Abstract
The hepatitis E virus (HEV) causes acute viral hepatitis, but its characterization is hampered by the lack of an efficient in vitro infection system that can be used to study the effects of HEV proteins on cellular processes. Previous studies suggest that the viral ORF3 protein (pORF3) is essential for infection in vivo and is likely to modulate the host response. Here, we report that pORF3 localizes to early and recycling endosomes and causes a delay in the postinternalization trafficking of epidermal growth factor receptor (EGFR) to late endosomes/lysosomes. The cytoplasmic phosphorylated signal transducer and activator of transcription 3 (pSTAT3) proteins require growth factor receptor endocytosis for their translocation from the cytoplasm to nucleus. Consequently, lower levels of pSTAT3 were found in the nuclei of ORF3-expressing Huh7 human hepatoma cells stimulated with EGF. This results in downregulation of the acute-phase response, a major determinant of inflammation in the host. We propose that through its effects on EGFR trafficking, pORF3 prolongs endomembrane growth factor signaling and promotes cell survival. The effects on STAT3 translocation would result in a reduced inflammatory response. Both of these events are likely to contribute positively to viral replication.
Hepatitis E virus (HEV) is the causative agent for hepatitis E, a waterborne disease that occurs sporadically and as focused outbreaks (25, 35). In areas of endemicity, HEV infections account for about one-third of all sporadic and almost all epidemic viral hepatitis cases. While the infection is generally acute and self-limiting, up to 20 to 30% mortality has been reported following HEV infection during pregnancy (21, 34). Recently classified as the only member of Hepevirus in the family Hepeviradae (9), HEV has a 7.2-kb capped and polyadenylated, positive-sense RNA genome that contains three open reading frames (ORFs). ORF1 encodes a viral nonstructural protein, ORF2 codes for the capsid protein, and ORF3 encodes a small protein whose functions are not fully understood. No efficient in vitro infection system or small-animal models, which can be used to study the effects of HEV proteins on cellular processes, exist thus far. We have therefore used a subgenomic expression strategy that in the past provided valuable clues for understanding the role of viral proteins in pathogenesis (18, 20, 24, 32, 49, 50).
The ORF3 of HEV codes for a protein of 123 amino acids ([aa] pORF3); a recent report proposed pORF3 to be translated from a bicistronic subgenomic RNA and to be 9 aa shorter (i.e., a protein of 114 aa) (16). pORF3 is phosphorylated at a single serine residue by the cellular mitogen-activated protein kinase (50), but as this serine residue is absent from some HEV strains, this phosphorylation is unlikely to have discernible effects on in vitro replication or on the ability to infect and cause hepatitis in primates. It contains two hydrophobic domains in its N-terminal half and two proline-rich regions in its C-terminal half, one of which contains the phosphorylated serine residue (50). The other proline-rich region contains a PXXPXXP (where X is any other residue) motif that was shown to bind several proteins with src-homology 3 domains (24). It was subsequently demonstrated that pORF3 activated the extracellularly regulated kinase, a member of the mitogen-activated protein kinase family of proteins, by binding and inhibiting its cognate phosphatase (20). No definite function has yet been assigned to pORF3. However, based on its cellular localization, binding partners, and observed effects, a role in cell survival signaling has been proposed (20).
Growth factor receptors control a wide variety of biological processes including cell proliferation, differentiation, survival, and migration (45). One such example is the epidermal growth factor receptor (EGFR) that plays important roles in many physiological and pathological processes in cells. Following ligand binding, EGFRs are rapidly internalized and initially delivered to early endosomes for sorting; these are then targeted to the late endosomal/lysosomal compartment for degradation (4, 47). On ligand binding, EGFRs initiate a series of biochemical events, including receptor activation and autophosphorylation, association and subsequent phosphorylation of cellular proteins, and the formation of protein-protein interaction networks that transduce growth factor signals to the nucleus (5, 11). The duration and strength of these signals are tightly regulated in the cell through various negative regulatory mechanisms. Signal termination is accomplished by degradation of activated receptors.
Growth factor receptors also serve as scaffolds for the trafficking of other signaling intermediates (11, 19, 27). One such protein is the signal transducer and activator of transcription 3 (STAT3), a transcription factor that is activated downstream of various cytokines, growth factors, or oncoproteins and plays critical roles in cell growth, survival, and the innate host response (3, 6). The STAT3 protein is recruited to activated receptors through an interaction between the Src homology 2 domain of STAT3 and phosphotyrosine (pTyr) docking sites on the receptors. Subsequently, STAT3 is phosphorylated on Tyr705 either directly by the receptor kinase or indirectly by an associated intermediary kinase, including members of the Janus kinase family. pSTAT3 dimerizes and colocalizes with receptor-ligand complexes on endocytic vesicles from the plasma membrane to the perinuclear region (2). A block in receptor-mediated endocytosis inhibits the nuclear transport of STAT3 (2). However, the effects of receptor sorting, recycling, and degradation on the transport of STAT3 from the cytoplasm to the nucleus are not fully understood.
Earlier known as the acute-phase response (APR) factor, STAT3 regulates the transcription of a number of acute-phase genes following interleukin-6 treatment (1). The acute-phase proteins (APPs) are expressed mainly by the liver and have a wide range of activities that contribute to host defense. For example, these directly neutralize inflammatory agents, help to minimize the extent of local tissue damage, and participate in tissue repair and regeneration (33). Depending upon whether the concentration of the APP in plasma increases or decreases following inflammation, it is classified as positive or negative (10).
Viruses have evolved various mechanisms to modulate host genome replication and transcription, protein translation, and posttranslational events to optimize their survival in the host (43). To accomplish this, viral proteins are targeted to appropriate subcellular compartments, either via translocation signals or through domain-specific binding to critical host proteins (12, 26, 31, 48). We show here the localization of HEV pORF3 to early and recycling endosomes and its effects on EGFR trafficking, STAT3 nuclear translocation, and transcription of STAT3-responsive targets. Our results support a role for pORF3 in promoting cell survival through diverse mechanisms.
MATERIALS AND METHODS
Plasmids, cell lines, and antibodies.
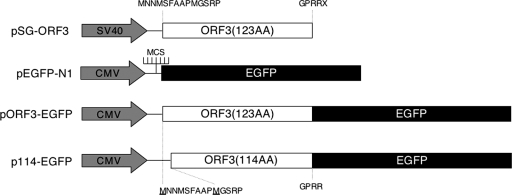
The ORF3 expression vectors, stable cell lines, and antibodies have been described earlier (18, 20, 24) and are schematically illustrated in Fig. 1. The 114-aa ORF3 was PCR amplified from pORF3-EGFP (where EGFP is enhanced green fluorescent protein) using the following primers: forward, GGAGCTAGCATGGGTTCGCGACCATGCG; and reverse, TAACTCGAGGCGGCGCGGCCCCAGC. The 360-bp PCR fragment was digested with NheI and XhoI and cloned into the same sites in plasmid pEGFP-N1. The clone was confirmed by restriction digestion and DNA sequencing.
FIG. 1.
Schematic illustration of expression vectors used in the study. The vector pSG-ORF3 has been described earlier (18). It carries the 123-aa ORF3 downstream of a simian virus 40 (SV40) promoter-enhancer unit. The N- and C-terminal amino acids of ORF3 are shown. The pEGFP-N1 vector is from Clontech and carries the EGFP gene downstream of a cytomegalovirus (CMV) promoter and a multiple cloning site (MCS). The vector pORF3-EGFP contains the 123-aa ORF3 lacking its stop codon cloned upstream of the EGFP gene. This vector produces a 123-aa ORF3-EGFP fusion protein. The vector p114-EGFP contains the 114-aa ORF3 lacking its stop codon cloned upstream of the EGFP gene. This vector produces a 114-aa ORF3-EGFP fusion protein. The N- and C-terminal amino acids of ORF3 are shown, and the starting methionines are underlined.
The sources of various plasmid constructs were as follows: pLucTKS3 and pLucTK (where TK is thymidine kinase) plasmids were from Gulam Waris (University of Colorado Medical School, CO); C-reactive protein (CRP)-chloramphenicol acetyltransferase (CAT), haptoglobin-CAT, and hemopexin-CAT were from Valeria Poli (University of Turin, Italy) (23); alpha-1-antitrypsin-CAT was from Gary Bulla (Eastern Illinois University); RA-Luc was from Cem Gabay (University of Geneva, Switzerland) (39); GFP-tagged Rabs were from Marino Zerial (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany); cyan fluorescent protein (CFP)-tagged TGN38 was from Antonius VanDongen (Duke University Medical Center, NC); and the HEV replicon plasmid pSK-E2 and its ORF3-null version pSK-E2ΔORF3 were from Suzanne Emerson (National Institutes of Health). The antibodies to the following were obtained from the indicated sources: STAT3 and phospho-STAT3 (pSTAT3) from Cell Signaling Technology (Beverly, MA), EGFR from Santa Cruz Biotechnology (Santa Cruz, CA), caveolin from BD Biosciences, and lysobisphosphatidic acid (LBPA) from T. Kobayashi (Hamamatsu University, Japan). Secondary antibodies were purchased from Jackson Immunoresearch and were labeled with Alexa dyes (Molecular Probes, Eugene, OR), according to the manufacturer's protocol. Rhodamine-EGF, DiI-low-density lipoprotein (LDL), and tetramethylrhodamine-conjugated dextran (TMR-dextran) for uptake assays were purchased from Molecular Probes. Human transferrin (Tf) was from Sigma Chemicals and was labeled with Alexa 647 after iron loading. Human hepatoma Huh7 cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS) and antibiotics, and cells were transfected using either Lipofectin (Invitrogen) or Fugene-6 (Roche) according to the supplier's protocol.
Replicon transfection and ORF3 expression.
Plasmids pSK-E2 and pSK-E2ΔORF3 were linearized at a unique BglII site located immediately downstream of the HEV poly(A) tract, and capped transcripts were synthesized as previously described (7, 8). Transcription mixtures were cooled on ice and then mixed with a liposome mixture consisting of 25 μl of DMRIE-C (1,2-dimyristyloxypropyl-3-dimethyl-hydroxy ethyl ammonium bromide and cholesterol; Invitrogen, Carlsbad, CA) in 1 ml of Opti-MEM. This was added to a T25 flask containing washed S10-3 cells at 40% confluence. The S10-3 cells are a subclone of the Huh7 line and were a kind gift of S. Emerson. The flasks were incubated at 34.5°C for 6 h, an additional 1 ml of Opti-MEM was added, and incubation was continued overnight. Cells were then trypsinized, split into two T25 flasks, and incubated in Dulbecco's modified Eagle's medium containing 10% FBS. On day 5 posttransfection, cells were again trypsinized and then transfected with red fluorescent protein (RFP)-tagged Rab5 expression vector or other compartment-specific markers. Immunostaining for ORF3 expression and colocalization was on day 8 postreplicon transfection.
Microscopy.
To study the subcellular localization of pORF3, Huh7 cells were transiently cotransfected with the appropriate expression vectors and imaged live in a CO2-independent buffer (M1 containing 0.1% bovine serum albumin and 0.1% glucose) using a confocal microscope (Zeiss LSM 510 meta imaging system). For endogenous LBPA and caveolin staining, cells transfected with pEGFP-ORF3 were fixed with 2% paraformaldehyde for 15 min, permeabilized at room temperature with 0.1% saponin for 10 min or 0.5% Tween 20 for 20 min, and stained using the appropriate primary and secondary antibodies. To study the effects of pORF3 on endocytic transport, Huh7 cells were transiently transfected with either pORF3-EGFP or the pEGFP-N1 control vector and serum starved overnight. Following this, cells were pulsed for the times indicated in Fig. 3 with 100 ng/ml rhodamine-EGF, 1 mg/ml TMR-dextran, or 10 μg/ml Alexa 647-Tf. To study the effects of pORF3 on STAT3 transport, Huh7 cells were transiently transfected as above and serum starved overnight. Following this, the cells were pulsed with EGF for 2 h and imaged live. Similarly transfected and EGF-treated cells were used to prepare lysates for Western blotting.
FIG. 3.
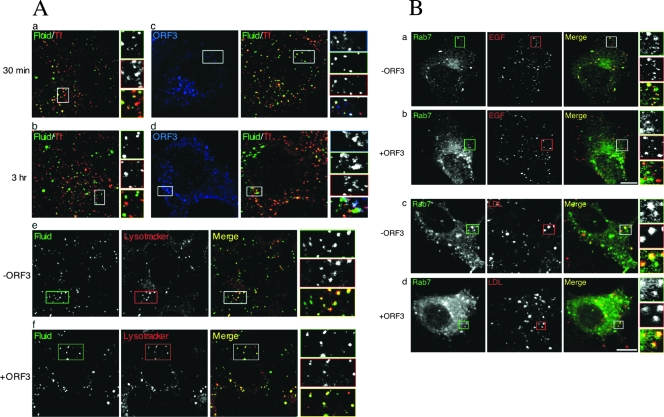
Effects of pORF3 on endocytosis and surface receptor trafficking. (A) Fluid uptake and TfR trafficking are not affected by pORF3. Huh7 cells were transfected with 0.5 μg each of either pEGFP-N1 (a and b) or pEGFP-ORF3 (c and d) for 36 h and then serum starved overnight. Cells were then pulsed with 1 mg/ml TMR-dextran and 10 μg/ml Alexa 647-Tf at 37°C for either 30 min (a and c) or 3 h (b and d) and imaged by confocal microscopy. Cells transfected with either pEGFP-N1 (e) or pEGFP-ORF3 (f) were similarly pulsed with Alexa 647-dextran and 5 μM Lysotracker red at 37°C for 2 h and then imaged. Monochrome and pseudocolored details of the boxed regions are shown and coded according to the color assigned for each entity. (B) pORF3 modulates EGFR trafficking. Huh7 cells were cotransfected with 0.5 μg each of pEGFP-Rab7 and either pECFP-N1 (a and c) or pECFP-ORF3 (b and d). After 36 h, the cells were serum starved overnight and then treated at 37°C with either 100 ng/ml rhodamine-EGF for 3 h (a and b) or 10 μg/ml LDL-Dil for 40 min (c and d). Live cells were imaged for the distribution of EGFR, LDLR, Rab7, and pORF3. The merged images as well as the monochromatic distributions from single confocal planes are shown. The pORF3 images are not shown. At least 50 cells that coexpressed pORF3 and the subcellular marker were imaged in each case. Representative images are shown that were observed in at least 70% of coexpressing cells.
Flow cytometry.
Transfected Huh7 cells were rinsed twice with ice-cold phosphate-buffered saline (PBS) to remove any unbound ligand. The cells were then chilled on ice to stop membrane trafficking, harvested, fixed using 2% paraformaldehyde, resuspended in PBS containing 1% FBS, and blocked for 45 min on ice. For total EGFR detection, the cells were permeabilized with 100% methanol at −20°C before the blocking step. The blocked cells were labeled with anti-EGFR antibodies followed by an Alexa 594-conjugated secondary antibody. The cells were washed with ice-cold PBS and analyzed by flow cytometry (CyanADP; DakoCytomation, Denmark). For each sample, 10,000 cells were analyzed, and the EGFP-expressing cells were gated for estimating the levels of cell surface and total EGFR.
Immunoprecipitation and Western blotting.
Cells were lysed with a buffer containing 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, and a protease inhibitor cocktail (Roche, Mannheim, Germany). The clarified supernatant was quantified for protein concentration by the Bradford assay (Bio-Rad Laboratories). For immunoprecipitation, 1 mg of total proteins in 500 μl of lysis buffer was incubated with 20 μl of protein A-agarose beads (Amersham Pharmacia Biotech, Uppsala, Sweden) for 1 h at 4°C. The precleared lysate was then incubated with 2 μg of the antibody overnight at 4°C, followed by 20 μl of protein A-agarose beads for 2 h at 4°C. After five washings in lysis buffer, the beads were boiled in Laemmli buffer, and the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. For Western blotting, proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis were transferred to a nitrocellulose membrane (Hybond ECL; Amersham Biosciences), and the membrane was blocked with Tris-buffered saline (TBS) containing 5% Blotto (Bio-Rad) for 1 to 2 h at room temperature and washed with TBST (TBS containing 0.1% Tween 20). The membrane was then incubated overnight at 4°C with the primary antibody appropriately diluted in TBST-5% bovine serum albumin, washed three times for 10 min each with TBST, and incubated with horseradish peroxidase-linked secondary antibodies diluted in TBST-5% Blotto for 1 h at room temperature. After the membrane was washed as described above, chemiluminescent detection of proteins was carried out using a Phototope detection system (Cell Signaling Technology, Beverly, MA) according to the supplier's protocol.
Electrophoretic mobility shift assay (EMSA).
Cells were serum starved overnight, treated with 100 ng/ml EGF for 3 h, rinsed, collected, and lysed in low-salt buffer containing 20 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5% Triton X-100, 0.5 mM dithiothreitol, and a protease inhibitor cocktail on ice for 20 min. The nuclear pellet obtained by centrifugation at 800 × g for 10 min at 4°C was resuspended in high-salt buffer containing 20 mM HEPES, 500 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.2% Triton X-100, 20% glycerol, 0.5 mM dithiothreitol, and a protease inhibitor cocktail for 1 h at 4°C. The nuclear extract was obtained by centrifugation at 15,700 × g for 10 min at 4°C and quantitated for protein content. Double-stranded oligonucleotides (Santa Cruz Biotechnology, Santa Cruz, CA) containing the consensus STAT3 binding site (GATCCTTCTGGGAATTCCTAGATC; binding site is underlined) or a mutant STAT3 binding site (AAT-to-CCG substitution) were labeled with [γ-32P]ATP using T4 polynucleotide kinase and purified on a Sephadex G50 spin column. About 50 fmoles of each 32P-labeled probe was then incubated separately with 10 μg of nuclear extract for 1 h on ice in the presence of excess unlabeled mutant probe to minimize nonspecific binding. The samples were resolved on a native 5% polyacrylamide gel using 0.5× Tris-borate-EDTA buffer, and the gel was then dried and subjected to autoradiography.
Luciferase assay.
Transfected cells at 36 h posttransfection were serum starved overnight, treated with 100 ng/ml EGF for 3 h, washed with PBS, and harvested. Preparation of cytosolic extracts and luciferase assays were carried out using a luciferase assay kit (Promega, Madison, WI), according to the supplier's protocol, and results were quantitated on a luminometer (Sirius, Berthold, Germany).
CAT assay.
Around 40 to 48 h posttransfection, ∼1.5 × 106 cells were rinsed twice with cold PBS, resuspended in 100 μl of 250 mM Tris-HCl, pH 7.8, and lysed by repeated freezing and thawing. To the clarified lysate containing 90 μg of protein, 20 μl of 4 mM S-acetyl coenzyme A and 40 μl of 600 μM chloramphenicol containing 2 μCi/ml [14C]chloramphenicol were added. The reactions were carried out at 37°C for 4 h before extraction with 1 ml of ethyl acetate. After vacuum drying, the residue was resuspended in 25 μl of ethyl acetate and applied to LK5D silica thin-layer chromatography plates (Whatman). The plates were developed in chloroform-methanol (95:5), dried, and subjected to autoradiography. The amount of acetylated [14C]chloramphenicol produced was quantitated by densitometry.
RESULTS
pORF3 protein localizes to early and recycling endosomes.
Since our earlier studies suggested a role for pORF3 in modulating cell signaling, we examined its subcellular localization in transfected Huh7 human hepatoma cells. Immunofluorescence detection of untagged pORF3 showed a punctate distribution in these cells, as did ORF3 fusion proteins with either EGFP or its red (DsRed) or cyan (enhanced CFP [ECFP]) versions, indicating that the different tags did not affect pORF3 localization (data not shown).
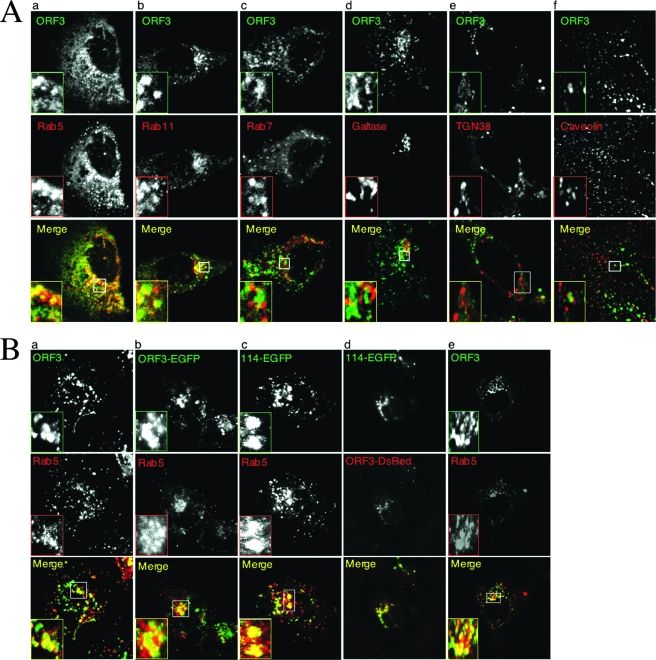
To identify the nature of punctate structures, Rab GTPases were employed as organelle markers (51). While Rab5 has been found on early endosomes, Rab4 and Rab11 are enriched on recycling endosomes, and Rab7 localizes to late endosomes. The subcellular localization of pORF3 was imaged in live Huh7 cells coexpressing ORF3-ECFP together with any one of the Rab-EGFP constructs at low expression levels. As shown in Fig. 2A, pORF3 colocalizes with Rab5 (panel a), Rab11 (panel b) and Rab 4 (not shown) but not with Rab7 (panel c), suggesting that it localizes to early and recycling but not to the late endosomes. On staining ORF3-EGFP-expressing cells for endogenous LBPA, which is another marker for late endosomes, no association of pORF3 with late endosomes was observed (data not shown). We also assessed the localization of pORF3 to cis- and trans-Golgi compartments using GFP-galtase and TGN38 as respective markers for these compartments. No colocalization was observed with these markers (Fig. 2A, panels d and e). The cells were also stained with anti-caveolin antibodies to visualize endogenous caveolin-rich compartments (caveosomes); no localization of pORF3 was observed in these compartments (Fig. 2A, panel f). At least 50 cells that coexpressed pORF3 and the subcellular marker were imaged in each case. The colocalization quantitation for pORF3 and the various markers were as follows: Rab5, ∼80%; Rab11, 100%; Rab7, ∼10%; galtase, 0%; TGN38, 0%; and caveolin, 0%. Collectively, our results suggest that cytosolic pORF3 localizes to the early and recycling endosomes.
FIG. 2.
The ORF3 proteins localize to early and recycling endosomes. (A) Huh7 cells were cotransfected with 0.5 μg of plasmid pECFP-ORF3 and 0.5 μg each of one of the following fluorescent fusion constructs to mark intracellular compartments: panel a, Rab5 (early endosomes); panel b, Rab11 (recycling endosomes); panel c, Rab7 (late endosomes); panel d, galtase (cis-Golgi compartment); or panel e, TGN38 (trans-Golgi compartment). Alternatively, Huh7 cells transfected with pECFP-ORF3 were stained with antibodies to caveolin (caveolae) using Alexa 488-conjugated secondary antibodies (panel f). The fixed cells were then imaged by confocal microscopy; the pORF3 and marker signals were pseudocolored green and red, respectively. The merged pictures of single confocal planes are shown. Details are also shown for the boxed regions. At least 50 cells that coexpressed pORF3 and the subcellular marker were imaged in each case. Representative images are shown that were observed in at least 70% of coexpressing cells. (B) Huh7 cells were cotransfected with 0.5 μg each of pRab5-RFP and one of the following: panel a, pSG-ORF3; panel b, pORF3-EGFP; or panel c, p114-EGFP. In panel d, Huh7 cells were cotransfected with p114-EGFP and pORF3-DsRed. The fixed cells were then either imaged directly for fluorescent constructs or stained with rabbit anti-ORF3 and anti-rabbit-Alexa 488 conjugate for the untagged pORF3. The cells were imaged by confocal microscopy. For panels a to c, the pORF3 and Rab5 signals were pseudocolored green and red, respectively. For panel d, the signals of the 114- and 123-aa forms of pORF3 were pseudocolored green and red, respectively. The merged pictures of single confocal planes are shown. Details are also shown for the boxed regions. At least 50 cells that coexpressed pORF3 and the subcellular marker were imaged in each case. For panel e, S10-3 cells were transfected with in vitro synthesized capped replicon transcript followed by the pRab5-RFP construct, as described in Materials and Methods. The cells were fixed and stained for pORF3 expression with rabbit anti-ORF3 and anti-rabbit-Alexa 488 conjugate and were imaged by confocal microscopy. The pORF3 and Rab5 signals were pseudocolored green and red, respectively. The merged pictures of single confocal planes are shown. Details are also shown for the boxed regions. In all cases, representative images are shown that were observed in at least 70% of coexpressing cells.
To confirm that EGFP fusion does not affect the subcellular distribution of pORF3, we studied the colocalization of untagged pORF3 and its EGFP fusions with the Rab5 marker and found no difference (Fig. 2B, panels a and b). Since a recent report proposed pORF3 to be 9 aa shorter, we also studied the subcellular distribution of the smaller protein, the 114-aa pORF3. This protein colocalized with the Rab5 marker (Fig. 2B, panel c) as well as with the 123-aa pORF3 (Fig. 2B, panel d). Finally, we also studied the subcellular distribution of pORF3 expressed from an HEV genomic replicon and also found it to colocalize with the Rab5 marker (Fig. 2B, panel e) but not the Rab7 marker (data not shown). In multiple experiments with the HEV replicon, we found about ∼10 to 15% of transfected S10-3 cells to express the ORF3 and ORF2 proteins; transfection of these cells with the ORF3-null replicon showed only ORF2 and no ORF3 staining (data not shown). These results support the use of EGFP and other tagged versions of the 123-aa pORF3 for its subcellular localization and subsequent studies.
pORF3 does not affect receptor recycling, fluid uptake, and transport.
In view of its subcellular distribution to early and recycling endosomes, it was of interest to determine whether pORF3 had any effects on endocytic transport and trafficking. For this, we chose to study the transport of Tf receptor (TfR) and dextran, which are prototypic markers of the recycling and lysosomal degradative pathways, respectively. TfRs located at the cell surface rapidly exchange with an intracellular pool of receptors whereas dextran is a high-molecular-weight polysaccharide that enters the cells by fluid-phase endocytosis and accumulates in lysosomes due to its resistance to intralysosomal hydrolysis. Alexa 647-conjugated Tf (Alexa 647-Tf) and TMR-dextran were used to mark these pathways. Huh7 cells transfected with the pORF3-EGFP expression plasmid (or an empty vector), after a 30 min treatment with Alexa 647-Tf and TMR-dextran, showed colocalization of these markers, indicating their presence in the same early endosomal compartments (Fig. 3A, panel a). After a 3-h pulse, Tf and dextran were predominantly localized to different compartments (Fig. 3A, panel b), likely to be the recycling and degradative compartments, respectively (36). Cells that expressed pORF3 also showed a pattern for the endosomal distribution of Tf and dextran similar to vector-transfected cells (Fig. 3A, panels c and d). There was no noticeable difference in fluid uptake or distribution with respect to Tf in ∼80% of cells. The remaining cells showed slightly reduced fluid uptake. The colocalization of TMR-dextran (or fluid) and Lysotracker was also compared in control (Fig. 3A, panel e) and ORF3-expressing cells (Fig. 3A, panel f). Again, the results showed dextran to accumulate in the lysosomal compartment in control as well as ORF3-expressing cells. The overall distribution of Lysotracker-positive compartments in control and ORF3-expressing cells remained unaltered in all of the observed cells. Together, these results demonstrated that pORF3 does not affect the normal cellular pathways of endocytic transport and trafficking of TfR and delivery of the fluid phase to late endosomes and lysosomes.
Delayed movement of EGFR to late endosome/lysosome in ORF3-expressing cells.
A well-established role of endocytosis is to internalize surface receptors, thereby downregulating the signaling event. In addition, accumulating evidence suggests that receptor signaling can take place from endosomes, and compartmentalized signaling could be physiologically important (13, 40). Since our earlier results suggested a prosurvival effect of pORF3 (18), we reasoned that this could be due to an alteration of endocytic dynamics of the EGFR and its ligand. Huh7 cells were cotransfected with pORF3-ECFP (or an empty vector) along with a vector encoding Rab7-EGFP. After serum starvation overnight, the cells were treated with rhodamine-labeled EGF at 37°C for 3 h, and live-cell imaging was done. The EGFR-ligand complex localized to the Rab7-positive compartment in ∼70% of the cells (Fig. 3B, panel a). In contrast, in ORF3-expressing cells, the EGFR-ligand complex was not found in the Rab7-positive compartment (Fig. 3B, panel b) and to some degree colocalized with pORF3 (data not shown). In the presence of pORF3, ∼60% of cells showed no colocalization of EGF and Rab7, while the remaining ∼40% of cells showed very weak colocalization. Since pORF3 was shown earlier to localize to early and recycling endosomes, these results suggest that while in control cells the EGFR-ligand complex enters the Rab7-positive late endosomal compartment destined for lysosomal degradation, the movement of this complex into the degradative compartment is delayed in ORF3-expressing cells. To check whether this effect was generic or growth factor receptor-specific, we similarly labeled the LDL receptor with Dil-LDL and observed its movement into the Rab7-positive compartment. No difference was observed between control and ORF3-expressing cells (Fig. 3B, panels c and d), with about 60% of cells in both cases showing very good colocalization of LDL and Rab7. Thus, pORF3 specifically delayed the movement of EGFR into the degradative compartment.
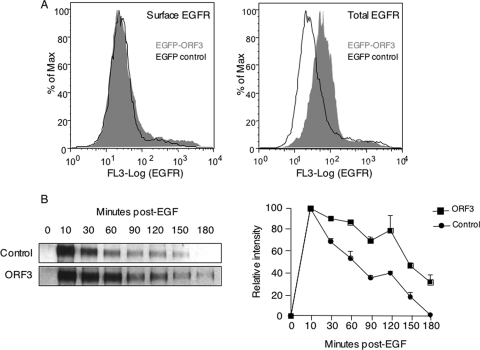
To confirm this result, we checked the steady-state levels of surface and total EGFR in control and ORF3-expressing cells. Huh7 cells were transfected to express the ORF3-EGFP fusion protein or only EGFP as a control. At 48 h posttransfection, the steady-state levels of surface or total EGFR were quantitated by antibody labeling and flow cytometry in cells that were gated for EGFP expression. While there was no change in the surface expression of EGFR, the total levels of EGFR were elevated in ORF3-expressing cells (Fig. 4A). From two independent experiments, the mean fluorescence intensities (MFIs) of EGFR were calculated to be the following: for surface expression, 34.4 ± 2.3 and 30.9 ± 2.0 in ORF3 and controls, respectively; for total expression, 68.8 ± 0.6 and 34.2 ± 2.5 in ORF3 and controls, respectively. On stimulation with growth factor, EGFR is phosphorylated, endocytosed, and eventually delivered to the lysosomal compartment for degradation and signal cessation. To compare the phospho-EGFR decay kinetics in ORF3-expressing versus control cells, transfected cells were serum starved and then stimulated with EGF. At various times in the presence of EGF, cells were harvested, and the lysates were immunoprecipitated with anti-EGFR followed by Western blotting with anti-pTyr. In control as well as ORF3-expressing cells, there was rapid EGFR phosphorylation, reaching a peak in 10 min (Fig. 4B). Subsequently, there was a slower loss of phospho-EGFR signal in ORF3-expressing cells than in control cells (Fig. 4B). There was a consistent increase in phospho-EGFR levels observed at around 2 h post-EGF treatment; this may be attributed to EGFR that was recycled back to the cell surface. Nevertheless, the flow cytometry and decay kinetics results supported delayed EGFR degradation in ORF3-expressing cells.
FIG. 4.
Increased EGFR levels in ORF3-expressing cells. (A) Flow cytometric analysis of surface or total EGFR in Huh7 cells transfected with 2 μg of either pEGFP-N1 (black line) or pEGFP-ORF3 (gray area). Single-parameter histograms are shown for EGFP-positive cells. From two independent experiments, the MFI values were the following: for surface expression, MFIs were 34.4 ± 2.3 and 30.9 ± 2.0 in ORF3 and control cells, respectively; for total expression, MFIs were 68.8 ± 0.6 and 34.2 ± 2.5 in ORF3 and the total cell population, respectively. The P values for the two sets, calculated using a Student's t test, were 0.282 for surface expression and 0.017 for total expression. (B) Huh7 cells were transfected as above and 40 h later serum starved for 12 h. The cells were then pulsed with 100 ng/ml EGF for the indicated times. Cell lysates containing equal amounts of total protein were immunoprecipitated with anti-EGFR and then Western blotted with anti-pTyr antibodies. The phospho-EGFR signals are shown. The graph on the right shows decay curves, taking the highest intensity (at 10 min post-EGF) to be 100%. The gel and decay curves are representative of three separate experiments. Max, maximum.
pORF3 affects transport of STAT3 to the nucleus.
Functional growth factor-mediated receptor endocytosis is required for the transport of STAT3 to the nucleus. Since pORF3 was found to delay the transport of EGFR to the degradative compartment, we tested whether this viral protein showed any effects on the levels and subcellular localization of STAT3. The subcellular distribution of STAT3 following EGF stimulation was examined in Huh7 cells transfected with expression vectors encoding either STAT3-EGFP alone or together with ORF3-DsRed. In control cells, STAT3 was found in the nuclei of ∼70% of cells following EGF stimulation (data not shown). However, in ORF3-expressing cells STAT3 showed a cytoplasmic perinuclear distribution, much like pORF3 itself, either with or without EGF stimulation in all of the observed cells (data not shown).
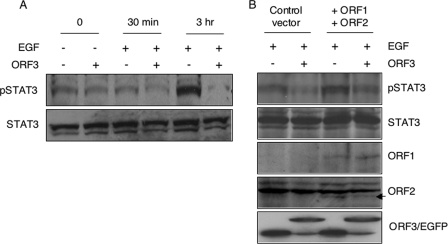
Next, we tested the nuclear accumulation of pSTAT3 and total STAT3 proteins in control and ORF3-expressing cells. Huh7 cells were transfected with either pORF3-EGFP or pEGFP-N1 plasmid. The cells were serum starved and treated with EGF for either 30 min or 3 h. Nuclear extracts prepared from these cells were Western blotted with either pSTAT3 or total STAT3 antibodies. Lower levels of pSTAT3 were observed in nuclear extracts prepared from EGF-stimulated ORF3-expressing cells than in control cells (Fig. 5A). While this difference was evident as early as 30 min following EGF stimulation, it was very significant at the late time. There were no significant differences in the levels of total STAT3 proteins in the nuclei of these cells. We also tested the nuclear levels of pSTAT3 in Huh7 cells cotransfected to express all three HEV proteins: ORF1, ORF2, and ORF3. Even in the presence of ORF1 and ORF2 expression, reduced levels of nuclear pSTAT3 were observed in ORF3-expressing cells (Fig. 5B). Since in our hands the HEV replicon was expressed in only ∼10 to 15% of transfected cells, it was not possible to carry out this experiment with the replicon.
FIG. 5.
Modulation of STAT3 nuclear transport by pORF3. (A) Huh7 cells were transfected with 2 μg of either pEGFP-N1 (−) or pEGFP-ORF3 (+). After 36 h the cells were serum starved overnight and either treated with 100 ng/ml EGF (+) or not (−) for the indicated times at 37°C. Nuclear lysates prepared from these cells were Western blotted with anti-pSTAT3 or anti-STAT3 antibodies. (B) Huh7 cells were transfected with 1.5 μg of either pEGFP-N1 (−) or pEGFP-ORF3 (+) along with 3 μg of pSGI (control vector) or a mixture of 1.5 μg each of pSG-ORF1 and pSG-ORF2, as indicated. After 36 h the cells were serum starved overnight and treated with 100 ng/ml EGF for 3 h at 37°C. Nuclear lysates prepared from these cells were Western blotted with anti-pSTAT3 or anti-STAT3 antibodies. As expression controls, whole-cell lysates from identically transfected and treated cells were Western blotted with anti-ORF1, anti-ORF2, or anti-EGFP antibodies. In the ORF2 plane, the arrow indicates the specific band; the nonspecific bands served as loading controls. In the ORF3/EGFP plane, the upper and lower bands represent the EGFP-ORF3 fusion protein and EGFP, respectively.
STAT3 target genes are downregulated in ORF3-expressing cells.
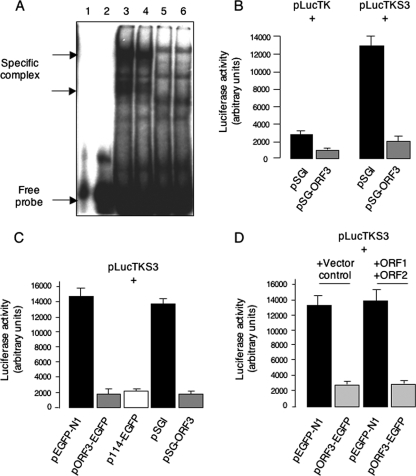
Impairment in the nuclear translocation of pSTAT3 is likely to affect the transcription of its target genes. We first checked for nuclear levels of DNA-binding-competent STAT3 protein by an EMSA. Nuclear extracts prepared from Huh7 cells transiently transfected with pSG-ORF3 or the control vector pSGI were incubated with a 32P-labeled STAT3 binding site oligonucleotide probe, and the resulting DNA-protein complexes were revealed by native gel electrophoresis. A labeled probe in which the STAT3 binding site had been mutated was used as a control. As shown in Fig. 6A, the STAT3 complex was diminished by about 40 to 50% when nuclear protein extracts from ORF3-expressing cells were used compared to those from control cells; the mutant probe did not reveal any specific complex. Similar EMSA results were also observed when nuclear lysates from an ORF3-expressing stable cell line (ORF3/4) and a vector control cell line (pCN) were used (data not shown).
FIG. 6.
pORF3 downregulates STAT3 transcriptional activity. (A) EMSA using 32P-labeled wild-type and mutant STAT3 probes and nuclear lysates prepared from Huh7 cells transfected with 2 μg of pSG-ORF3 or the control vector pSGI. Lane 1, mutant probe; lane 2, wild-type probe; lane 3, wild-type probe and control lysate; lane 4, wild-type probe and ORF3 lysate; lane 5, mutant probe and control lysate; lane 6, mutant probe and ORF3 lysate. The positions of free probe and specific complexes are indicated. (B) Huh7 cells were cotransfected with plasmids pLucTK (1 μg) or pLucTKS3 (1 μg) together with either plasmid pSGI (2 μg) or pSG-ORF3 (2 μg). Cell lysates containing equal amounts of total protein were quantitated for luciferase activity as described in Materials and Methods. (C) Huh7 cells were cotransfected with plasmid pLucTKS3 (1 μg) together with 2 μg each of one of the indicated plasmids: pEGFP-N1 (control), pORF3-EGFP (123-aa EGFP-fused ORF3), p114-EGFP (114-aa EGFP-fused ORF3), pSGI (control), or pSG-ORF3 (untagged ORF3). Cell lysates containing equal amounts of total protein were quantitated for luciferase activity. (D) Huh7 cells were cotransfected with plasmid pLucTKS3 (1 μg) together with pEGFP-N1 or pORF3-EGFP (1.5 μg) and either pSGI (3 μg) or a mixture of pSG-ORF1 and pSG-ORF2 (1.5 μg), as indicated. Cell lysates containing equal amounts of total protein were quantitated for luciferase activity. For panels B to D, each transfection was in triplicate. The results from a single experiment are shown and are typical of three independent experiments.
To test the impact on transcription of STAT3 target genes, we used plasmid pLucTKS3 in which a cis-element containing a minimal TK promoter and seven STAT3 response elements is used to drive luciferase expression. When this construct was transfected into the pCN and ORF3/4 cells, a strong inhibition of reporter expression was observed in the latter (not shown). In Huh7 cells that were transiently transfected with pLucTKS3 in the presence or absence of pSG-ORF3, reduced luciferase levels were observed in ORF3-expressing cells (Fig. 6B). We also noticed a small inhibitory effect of pORF3 on the minimal TK promoter (plasmid pLucTK) in the same experiment. That these differences in luciferase expression were not due to differences in transfection efficiencies of the cells being compared was confirmed by cotransfection of other promoter-reporter constructs such as pCH110 (simian virus 40-β-galactosidase) and pEGFP-N1 (cytomegalovirus-EGFP) followed by either a β-galactosidase assay from the same cell lysates or quantitation of EGFP-positive cells by microscopy (data not shown). To further demonstrate that tagged or EGFP-tagged ORF3 or the 123-aa and 114-aa versions of ORF3 behaved similarly, pLucTKS3 was cotransfected with plasmids pORF3-EGFP, p114-EGFP, or pSG-ORF3 with appropriate controls. All three versions of pORF3 showed inhibition of STAT3-driven reporter activity (Fig. 6C), confirming that these proteins had similar downstream effects. Finally, on coexpression of the other HEV proteins, pORF1 and pORF2, there was similar pORF3-mediated inhibition of STAT3 promoter activity (Fig. 6D). These results collectively demonstrate that a block in the cytoplasm to nuclear transport of STAT3 takes place in ORF3-expressing cells, resulting in reduced transcriptional activity of STAT3 in these cells.
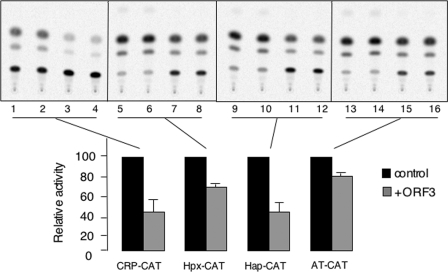
The transcription factor STAT3 activates a number of APR genes. Since we observed reduced nuclear levels and transcriptional activity of STAT3 in ORF3-expressing cells, it was of interest to determine the effects of pORF3 on the expression of acute-phase genes. For this we used reporter constructs containing the CAT gene driven by promoters for the CRP, haptoglobin, hemopexin, and alpha-1-antitrypsin genes. Huh7 cells were cotransfected with pSG-ORF3 or the control vector together with one of the above reporter constructs. The activities of all these acute-phase promoters were found to be downregulated in ORF3-expressing cells, albeit to different levels (Fig. 7). An interleukin-1 receptor antagonist promoter-luciferase construct was similarly tested, but no significant differences were observed between control and ORF3-expressing cells (data not shown). Thus, pORF3 was found to affect the transcriptional activation of a number of STAT3-responsive acute-phase genes.
FIG. 7.
pORF3 downregulates the acute-phase genes. Huh7 cells were cotransfected with 0.5 μg each of the indicated acute-phase gene promoter-reporter construct and 2 μg of either the pSGI (control) or the pSG-ORF3 construct. After 48 h, the cell lysates were prepared, and equal amounts of protein were used for CAT assays. Each transfection is shown in duplicate for control (lanes 1, 2, 5, 6, 9, 10, 13, and 14) and ORF3-transfected (lanes 3, 4, 7, 8, 11, 12, 15, and 16) cells. The results shown are representative of two separate experiments. The bar graph shown below the gel reflects the average percent CAT activity in the presence of ORF3, taking the activity in its absence as 100%.
DISCUSSION
Previous studies have demonstrated that pORF3 activates the cellular extracellularly regulated kinase pathway (20, 24), which may result in a cell survival effect, preventing premature cell death and, in turn, increasing the efficiency of viral replication (20). pORF3 was also shown to promote the secretion of alpha-1-microglobulin, a protein with immunosuppressive properties (44) that is proposed to protect HEV-infected cells in the milieu of liver tissue. We have recently observed a protective effect of pORF3 on mitochondrial depolarization and death as well (32). Thus, pORF3 appears to be a regulatory protein that is likely to influence multiple pathways toward the establishment and propagation of HEV infection. Although ORF3 was required for viral infection following intrahepatic inoculation of HEV genomic RNA directly into monkey liver (14, 15), it was found to be dispensable when HEV genomic RNA was tested by transfection of cells in vitro (8). Due to the lack of traditional in vitro viral infection systems, small-animal models, or efficient replicon systems which can be used to study the effects of HEV proteins on cellular processes, our strategy is mainly based on subgenomic expression of pORF3 (18, 49, 50). We did, however, study the subcellular distribution of pORF3 when it was either singly expressed as the 123- or 114-aa protein, as an EGFP fusion, or from the HEV genomic replicon. In all cases, pORF3 was found to localize to Rab5-positive compartments, suggesting that this is its likely distribution in HEV-infected cells and that the approach is biologically relevant. While we could utilize the HEV genomic replicon for studying the subcellular distribution of pORF3, its expression in only ∼10 to 15% of transfected cells in our hands prevented any downstream pathway analysis with the replicon. In some instances, therefore, we have coexpressed by transient transfection the other two HEV proteins, pORF1 and pORF2, along with pORF3 to study the latter's downstream effects.
Many viruses utilize endocytosis to gain entry into cells and to regulate the host cell environment (17, 38). Viruses encode proteins with multiple functions that are often involved in extensive interactions with host proteins at various subcellular locations, and compartmentalization plays a critical role in signal outcome (12, 26, 31, 48). We show here that pORF3 associated with early and recycling endosomes but not with late endosomes, lysosomes, the Golgi compartment, or caveosomes. This localization to early and recycling endosomes modulates trafficking of specific receptors through these compartments. While pORF3 showed no effect on TfR or fluid trafficking, it retarded the movement of EGFR to late endosomes. Endocytosis and lysosomal targeting of EGFR are normal consequences of receptor activation (4, 47). Because degradation will inevitably terminate receptor signaling, trafficking of EGFR has traditionally been viewed in the context of attenuation, while inhibition of receptor degradation will enhance signaling. There are reports suggesting compartmentalized growth factor signaling from endosomes (13, 40). An endosomal compartment important for signaling was uncovered in the search for Rab5 effectors and interactors that have roles in cell signaling (30, 51). We propose that this delay in EGFR trafficking by pORF3 would contribute to endomembrane growth factor signaling and survival of HEV-infected hepatocytes.
Growth factor receptor endocytosis can also affect the activity and localization of signaling intermediates. The STAT3 protein colocalizes with growth factor receptors in endosomes, and inhibition of endocytosis inhibits its transport from the cytoplasm to the nucleus and thereby its transcriptional activity on target genes (2). Independently, a role for the endosome regulator HRS (hepatocyte growth factor-regulated tyrosine kinase substrate) has also been shown in EGF/STAT3 signaling (37). However, the role of endocytic trafficking processes in STAT3 transport has not been characterized. We found the nuclear levels of pSTAT3 were significantly lower in ORF3-expressing cells. As pORF3 retarded the movement of EGFR from early to late endocytic compartments, this suggested a link between retarded trafficking of EGFR and reduced nuclear translocation of pSTAT3. The nuclear accumulation of unphosphorylated STAT3 protein is independent of endocytosis; in these cases STAT3 is not recruited to any plasma membrane receptor and is directly transported into the nucleus by importin α (28). However, the phosphorylation of STAT3 by Janus kinases takes place once it is bound to activated receptors; this association at the plasma membrane is maintained during endocytic vesicle formation and trafficking and is proposed to be important for delivering pSTAT3 to the perinuclear region (2). In late endosomes, EGF is released from its receptor before degradation (4). This provides a possible mechanism by which pSTAT3 is transported to the nucleus. pORF3 retards movement of EGFR to late endosomes and thus also prolongs the association of pSTAT3 to the growth factor endocytic vesicles. This association is likely to result in lower levels of free pSTAT3 for transport into the nucleus.
The STAT3 transcription factor is a major regulator of the APR in liver (1). Promoter-reporter expression studies suggest that pORF3 downregulates the expression of important APPs in liver cells. Many of the APPs have the potential to influence one or more stages of inflammation. CRP has a number of biological activities related to nonspecific host defense (22, 41, 46). The increased plasma levels of some metal-binding APPs like haptoglobin and hemopexin help by preventing iron loss during infection and injury along with minimizing the level of iron available and by acting as scavengers for potentially damaging oxygen free radicals (42). Protease inhibitor acute-phase reactants like alpha-1-antitrypsin neutralize the lysosomal proteases released following the infiltration of activated neutrophils and macrophages, thus controlling the activity of the proinflammatory enzyme cascades (29). The interactions of APPs with other well-defined defense systems suggest a critical requirement for these proteins very early in the establishment of host defense and are likely to be of considerable clinical importance. Thus, by downregulating the transcription of APP genes, pORF3 can potentially attenuate inflammatory responses and create an environment for increased viral replication and survival. In agreement with this hypothesis, we have found reduced levels of these and other APPs in the plasma of hepatitis E patients compared to healthy volunteers (S. Taneja and S. Jameel, unpublished data).
We show here that pORF3 of HEV delays the degradation of EGFR possibly in early and recycling endosomes. This blocks pSTAT3 nuclear transport and results in reduced transcriptional activity of APP genes. A recent report proposed pORF3 to be translated from a bicistronic subgenomic RNA and to be shortened by 9 aa (16). We also tested the 114-aa pORF3 for two properties, one upstream and one downstream in the proposed mechanistic details. Both the 114- and the 123-aa versions of pORF3 localized to the early endosomes but not late endosomes, and both inhibited STAT3-mediated transcription to similar extents. We also found pORF3 expressed from an HEV genomic replicon to distribute similarly and coexpression of other HEV proteins not to have any apparent effects on pORF3-mediated pSTAT3 nuclear translocation and transcription factor activity.
Collectively, our data provide a novel example of a viral protein that blocks growth factor receptor degradation for prolonged endomembrane signaling and an attenuated APR. Both these effects would promote the survival of infected cells and therefore aid in the replication and pathogenesis of these small hepatotropic viruses. Our results also support a critical role of EGFR degradation for pSTAT3 nuclear transport. Current experiments are aimed at understanding the molecular mechanisms of this novel effect of HEV pORF3.
Acknowledgments
We thank H. Krishnamurthy and the Wellcome Trust-aided imaging facility at NCBS for help with confocal imaging and Suzanne Emerson (NIH) for the generous gift of HEV replicons and S10-3 cells.
An International Senior Research Fellowship of the Wellcome Trust to S.J. supported this work. V.C. and S.K. are supported by predoctoral fellowships from the Indian Council for Medical Research and the Council for Scientific and Industrial Research, respectively. Work in S.M.'s laboratory is supported by intramural funds from NCBS and a J. C. Bose fellowship. ICGEB receives core funding from the Department of Biotechnology (Government of India).
Footnotes
Published ahead of print on 30 April 2008.
REFERENCES
- 1.Alonzi, T., D. Maritano, B. Gorgoni, G. Rizzuto, C. Libert, and V. Poli. 2001. Essential role of STAT3 in the control of the acute-phase response as revealed by inducible gene inactivation [correction of activation] in the liver. Mol. Cell. Biol. 211621-1632. (Erratum, 21:2967.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bild, A. H., J. Turkson, and R. Jove. 2002. Cytoplasmic transport of Stat3 by receptor-mediated endocytosis. EMBO J. 213255-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowman, T., R. Garcia, J. Turkson, and R. Jove. 2000. STATs in oncogenesis. Oncogene 192474-2488. [DOI] [PubMed] [Google Scholar]
- 4.Burke, P., K. Schooler, and H. S. Wiley. 2001. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol. Biol. Cell 121897-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter, G. 2000. The EGF receptor: a nexus for trafficking and signaling. Bioessays 22697-707. [DOI] [PubMed] [Google Scholar]
- 6.Darnell, J. E., Jr. 1997. STATs and gene regulation. Science 2771630-1635. [DOI] [PubMed] [Google Scholar]
- 7.Emerson, S. U., H. Nguyen, J. Graff, D. A. Stephany, A. Brockington, and R. H. Purcell. 2004. In vitro replication of hepatitis E virus (HEV) genomes and of an HEV replicon expressing green fluorescent protein. J. Virol. 784838-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emerson, S. U., H. Nguyen, U. Torian, and R. H. Purcell. 2006. ORF3 protein of hepatitis E virus is not required for replication, virion assembly, or infection of hepatoma cells in vitro. J. Virol. 8010457-10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emerson, S. U., D. Anderson, A. Arankalle, X. J. Meng, M. Purdy, G. G. Schlauder, and S. A. Tsarev. 2004. Hepevirus, p. 851-855. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: classification and nomenclature of viruses. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 10.Fey, G. H., and J. Gauldie. 1990. The acute phase response of the liver in inflammation. Prog. Liver Dis. 989-116. [PubMed] [Google Scholar]
- 11.Fu, X. Y., and J. J. Zhang. 1993. Transcription factor p91 interacts with the epidermal growth factor receptor and mediates activation of the c-fos gene promoter. Cell 741135-1145. [DOI] [PubMed] [Google Scholar]
- 12.Gershburg, E., M. Marschall, K. Hong, and J. S. Pagano. 2004. Expression and localization of the Epstein-Barr virus-encoded protein kinase. J. Virol. 7812140-12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Gaitan, M., and H. Stenmark. 2003. Endocytosis and signaling: a relationship under development. Cell 115513-521. [DOI] [PubMed] [Google Scholar]
- 14.Graff, J., H. Nguyen, C. Kasorndorkbua, P. G. Halbur, M. St Claire, R. H. Purcell, and S. U. Emerson. 2005. In vitro and in vivo mutational analysis of the 3′-terminal regions of hepatitis E virus genomes and replicons. J. Virol. 791017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graff, J., H. Nguyen, C. Yu, W. R. Elkins, M. St Claire, R. H. Purcell, and S. U. Emerson. 2005. The open reading frame 3 gene of hepatitis E virus contains a cis-reactive element and encodes a protein required for infection of macaques. J. Virol. 796680-6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graff, J., U. Torian, H. Nguyen, and S. U. Emerson. 2006. A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J. Virol. 805919-5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruenberg, J., and F. G. van der Goot. 2006. Mechanisms of pathogen entry through the endosomal compartments. Nat. Rev. Mol. Cell Biol. 7495-504. [DOI] [PubMed] [Google Scholar]
- 18.Jameel, S., M. Zafrullah, M. H. Ozdener, and S. K. Panda. 1996. Expression in animal cells and characterization of the hepatitis E virus structural proteins. J. Virol. 70207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jans, D. A., and G. Hassan. 1998. Nuclear targeting by growth factors, cytokines, and their receptors: a role in signaling? Bioessays 20400-411. [DOI] [PubMed] [Google Scholar]
- 20.Kar-Roy, A., H. Korkaya, R. Oberoi, S. K. Lal, and S. Jameel. 2004. The hepatitis E virus open reading frame 3 protein activates ERK through binding and inhibition of the MAPK phosphatase. J. Biol. Chem. 27928345-28357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khuroo, M. S., M. R. Teli, S. Skidmore, M. A. Sofi, and M. I. Khuroo. 1981. Incidence and severity of viral hepatitis in pregnancy. Am. J. Med. 70252-255. [DOI] [PubMed] [Google Scholar]
- 22.Kilpatrick, J. M., and J. E. Volanakis. 1985. Opsonic properties of C-reactive protein. Stimulation by phorbol myristate acetate enables human neutrophils to phagocytize C-reactive protein-coated cells. J. Immunol. 1343364-3370. [PubMed] [Google Scholar]
- 23.Kim, H., and H. Baumann. 1997. The carboxyl-terminal region of STAT3 controls gene induction by the mouse haptoglobin promoter. J. Biol. Chem. 27214571-14579. [DOI] [PubMed] [Google Scholar]
- 24.Korkaya, H., S. Jameel, D. Gupta, S. Tyagi, R. Kumar, M. Zafrullah, M. Mazumdar, S. K. Lal, L. Xiaofang, D. Sehgal, S. R. Das, and D. Sahal. 2001. The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J. Biol. Chem. 27642389-42400. [DOI] [PubMed] [Google Scholar]
- 25.Krawczynski, K. 1993. Hepatitis E. Hepatology 17932-941. [PubMed] [Google Scholar]
- 26.Lilley, B. N., and H. L. Ploegh. 2004. A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429834-840. [DOI] [PubMed] [Google Scholar]
- 27.Lin, S. Y., K. Makino, W. Xia, A. Matin, Y. Wen, K. Y. Kwong, L. Bourguignon, and M. C. Hung. 2001. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 3802-808. [DOI] [PubMed] [Google Scholar]
- 28.Liu, L., K. M. McBride, and N. C. Reich. 2005. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc. Natl. Acad. Sci. USA 1028150-8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lomas, D. A., and R. Mahadeva. 2002. Alpha1-antitrypsin polymerization and the serpinopathies: pathobiology and prospects for therapy. J. Clin. Investig. 1101585-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miaczynska, M., S. Christoforidis, A. Giner, A. Shevchenko, S. Uttenweiler-Joseph, B. Habermann, M. Wilm, R. G. Parton, and M. Zerial. 2004. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell 116445-456. [DOI] [PubMed] [Google Scholar]
- 31.Miyazaki, Y., T. Takamatsu, T. Nosaka, S. Fujita, T. E. Martin, and M. Hatanaka. 1995. The cytotoxicity of human immunodeficiency virus type 1 Rev: implications for its interaction with the nucleolar protein B23. Exp. Cell Res. 21993-101. [DOI] [PubMed] [Google Scholar]
- 32.Moin, S. M., M. Panteva, and S. Jameel. 2007. The hepatitis E virus Orf3 protein protects cells from mitochondrial depolarization and death. J. Biol. Chem. 28221124-21133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moshage, H. 1997. Cytokines and the hepatic acute phase response. J. Pathol. 181257-266. [DOI] [PubMed] [Google Scholar]
- 34.Nayak, N. C., S. K. Panda, R. Datta, A. J. Zuckerman, D. K. Guha, N. Madanagopalan, and K. Buckshee. 1989. Aetiology and outcome of acute viral hepatitis in pregnancy. J. Gastroenterol. Hepatol. 4345-352. [DOI] [PubMed] [Google Scholar]
- 35.Ramalingaswami, V., and R. H. Purcell. 1988. Waterborne non-A, non-B hepatitis. Lancet 1571-573. [DOI] [PubMed] [Google Scholar]
- 36.Sabharanjak, S., P. Sharma, R. G. Parton, and S. Mayor. 2002. GPI-anchored proteins are delivered to recycling endosomes via a distinct cdc42-regulated, clathrin-independent pinocytic pathway. Dev. Cell 2411-423. [DOI] [PubMed] [Google Scholar]
- 37.Scoles, D. R., V. D. Nguyen, Y. Qin, C. X. Sun, H. Morrison, D. H. Gutmann, and S. M. Pulst. 2002. Neurofibromatosis 2 (NF2) tumor suppressor schwannomin and its interacting protein HRS regulate STAT signaling. Hum. Mol. Genet. 113179-3189. [DOI] [PubMed] [Google Scholar]
- 38.Sieczkarski, S. B., and G. R. Whittaker. 2002. Dissecting virus entry via endocytosis. J. Gen. Virol. 831535-1545. [DOI] [PubMed] [Google Scholar]
- 39.Smith, M. F., Jr., D. Eidlen, M. T. Brewer, S. P. Eisenberg, W. P. Arend, and A. Gutierrez-Hartmann. 1992. Human IL-1 receptor antagonist promoter. Cell type-specific activity and identification of regulatory regions. J. Immunol. 1492000-2007. [PubMed] [Google Scholar]
- 40.Sorkin, A., and M. Von Zastrow. 2002. Signal transduction and endocytosis: close encounters of many kinds. Nat. Rev. Mol. Cell Biol. 3600-614. [DOI] [PubMed] [Google Scholar]
- 41.Sui, S. F., Y. T. Sun, and L. Z. Mi. 1999. Calcium-dependent binding of rabbit C-reactive protein to supported lipid monolayers containing exposed phosphorylcholine group. Biophys. J. 76333-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolosano, E., S. Fagoonee, E. Hirsch, F. G. Berger, H. Baumann, L. Silengo, and F. Altruda. 2002. Enhanced splenomegaly and severe liver inflammation in haptoglobin/hemopexin double-null mice after acute hemolysis. Blood 1004201-4208. [DOI] [PubMed] [Google Scholar]
- 43.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18861-926. [DOI] [PubMed] [Google Scholar]
- 44.Tyagi, S., M. Surjit, A. K. Roy, S. Jameel, and S. K. Lal. 2004. The ORF3 protein of hepatitis E virus interacts with liver-specific alpha1-microglobulin and its precursor alpha1-microglobulin/bikunin precursor (AMBP) and expedites their export from the hepatocyte. J. Biol. Chem. 27929308-29319. [DOI] [PubMed] [Google Scholar]
- 45.Ullrich, A., and J. Schlessinger. 1990. Signal transduction by receptors with tyrosine kinase activity. Cell 61203-212. [DOI] [PubMed] [Google Scholar]
- 46.Volanakis, J. E., and M. H. Kaplan. 1971. Specificity of C-reactive protein for choline phosphate residues of pneumococcal C-polysaccharide. Proc. Soc Exp. Biol. Med. 136612-614. [DOI] [PubMed] [Google Scholar]
- 47.Waterman, H., and Y. Yarden. 2001. Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett. 490142-152. [DOI] [PubMed] [Google Scholar]
- 48.Wiertz, E. J., D. Tortorella, M. Bogyo, J. Yu, W. Mothes, T. R. Jones, T. A. Rapoport, and H. L. Ploegh. 1996. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384432-438. [DOI] [PubMed] [Google Scholar]
- 49.Zafrullah, M., M. H. Ozdener, R. Kumar, S. K. Panda, and S. Jameel. 1999. Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. J. Virol. 734074-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zafrullah, M., M. H. Ozdener, S. K. Panda, and S. Jameel. 1997. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J. Virol. 719045-9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zerial, M., and H. McBride. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2107-117. [DOI] [PubMed] [Google Scholar]