we wanted to stringently exclude the possibility that low levels of spreading viruses are created during HCVTCP preparation in the packaging cell lines. We therefore generated concentrated HCVTCP preparations carrying either the Luc-NS3-5B genome (Luc-SG) or the Jc1Δp7 RNA, an assembly-defective HCV genome (57) that is complemented with highest efficiency in the packaging cells (see below and Fig. 9). Using these preparations, we inoculated naïve Huh7.5 cells (to our knowledge the most permissive cell line for HCV infection in vitro) and the Huh7.5[CE1][E2p7NS2] packaging cell line in parallel. Seventy-two hours later, we fixed these cells for detection of HCV NS5A by immunofluorescence and of cell-associated HCV RNA by quantitative RT-PCR (Fig. 8). In addition, we measured the infectivity in the culture fluid of the cells (Fig. 8B, left) and passed the cell-free culture fluid of these cells to naïve Huh7.5 and the Huh7.5 packaging cells for initiation of a second round of infection. Seventy-two hours later, these cells were again processed as described above.
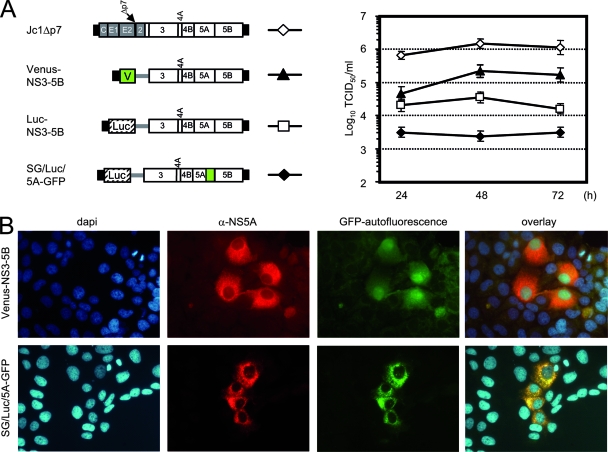
FIG. 9.
Packaging efficiency of various HCV genomes. (A) HCV constructs given on the left were transfected into Huh7.5[CE1][E2p7NS2] packaging cells. Culture fluid of the transfected cells was harvested 24, 48, and 72 h later, and infectious HCVTCP were titrated using a limiting dilution assay. C, core. (B) Expression of the fluorescent proteins Venus-GFP (V) and NS5 fused with GFP (50) was analyzed with fluorescence microscopy. From left to right, nuclear staining (DAPI; blue), anti-NS5A (red), GFP-autofluorescence (green), and the overlay of all three channels are given. α, anti.
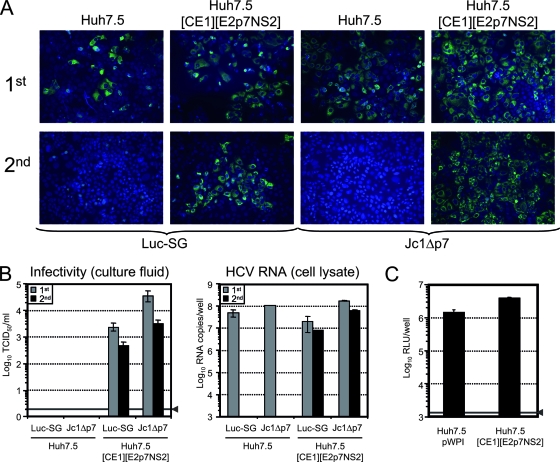
FIG. 8.
Serial passage of HCVTCP with Luc-NS3-5B (Luc-SG) or Jc1Δp7 genome on Huh7.5 and Huh7.5[CE1][E2p7NS2] packaging cells. (A) Cells were inoculated with a high dose of the respective HCVTCP to initiate the first round of infection (1st) and cultured for 72 h before they were fixed and stained using NS5A-specific monoclonal antibodies (green). Nuclei were counterstained with DAPI (blue). Cell-free culture fluid of these cells was harvested and passed to naïve Huh7.5 cells (second round of infection) or naive packaging cells, which were again fixed and stained 72 h later. (B) Infectivity of the culture fluids harvested 72 h after the first and second infections (left) as well as the quantity of HCV RNA in the lysate (right) of the cells at this time point was determined. The solid line (arrowhead) represents the detection limit of the limiting dilution assay. (C) Absence of superinfection exclusion in Huh7.5[CE1][E2p7NS2] packaging cells. Huh7.5[CE1][E2p7NS2] cells and control cells transduced with the empty lentiviral vector (Huh7.5 pWPI) were inoculated with Luc-Jc1 reporter viruses and harvested 72 h later. Infection efficiency was quantified by determining cell-associated reporter gene activity. Mean values of quadruplicate measurements and the standard errors of the means are given. The solid line (arrowhead) represents the background level of luciferase activity measured in uninfected cells.
In the first round of inoculation for both HCVTCP preparations, we detected no difference in the efficiency of infection between Huh7.5 cells and the packaging cells, as evidenced by similar numbers of NS5A-positive cells and comparable quantities of cell-associated HCV RNA (Fig. 8A, top panels, and B, right panel, gray bars). Not surprisingly, due to a higher infection dose (data not shown), Jc1Δp7 HCVTCP infected somewhat higher cell numbers than the Luc-SG HCVTCP, resulting in proportionally elevated levels of HCV RNA in the cells. Importantly, only the packaging cells released detectable quantities of infectious particles. In agreement with this, after the second passage we detected high numbers of infected cells when HCVTCP were propagated on the packaging cell line, whereas not a single infection event was noted in case of passaging in naïve Huh7.5 cells (Fig. 8A, bottom panel). To strictly rule out contamination of the HCVTCP preparation with a fully replication-competent virus that may have arisen due to recombination, we quantified cell-associated HCV RNA in the second-round-infected cells using an RT-PCR assay with a detection limit of ca. 0.001 HCV RNA copies per cell. Even with this highly sensitive method, we were unable to detect HCV RNA in the cells inoculated with the culture fluid of Huh7.5 cells that had previously been infected with either the Luc-SG or the Jc1Δp7 HCVTCP. To further rule out that a low-level contamination with a replication-competent recombinant virus only evident after an additional passage had occurred, we passaged the second-round-infected cells one more time. However, the cells remained RT-PCR negative (data not shown).
On one hand, these data indicate that the concentrated HCVTCP preparations did not contain detectable amounts of recombinant full-length HCV capable of spreading in culture; on the other hand, these results show that HCVTCP can efficiently infect the packaging cells and thus be propagated. This excludes the possibility that stable expression of the structural proteins including p7 and NS2 leads to superinfection exclusion, a state when infection of the cell by the autologous virus is prevented by previous infection or viral protein expression. In line with this conclusion, Luc-Jc1 HCVcc particles entered the Huh7.5[CE1][E2p7NS2] packaging cells with an efficiency indistinguishable from infection of Huh7.5 pWPI cells transduced with the empty lentiviral vector (Fig. 8C).
Versatility of HCV packaging cell lines.
To investigate the flexibility and efficiency of the HCVTCP packaging system in more detail, we transfected Huh7.5[CE1][E2p7NS2] cells with a set of HCV reporter replicons and a Jc1 p7 deletion mutant recently shown to be assembly deficient (57); we assessed HCVTCP production and, where applicable, transgene expression in the target cells (Fig. 9). Depending on the genome transfected, we observed various HCVTCP titers ranging from ca. 3 × 103 TCID50/ml in the case of SG/Luc/5A-GFP to more than 1 × 106 TCID50/ml for Jc1Δp7. In the case of Venus-NS3-5B and SG/Luc/5A-GFP (50), cells expressing the respective fluorescent protein (Venus-GFP or NS5A-GFP, respectively) were readily detected by fluorescence microscopy (Fig. 9B). Importantly, in the infected cells both proteins displayed their typical subcellular localization, which is cytoplasmic and nuclear for Venus-GFP but cytoplasmic and mostly dot-like for the NS5A-GFP fusion protein (indicating the membranous web) (40, 50). Together, these data show that a large spectrum of HCV replicons with various configurations and with different transgenes can be efficiently packaged into HCVTCP.
DISCUSSION
In this study, we analyzed to what extent HCV genomes with defects that inactivate viral functions essential for autonomous virus assembly and release can be rescued by expression of these functions in trans. This kind of flexibility in terms of virus assembly and genome packaging is commonly observed for plus-strand RNA viruses and often exploited for gene delivery applications and viral vector-based immunization approaches but has not previously been observed for HCV. In a first series of experiments, we documented that full-length JFH1 RNA, when cotransfected together with subgenomic JFH1 replicons, functions as helper virus and trans-packages assembly-defective HCV genomes into virus-like particles. Importantly, we have demonstrated that these particles, like HCVcc particles, penetrate target cells in a CD81-dependent fashion, thus documenting HCV receptor-dependent entry rather than nonspecific uptake of these particles into host cells. By analyzing infection events at the single-cell level, we observed foci expressing E2 and NS3 simultaneously and other nests expressing NS3 only. These data indicate that HCV particles carrying a full-length HCV genome in addition to particles comprising the subgenomic RNA had been generated and had then initiated the infection, respectively.
Since we could readily delete the entire core-to-NS2 coding region without loss of trans-packaging, this portion of the viral genome does not contain crucial cis-active elements required for packaging. Hence, if the HCV genome comprises essential packaging signals as, for instance, retroviruses do (reviewed in reference [7]), which may be recognized by the core protein in the course of assembly, these must reside within the coding region of NS3 to NS5B and/or the NTRs. Given the extensive RNA secondary structures observed within the NTRs of the HCV genome, these stretches may mediate a specific interaction with HCV core protein and thus initiate packaging. Although not undisputed (67), various reports identified a direct and specific binding of HCV core to the IRES modulating its activity (5, 53, 54). The latter findings are in principal in line with the hypothesis that the HCV RNA comprises specific packaging signals and further suggest that the interaction between core and these sequences may be regulated to permit translation by ribosomes, replication by HCV NS proteins, and packaging by core in a coordinated manner. Alternatively, a model can be envisioned wherein specific HCV RNA packaging is accomplished by specific protein-protein rather protein-RNA interactions, ensuring loading of nascent HCV particles with only the cognate HCV RNA. In this regard it is noteworthy that Miyanari and colleagues recently described recruitment of HCV NS proteins and replication complexes to lipid droplets and that this process was essential for virus production (39). On the basis of this finding, it is tempting to speculate that HCV RNA destined for packaging may be shuttled to core protein via a direct interaction with viral NS protein(s). In this scenario, particular cis-active RNA packaging elements would not necessarily be required—except for cis-active signals that mediate RNA replication and recognition by the HCV replicase complex. Although our data cannot distinguish between these models, the technology described in this article should be instrumental in further defining the requirements for HCV genome packaging.
It is interesting that the Luc-ΔE1/E2 construct was rescued by the JFH1 helper virus with much lower efficiency than the Luc-NS3-5B replicon. This is likely due to gross differences in replication capacity between helper virus and the Luc-ΔE1/E2 genome. It is known that Luc-ΔE1/E2 replicates with substantially delayed kinetics compared to JFH1 (30). As a consequence, efficient replication of the helper virus displaces the Luc-ΔE1/E2 RNA due to competition for limiting cellular factors essential for HCV genome amplification, as described recently (11, 35). Due to this interference, trans-packaging of the assembly-defective RNA is restricted by limited availability of the replicon RNA. When lower quantities of the helper are transfected, Luc-ΔE1/E2 replicates with high efficiency; however, presumably due to low levels of the helper virus and therefore lack of sufficient quantities of structural proteins, HCVTCP production is inefficient. To some extent these in vitro transient coreplication experiments reflect the interplay between a defective interfering (DI) particle and its cognate helper virus. The occurrence of DI particles is a well-characterized phenomenon for many viruses (51). Such particles correspond to viruses that contain genomes with large deletions of essential functions but which nevertheless retain critical elements for genome encapsidation and replication. In the presence of a helper virus which provides the lacking function in trans, these DI genomes are packaged and compete with the helper for cellular and viral resources, thus interfering with helper replication. For instance, in the case of the bovine viral diarrhea virus, a close relative of HCV from the Pestivirus genus within the family Flaviviridae, the natural occurrence of DI particles that dramatically influence pathogenesis and comprise a genome with an in-frame deletion of the entire structural protein-encoding region is well documented (58). Interestingly, Yagi and colleagues recently observed HCV subgenomes with large in-frame deletions of E1-E2 and portions of NS2 in samples from chronically infected patients (69), indicating that DI particles arise during chronic HCV infection. Given the competition between the defective RNA and the helper virus observed in vitro, it is conceivable that production of DI particles in vivo may reduce virus fitness and thus support persistence. Future studies using the trans-complementation system described in this study could help to understand this interplay and its consequences on virus replication in more detail.
To open the way for HCV assembly and infection studies with less stringent biosafety measures, we wished to develop a flexible and safe system for the preparation of high-titer HCVTCP. To this end, we dissected the HCV genome into three independent genetic modules. These are the core-E1 region and the E2p7NS2 genes inserted into two lentiviral vectors and stably integrated into the genome of Huh7-Lunet or Huh7.5 cells and an assembly-deficient HCV replicon, which in the minimal case carries only those elements necessary for autonomous replication. Due to this configuration, recombination and regeneration of fully replication-competent and spreading HCV are largely excluded. In line with this notion, we did not detect any spreading HCV particles in concentrated HCVTCP preparations carrying either the Luc-NS3-5B subgenome or the Jc1Δp7 genome, which harbors only a small in-frame deletion of 29 codons of the p7 coding region, thus documenting the improved safety of the novel system.
Further characterization of the packaging cells revealed the absence of superinfection exclusion, a state wherein virus-infected cells are refractory to secondary virus infection with the homologous virus. Although this phenomenon has recently been observed for Huh7 cells previously infected with HCVcc or stably replicating a selectable HCV replicon, the interference was found to be due to competition at the level of RNA translation and replication and not at the level of infection (50, 64). In agreement, we found that the mere expression of the core-to-NS2 region is apparently not sufficient to render the cells noninfectable. This finding also has practical implications since reinfection of the packaging cells by HCVTCP permits passage of the otherwise only single-round-infected particles in these cells and thus allows simple production of large-scale HCVTCP stocks. It is worth noting here that due to limited CD81 expression in Huh7-Lunet cells, the most efficient propagation of HCVTCP can be achieved by using the Huh7.5[CE1][E2p7NS] packaging cell line which expresses high levels of this crucial receptor (29).
Analyzing the biophysical properties of HCVTCP, we observed a density profile comparable to HCVcc, which was characterized by a large number of particles displaying low and very low densities. These data suggest that HCVTCP, like HCVcc particles, are associated with various types of lipoproteins, which likely account for the heterogeneous density of HCV particles (6, 62, 63). Despite this congruence, HCVTCP prepared with the Luc-NS3-5B genome displayed somewhat reduced specific infectivity compared to HCVcc. Although the reason for this difference is currently unknown, it is conceivable that genome packaging or saturation of the particles with glycoproteins or host factors (e.g., lipoproteins) may be slightly less efficient, resulting in an elevated proportion of noninfectious particles.
Given the flexibility of trans-encapsidation, the described packaging cells represent a versatile platform for preparation of HCVTCP customized for the required application and carrying various reporter genes and even directly labeled viral proteins (e.g., NS5A). Since these viruses (when applied to naïve Huh7 cells) cannot spread in cell culture and are unable to produce infectious progeny, their utilization permits strictly synchronous infections, thus facilitating kinetic studies of virus entry. Depending on the type of assembly-defective genome transfected, we observed production of infectious HCVTCP titers ranging from ca. 3 × 103 TCID50/ml in the case of SG/Luc/5A-GFP to more than 1 × 106 TCID50/ml for Jc1Δp7. As we have previously observed that insertion of GFP into NS5A does not affect RNA replication but decreases the efficiency of HCVcc production (50), it is not surprising that the SG/Luc/5A-GFP was packaged into HCVTCP with the least efficiency. Virus production of Luc-NS3-5B and Venus-NS3-5B in Huh7.5[CE1][E2p7NS] cells was substantially more efficient, probably due to the availability of an intact full-length NS5A in both cases and, for the latter, also a shorter reporter genome, which likely replicates more efficiently and thus sustains higher numbers of HCVTCP production. It is interesting that Jc1Δp7 was packaged with by far the highest efficiency, reaching a level of infectivity comparable to Jc1 in the range of 106 TCID50/ml when transfected into the packaging cell line. This level of HCVTCP production, which is much superior compared to HCV subgenomes expressing only viral proteins NS3 to NS5B, suggests that either the quantity of expressed core-to-NS2 proteins in these cell lines limits HCVTCP assembly in the case of the latter and/or that expression of core-to-NS2 proteins in cis facilitates virus production. Additional experiments are needed to distinguish between these two possibilities.
In summary, we have analyzed requirements for trans-packaging of assembly-defective subgenomic HCV RNA molecules into infectious virus-like particles. Our data show that the coding region of core, E1, E2, p7, and NS2 does not contain essential packaging signals for incorporation into infectious virions. Taking advantage of this flexibility, we have established stable packaging cell lines for production of high-titer HCVTCP preparations. This novel trans-complementation system should facilitate HCV infection studies due to its flexibility and improved biosafety; in addition, it should be instrumental for characterizing key cis-active functions essential for virus assembly.
Acknowledgments
We are grateful to Takaji Wakita for the gift of the JFH1 isolate and to Jens Bukh for the J6CF strain, to Didier Trono for providing the pWPI and pCMVΔR8.74 constructs, to Chris Brown for Venus-GFP, to Charles Rice for Huh7.5 cells and the E9E10 monoclonal antibody, to Arvind Patel for the AP33 antibody, and to Darius Moradpour for the C7/50 antibody. Moreover, we thank Ulrike Herian and Uwe Herzig for excellent technical assistance, Sibylle Haid for support with the density gradient centrifugation, and Ilona Glowacka for assistance with fluorescence microscopy.
This work was supported by a stipend from the Hannover Biomedical Research School funded by the Excellence Initiative of the German Government awarded to C.B., by an Emmy Noether fellowship from the Deutsche Forschungsgemeinschaft to T.P. (PI 734/1-1), by a grant from the Deutsche Forschungsgemeinschaft to R.B. (BA1505/2-1), the Sonderforschungsbereich 638 (Teilprojekt A5) to R.B., and by grants from the Helmholtz Association SO-024 to T.P.
Footnotes
Published ahead of print on 14 May 2008.
REFERENCES
- 1.Appel, N., U. Herian, and R. Bartenschlager. 2005. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J. Virol. 79896-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartosch, B., and F. L. Cosset. 2006. Cell entry of hepatitis C virus. Virology 3481-12. [DOI] [PubMed] [Google Scholar]
- 3.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 7613001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boni, S., J. P. Lavergne, S. Boulant, and A. Cahour. 2005. Hepatitis C virus core protein acts as a trans-modulating factor on internal translation initiation of the viral RNA. J. Biol. Chem. 28017737-17748. [DOI] [PubMed] [Google Scholar]
- 6.Chang, K. S., J. Jiang, Z. Cai, and G. Luo. 2007. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 8113783-13793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Souza, V., and M. F. Summers. 2005. How retroviruses select their genomes. Nat. Rev. Microbiol. 3643-655. [DOI] [PubMed] [Google Scholar]
- 8.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dull, T., R. Zufferey, M. Kelly, R. J. Mandel, M. Nguyen, D. Trono, and L. Naldini. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 728463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 765974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans, M. J., C. M. Rice, and S. P. Goff. 2004. Genetic interactions between hepatitis C virus replicons. J. Virol. 7812085-12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friebe, P., J. Boudet, J. P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in huh-7 cells harboring subgenomic replicons. J. Virol. 775487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottwein, J. M., T. K. Scheel, A. M. Hoegh, J. B. Lademann, J. Eugen-Olsen, G. Lisby, and J. Bukh. 2007. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology 1331614-1626. [DOI] [PubMed] [Google Scholar]
- 15.Grassmann, C. W., O. Isken, N. Tautz, and S. E. Behrens. 2001. Genetic analysis of the pestivirus nonstructural coding region: defects in the NS5A unit can be complemented in trans. J. Virol. 757791-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffin, S. D., L. P. Beales, D. S. Clarke, O. Worsfold, S. D. Evans, J. Jaeger, M. P. Harris, and D. J. Rowlands. 2003. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 53534-38. [DOI] [PubMed] [Google Scholar]
- 17.Harvey, T. J., W. J. Liu, X. J. Wang, R. Linedale, M. Jacobs, A. Davidson, T. T. Le, I. Anraku, A. Suhrbier, P. Y. Shi, and A. A. Khromykh. 2004. Tetracycline-inducible packaging cell line for production of flavivirus replicon particles. J. Virol. 78531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helle, F., and J. Dubuisson. 2008. Hepatitis C virus entry into host cells. Cell Mol. Life Sci. 65100-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heller, T., S. Saito, J. Auerbach, T. Williams, T. R. Moreen, A. Jazwinski, B. Cruz, N. Jeurkar, R. Sapp, G. Luo, and T. J. Liang. 2005. An in vitro model of hepatitis C virion production. Proc. Natl. Acad. Sci. USA 1022579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoofnagle, J. H. 2002. Course and outcome of hepatitis C. Hepatology 36S21-S29. [DOI] [PubMed] [Google Scholar]
- 21.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 1007271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, C. T., C. L. Murray, D. K. Eastman, J. Tassello, and C. M. Rice. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 818374-8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalajzic, I., M. L. Stover, P. Liu, Z. Kalajzic, D. W. Rowe, and A. C. Lichtler. 2001. Use of VSV-G pseudotyped retroviral vectors to target murine osteoprogenitor cells. Virology 28437-45. [DOI] [PubMed] [Google Scholar]
- 24.Kärber, G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Archiv für experimentelle Pathologie und Pharmakologie 162480-487. [Google Scholar]
- 25.Kato, T., T. Matsumura, T. Heller, S. Saito, R. K. Sapp, K. Murthy, T. Wakita, and T. J. Liang. 2007. Production of infectious hepatitis C virus of various genotypes in cell cultures. J. Virol. 814405-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaul, A., I. Woerz, P. Meuleman, G. Leroux-Roels, and R. Bartenschlager. 2007. Cell culture adaptation of hepatitis C virus and in vivo viability of an adapted variant. J. Virol. 8113168-13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 2000. cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 743253-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khromykh, A. A., A. N. Varnavski, and E. G. Westaway. 1998. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J. Virol. 725967-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koutsoudakis, G., E. Herrmann, S. Kallis, R. Bartenschlager, and T. Pietschmann. 2007. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J. Virol. 81588-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koutsoudakis, G., A. Kaul, E. Steinmann, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 805308-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemon, S. M., C. M. Walker, M. J. Alter, and M. Yi. 2007. Hepatitis C virus, p. 1253-1304. In B. N. Fields, D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott, Williams and Wilkins, Philadelphia, PA.
- 32.Liljestrom, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology 91356-1361. [DOI] [PubMed] [Google Scholar]
- 33.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309623-626. [DOI] [PubMed] [Google Scholar]
- 34.Lindenbach, B. D., H. J. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In B. N. Fields, D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott, Williams and Wilkins, Philadelphia, PA.
- 35.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 773007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohmann, V., F. Körner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285110-113. [DOI] [PubMed] [Google Scholar]
- 37.Lundstrom, K. 2005. Biology and application of alphaviruses in gene therapy. Gene Ther. 12(Suppl. 1)S92-S97. [DOI] [PubMed] [Google Scholar]
- 38.Manns, M. P., H. Wedemeyer, and M. Cornberg. 2006. Treating viral hepatitis C: efficacy, side effects, and complications. Gut 551350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 91089-1097. [DOI] [PubMed] [Google Scholar]
- 40.Moradpour, D., M. J. Evans, R. Gosert, Z. Yuan, H. E. Blum, S. P. Goff, B. D. Lindenbach, and C. M. Rice. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 787400-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagai, T., K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba, and A. Miyawaki. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2087-90. [DOI] [PubMed] [Google Scholar]
- 42.Owsianka, A., R. F. Clayton, L. D. Loomis-Price, J. A. McKeating, and A. H. Patel. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 821877-1883. [DOI] [PubMed] [Google Scholar]
- 43.Pavlovic, D., D. C. Neville, O. Argaud, B. Blumberg, R. A. Dwek, W. B. Fischer, and N. Zitzmann. 2003. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. USA 1006104-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pham, H. M., E. R. Arganaraz, B. Groschel, D. Trono, and J. Lama. 2004. Lentiviral vectors interfering with virus-induced CD4 down-modulation potently block human immunodeficiency virus type 1 replication in primary lymphocytes. J. Virol. 7813072-13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pietschmann, T., and R. Bartenschlager. 2001. The hepatitis C virus replicon system and its application to molecular studies. Curr. Opin. Drug. Discov. Devel. 4657-664. [PubMed] [Google Scholar]
- 46.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 1037408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282938-941. [DOI] [PubMed] [Google Scholar]
- 48.Porter, D. C., D. C. Ansardi, and C. D. Morrow. 1995. Encapsidation of poliovirus replicons encoding the complete human immunodeficiency virus type 1 gag gene by using a complementation system which provides the P1 capsid protein in trans. J. Virol. 691548-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pushko, P., M. Parker, G. V. Ludwig, N. L. Davis, R. E. Johnston, and J. F. Smith. 1997. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology 239389-401. [DOI] [PubMed] [Google Scholar]
- 50.Schaller, T., N. Appel, G. Koutsoudakis, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2007. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 814591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schlesinger, S. 1988. The generation and amplification of defective interfering RNAs, p. 167-185. In E. Domingo, J. J. Holland, and P. Ahlquist (ed.), RNA genetics. CRC Press, Inc., Boca Raton, FL.
- 52.Shavinskaya, A., S. Boulant, F. Penin, J. McLauchlan, and R. Bartenschlager. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 28237158-37169. [DOI] [PubMed] [Google Scholar]
- 53.Shimoike, T., C. Koyama, K. Murakami, R. Suzuki, Y. Matsuura, T. Miyamura, and T. Suzuki. 2006. Down-regulation of the internal ribosome entry site (IRES)-mediated translation of the hepatitis C virus: critical role of binding of the stem-loop IIId domain of IRES and the viral core protein. Virology 345434-445. [DOI] [PubMed] [Google Scholar]
- 54.Shimoike, T., S. Mimori, H. Tani, Y. Matsuura, and T. Miyamura. 1999. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 739718-9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shustov, A. V., P. W. Mason, and I. Frolov. 2007. Production of pseudoinfectious yellow fever virus with a two-component genome. J. Virol. 8111737-11748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spearman, C. 1908. The method of “right and wrong cases” (“constant stimuli”) without Gauss's formulae. Br. J. Psychol. 2227-242. [Google Scholar]
- 57.Steinmann, E., F. Penin, S. Kallis, A. H. Patel, R. Bartenschlager, and T. Pietschmann. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS. Pathog. 3e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tautz, N., H. J. Thiel, E. J. Dubovi, and G. Meyers. 1994. Pathogenesis of mucosal disease: a cytopathogenic pestivirus generated by an internal deletion. J. Virol. 683289-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tellinghuisen, T. L., M. J. Evans, T. von Hahn, S. You, and C. M. Rice. 2007. Studying hepatitis C virus: making the best of a bad virus. J. Virol. 818853-8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teterina, N. L., W. D. Zhou, M. W. Cho, and E. Ehrenfeld. 1995. Inefficient complementation activity of poliovirus 2C and 3D proteins for rescue of lethal mutations. J. Virol. 694245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thiel, V., N. Karl, B. Schelle, P. Disterer, I. Klagge, and S. G. Siddell. 2003. Multigene RNA vector based on coronavirus transcription. J. Virol. 779790-9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomssen, R., S. Bonk, C. Propfe, K. H. Heermann, H. G. Kochel, and A. Uy. 1992. Association of hepatitis C virus in human sera with beta-lipoprotein. Med. Microbiol. Immunol. (Berl.) 181293-300. [DOI] [PubMed] [Google Scholar]
- 63.Thomssen, R., S. Bonk, and A. Thiele. 1993. Density heterogeneities of hepatitis C virus in human sera due to the binding of beta-lipoproteins and immunoglobulins. Med. Microbiol. Immunol. (Berl.) 182329-334. [DOI] [PubMed] [Google Scholar]
- 64.Tscherne, D. M., M. J. Evans, T. von Hahn, C. T. Jones, Z. Stamataki, J. A. McKeating, B. D. Lindenbach, and C. M. Rice. 2007. Superinfection exclusion in cells infected with hepatitis C virus. J. Virol. 813693-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van den Hoff, M. J., V. M. Christoffels, W. T. Labruyere, A. F. Moorman, and W. H. Lamers. 1995. Electrotransfection with “intracellular” buffer. Methods Mol. Biol. 48185-197. [DOI] [PubMed] [Google Scholar]
- 66.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, T. H., R. C. Rijnbrand, and S. M. Lemon. 2000. Core protein-coding sequence, but not core protein, modulates the efficiency of cap-independent translation directed by the internal ribosome entry site of hepatitis C virus. J. Virol. 7411347-11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiong, C., R. Levis, P. Shen, S. Schlesinger, C. M. Rice, and H. V. Huang. 1989. Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science 2431188-1191. [DOI] [PubMed] [Google Scholar]
- 69.Yagi, S., K. Mori, E. Tanaka, A. Matsumoto, F. Sunaga, K. Kiyosawa, and K. Yamaguchi. 2005. Identification of novel HCV subgenome replicating persistently in chronic active hepatitis C patients. J. Med. Virol. 77399-413. [DOI] [PubMed] [Google Scholar]
- 70.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262250-263. [DOI] [PubMed] [Google Scholar]
- 71.Yi, M., Y. Ma, J. Yates, and S. M. Lemon. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81629-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 1029294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]