Abstract
To examine the pathway of the coreceptor switching of CCR5-using (R5) virus to CXCR4-using (X4) virus in simian-human immunodeficiency virus SHIVSF162P3N-infected rhesus macaque BR24, analysis was performed on variants present at 20 weeks postinfection, the time when the signature gp120 V3 loop sequence of the X4 switch variant was first detected by PCR. Unexpectedly, circulating and tissue variants with His/Ile instead of the signature X4 V3 His/Arg insertions predominated at this time point. Phylogenetic analysis of the sequences of the C2 conserved region to the V5 variable loop of the envelope (Env) protein showed that viruses bearing HI insertions represented evolutionary intermediates between the parental SHIVSF162P3N and the final X4 HR switch variant. Functional analyses demonstrated that the HI variants were phenotypic intermediates as well, capable of using both CCR5 and CXCR4 for entry. However, the R5X4 intermediate virus entered CCR5-expressing target cells less efficiently than the parental R5 strain and was more sensitive to both CCR5 and CXCR4 inhibitors than either the parental R5 or the final X4 virus. It was also more sensitive than the parental R5 virus to antibody neutralization, especially to agents directed against the CD4 binding site, but not as sensitive as the late X4 virus. Significantly, the V3 loop sequence that determined CXCR4 use also conferred soluble CD4 neutralization sensitivity. Collectively, the data illustrate that, similar to human immunodeficiency virus type 1 (HIV-1) infection in individuals, the evolution from CCR5 to CXCR4 usage in BR24 transitions through an intermediate phase with reduced virus entry and coreceptor usage efficiencies. The data further support a model linking an open envelope gp120 conformation, better CD4 binding, and expansion to CXCR4 usage.
Entry of human immunodeficiency virus type 1 (HIV-1) into target cells requires the CD4 receptor and one of two coreceptors, CCR5 or CXCR4 (2). CCR5-using (R5) virus predominates early in infection, but in about 50% of subtype B-infected individuals, CXCR4-tropic (X4) virus appears and coexists with R5 viruses, and this is associated with more rapid decline of CD4+ T cells and poorer prognosis (3, 5, 11, 12, 58, 66). The basis for X4 emergence late in infection remains ill defined, but among the hypotheses proposed are mutation by chance, CCR5 bearing target cell limitation, and differential immune recognition of X4 and R5 viruses (43, 53). Furthermore, it is unclear whether X4 viruses evolve during the course of infection or were present at time of transmission but preferentially suppressed early in infection.
In HIV-1-infected individuals and in tissue culture systems, the pathway to coreceptor switching transitions through intermediates with the ability to use CXCR4 in addition to CCR5 (12, 50, 57, 60, 61). Compared to the early or inoculating R5 viruses, these R5X4 dual-tropic viruses often display a loss in replicative fitness as well as less efficient use of the CCR5 coreceptor in vitro (30, 50). It has been suggested that the fitness disadvantage of the intermediates compared with the initial R5 virus constitutes one of the blockades to coreceptor switching, explaining the late appearance of X4 viruses (50). Additionally, recently emerged R5X4 and X4 viruses in humans are found to be more sensitive to antibody neutralization than coexisting R5 viruses, implicating antiviral antibody response as another obstacle to coreceptor switching (6).
We recently described the first case of a coreceptor switch in rhesus macaque BR24 that was infected with the late R5 simian-human immunodeficiency virus SHIVSF162P3N isolate (23). Animal BR24 progressed to disease rapidly after transient seroconversion. Virus recovered at end-stage disease (28 weeks postinfection) was shown to use CXCR4 exclusively and, compared to the inoculating virus, was highly susceptible to antibody neutralization, in particular, to agents such as soluble CD4 (sCD4) and the monoclonal antibody (MAb) immunoglobulin G1b12 (IgG1b12) directed at the CD4 binding site (CD4BS). Furthermore, similar to cases reported in humans (10, 46), X4 emergence lagged rather than preceded or coincided with the onset of a precipitous CD4+ T-cell decline in macaque BR24, lending support to the notion that X4 emergence is the result, rather than the cause, of immune failure. The goal of the present study is to reconstruct the pathway to coreceptor switching in macaque BR24 and determine the consequences for envelope (Env) protein functions associated with evolution to CXCR4 usage. We seek to identify transitional intermediates and to assess the costs and benefits of, and reasons for, coreceptor switching in a nonhuman primate model of HIV/AIDS.
MATERIALS AND METHODS
Cells.
293T cells and TZM-bl cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin, streptomycin, and l-glutamine. The latter expressed CD4, CCR5, and CXCR4 and contained integrated reporter genes for firefly luciferase and β-galactosidase under control of the HIV-1 long terminal repeat. U87 glioma cell lines stably expressing CD4 and one of the chemokine receptors were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, antibiotics, 1 μg/ml puromycin (Sigma-Aldrich, St. Louise, MO), and 300 μg/ml G418 (geneticin; Invitrogen, Carlsbad, CA).
RNA/DNA extraction, sequencing, and analysis.
Viral RNA was prepared from 500 μl of BR24 week 20 plasma using a commercially available RNA extraction kit (Qiagen, Chatsworth, CA), and reverse transcribed with Superscript III RT (Invitrogen) and random hexamer primers (Amersham Pharmacia, Piscataway, NJ). For DNA extraction, cells from axillary lymph node (LN) biopsy sampled at 20 weeks postinfection (wpi) or from tissues collected at necropsy were used. The V1 to V5 region of gp120 was amplified from the reverse transcription products or extracted proviral DNA by Taq DNA polymerase (Qiagen) with primers ED5 and ED12 or ES7 and ES8 as previously described (14). PCR products were cloned with the TOPO TA cloning kit (Invitrogen) per the manufacturer's instructions, followed by direct automated sequencing of cloned gp120 amplicons (SeqWright; Fisher Scientific, Houston, TX). Nucleotide sequences were aligned using the CLUSTAL X program, version 1.81, and further adjusted manually (28).
Env gp160 subcloning and pseudotype virus production.
The construction of Env expression plasmids for the inoculating R5 SHIVSF162P3 (EnvP3N), the final X4 SHIVBR24N (EnvBR24N), and EnvP3N expressing the V3 variable loop of EnvBR24N [EnvP3N(HR-V3)] has been previously described (23). For expression of the full-length gp160 coding sequence of viruses predominating at 20 wpi in macaque BR24 (EnvHI20, week 20 virus bearing the HI insertion in Env gp160), DNA was extracted from LN cells, amplified with primers SH52 (5′-TAG ATC GAA TTC TAG AGC CCT GGA AGC ATC CAG GAA GTC AGC CTA-3′) and SH53 (5′-AGA GAG GGA TCC TCC AGT CCC CCC TTT TCT TTT AAA AA-3′), and subcloned into the pCAGGS expression plasmid. To generate P3N recombinant Env carrying the V3 loop of EnvHI20, PCR-based overlapping extension methodology was employed (21). Briefly, with EnvP3N serving as a template for PCR amplification, the outer primers used were SH43 (5′-AAGACAGAATTCATGAGAGTGAAGGGGATCAGGAAG-3′) and SH44 (5′-AGAGAGGGATCCTTATAGCAAAGCCCTTTC AAAGCCCT-3′), and the inner primers were SH66 (5′-AATAATACAAGAAAAAGTATACGTATACATATAGGACCGGGGAGAGCATTTTATGC-3′) and SH67 (5′-TATATGTATACGTATACTTTTTCTTGTATTATTGTTAGGTCTTGTACAATTAATTTCTAC-3′), which encompassed the amino acid changes found in the V3 loop of EnvHI20 (underlined). The resulting amplified fragment was verified by sequencing and subcloned back into pCAGGS. Single-cycle replication-competent luciferase reporter viruses were prepared by transfecting 293T cells with the NL4.3-Luc-E− R+ vector and the corresponding Env expression plasmid with polyethylenimine (Polyscience, Warrington, PA). Supernatants were harvested 72 h posttransfection and quantified for p24Gag antigen content by antigen capture assay (Alliance HIV-1 p24 antigen enzyme-linked immunosorbent assay kit; Perkin Elmer, Waltham, MA).
Entry and blocking assays.
Single-cycle infectivity assays were used to measure the entry efficiency of various Env proteins. Briefly, 104 U87.CD4.CCR5 or U87.CD4.CXCR4 cells were seeded in 96-well plates 24 h before use. Cells were infected with 5 ng of a p24Gag-equivalent of the indicated pseudotyped viruses, followed by incubation for 72 h at 37°C. At the end of the incubation period, cells were harvested, lysed, and measured for luciferase activity according to the manufacturer's instructions (Luciferase Assay System; Promega, Madison, WI). Entry, as quantified by luciferase activity, was measured with a Dynex MLX microtiter plate luminometer (Dynex Technologies, Inc., Chantilly, VA). For the entry-blocking assays, U87.CD4 indicator or TZM-bl cells were pretreated with various concentrations of PSC-RANTES (gift of Oliver Hartley, University of Zurich), TAK-779, or AMD3100 for 30 min at 37°C before infection. The percentage of entry blocking was calculated by the amount of entry in the presence of the inhibitor relative to that in the absence of the inhibitor, and 50% and 90% inhibitory concentrations (IC50 and IC90, respectively) were determined using Prism 4 software (GraphPad, San Diego, CA).
Neutralization assay and antibodies.
Virus neutralization sensitivity was assessed using TZM-bl or U87.CD4 indicator cells in 96-well plates. Briefly, equal volumes (50 μl) of pseudotyped viruses (5 ng of p24Gag equivalent) and serial dilutions of heat-inactivated macaque serum or MAbs were incubated for 30 min at 37°C and then added to cells, in duplicate wells, for an additional 2 to 3 h at 37°C. A total of 100 μl of medium was then added to each well, and the virus-MAb cultures were maintained for 72 h. At the end of the culture period, the cells were lysed and processed for β-galactosidase (Galacto-Star System; Applied Biosystems, Bedford, MA) or luciferase activity. A neutralization curve was generated by plotting the percentage of neutralization versus serum or MAb dilution, with IC50 and IC90 values calculated as described above. Control cultures received virus in the absence of antibodies. The human MAb IgG1b12 was kindly supplied by Dennis Burton (Scripps Research institute, La Jolla, CA), CD4-IgG2 (PRO 542) was from William Olsen (Progenics Pharmaceuticals, Tarrytown, NY), and the anti-V3 antibody 447-52D was from Susan Zolla-Pazner. MAbs 2G12, 4E10, and 2F5 were obtained through the NIH AIDS Research and Reference Reagent Program.
RESULTS
Variants with HI insertions predominate at the time of first X4 virus appearance.
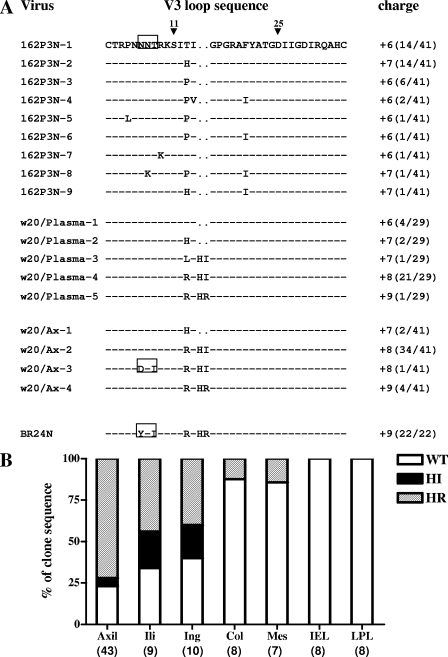
We recently reported that mutations within the gp120 V3 loop, notably an insertion of two basic amino acids (HR) immediately upstream of the tip of the GPGR crown of the R5 SHIVSF162P3N virus, were responsible for the change in coreceptor preference of the virus replicating in infected rhesus macaque BR24 at the time of death (23). Variants bearing HR sequences were first detected by PCR in the plasma and peripheral blood mononuclear cells of BR24 at 20 wpi, 8 weeks prior to euthanasia. To determine the spectrum of X4 variants at the time of their first emergence, as well as to reconstruct the evolutionary pathway to the phenotypic switch from R5 to X4 in an SHIV model, the Env V1 to V5 sequences of week 20 circulating viruses were cloned, determined, and compared. V3 loop sequence analysis showed that the predominating virus (22 of 29 clones sequenced) had HI instead of HR insertions, with a net positive charge of +7 or +8 instead of +9 for this gp120 region (Fig. 1A). In contrast to the final X4, but similar to the initial R5 virus, the highly conserved potential N-linked glycosylation (PNG) site at the base of the V3 loop was intact for all 29 sequences of week 20 Env examined. Variants with HI insertions also predominated in the axillary LN sampled at week 20. Of 41 Env clones obtained from this tissue site, 35 harbored HI insertions, 4 had HR insertions, while the remaining 2 clones retained wild-type (WT) R5 sequences. One of the 35 HI-bearing clones from the axillary LN lacked the V3 PNG site.
FIG. 1.
(A) Comparison of V3 loop sequences of the parental R5 SHIVSF162P3N isolate, the recovered X4 virus SHIVBR24N, and viruses present in plasma and axillary (Ax) LN of macaque BR24 at 20 wpi. Dashes denote similarity in sequences, and gaps are indicated as dots. The net positive charge of this region is indicated in the right column. Positions 11 and 25 within the V3 loop that frequently distinguished HIV-1 X4 and R5 viruses (16) are designated by arrowheads, and the highly conserved PNG site at the V3 base is boxed. The numbers in parentheses represent the number of clones with the indicated V3 loop sequence reported relative to the total number of clones sequenced. (B) Representation of SHIV viral variants without (WT) or with HI or HR insertions in various tissue sites of macaque BR24 at the time of death (28 wpi). Percentages of Env clones present in axillary (axil), iliac (ili), inguinal (ing), colonic (col), and mesenteric (mes) LNs, as well as intraepithelial lymphocytes (IEL) and LPL of the gut with the indicated signature V3 loop sequences are shown. Numbers in parenthesis indicate the number of gp120 clones sequenced from each of the tissue sites.
The tissue reservoirs of viruses with HI or HR insertion sequences were further examined using samples obtained at necropsy (week 28). Results showed that these sequences were poorly represented in the gut (intraepithelial and laminar propria lymphocytes [LPL]) and intestinal LNs (colonic and mesenteric LNs) but predominated in peripheral LNs such as the axillary, iliac, and inguinal LNs (Fig. 1B). Within the latter tissue sites, variants with HR insertions now dominated over those with HI insertions. For example, of 43 Env clones sequenced from the axillary LNs at time of death, 31 harbored HR insertions, 10 had WT sequences, and only 2 contained HI insertions. The change in dominance over time from variants with HI to those with HR insertions in axillary LNs suggests that the former are less fit than the late X4 variants.
HI-bearing variants are intermediates in the evolutionary pathway from CCR5 to CXCR4 use.
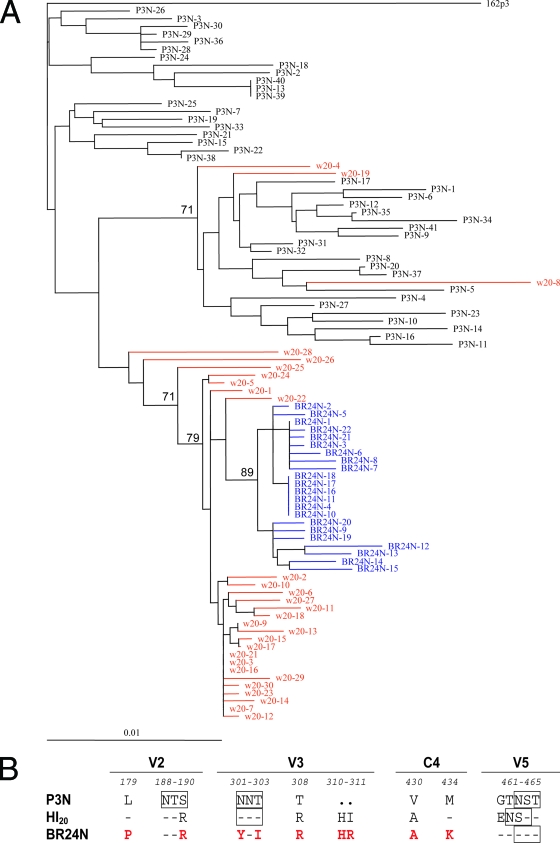
Neighbor-joining phylogenetic analysis of sequences of Env V1 to V5 supports a progressive mutational process leading to phenotypic switching in macaque BR24, with HI-bearing sequences present at 20 wpi representing evolutionary intermediates to HR-bearing sequences (Fig. 2A). Although minor week 20 viral populations sitting in various locations in the tree were noted, it is clear, based on the genetic distance, that the closest relatives of SHIVBR24N sequences were among the week 20 sequences. We previously reported that, compared to the inoculating R5 SHIVSF162P3N isolate, the final X4 SHIVBR24N variant harbored nine unique amino acid changes in V2 and V3 and in a C4 region of gp120 implicated in CCR5 binding (23). gp120 sequence of viruses bearing HI insertions differed from SHIVSF162P3N as well as SHIVBR24N in five of the nine amino acids (Fig. 2B). In addition, the HI variants differed from both the parental R5 and final X4 SHIV virus in the V5 hypervariable region, where a charge substitution (change of G to E) and repositioning of a PNG site (change of T to N) were noted. Besides the V3 loop (17, 20), several regions of gp120, including the V1-V2 loop (9, 17, 29, 72), are known to influence coreceptor choice. Furthermore, the C4 domain interacts with the V3 loop (44, 74) and influences the binding and fusion steps of the entry process (63). Combined, the data show that viruses predominating at week 20 are evolutionary intermediates, differing from the initial R5 SHIV and final X4 variant in Env regions known to modulate the efficiency of coreceptor use.
FIG. 2.
(A) Phylogenetic tree showing the relationship between the Env V1 to V5 sequences of the parental R5 SHIV162P3N, the week 20 variants, and the final X4 SHIVBR24N. A neighbor-joining tree was generated, with the SHIVSF162P3 sequence serving as an outgroup. Week 20 plasma sequences are marked in red while those of the X4 SHIVBR24N isolate are in blue. The scale bar indicates genetic distances along the horizontal branches. The values on the branches represent the percentage of bootstrapped trees out of 1,000 replicates. Sequence gaps are excluded for analysis. (B) Comparison of the predicted V2, V3, C4, and V5 gp120 amino acid sequences of SHIVSF162P3N, SHIVBR24N, and week 20 Envs. The nine amino acids in X4 SHIVBR24N that differed from R5 SHIVSF162P3N Env gp120 are indicated in red. Dashes denote similarity in sequence, dots indicate gaps, and the PNG sites are boxed. Residues that differ between the viruses are numbered based on their relative positions in the HXB2 sequence. w, week.
Variants with HI insertions use both CCR5 and CXCR4 for entry.
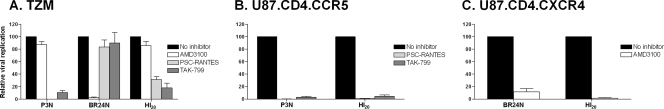
To determine the tropism of intermediate week 20 viruses bearing the HI insertions, the predominant HI Env gp160 sequences (designated EnvHI20) were used to generate pseudotype viruses and tested for coreceptor usage. Infection with viruses bearing the Env of the parental R5 SHIVSF162P3N (EnvP3N) and the final X4 variant SHIVBR24N (EnvBR24N) served as controls. In single-round infectivity assays, infection of TZM-bl cells that express both coreceptors with virus pseudotypes bearing EnvHI20 was efficiently blocked by the CCR5 inhibitors TAK-779 and PSC-RANTES (∼80%) but only minimally by the CXCR4 antagonist AMD3100 (∼20%), mostly likely because the virus is using the CCR5 coreceptor in these cells for entry (Fig. 3A). Infection and blocking experiments in the U87.CD4 indicator cells, however, demonstrated the dual-tropic nature of HI20 Env-bearing viruses, which were able to infect cells expressing the CCR5 (Fig. 3B) or CXCR4 coreceptor (Fig. 3C) and were potently inhibited by cognate coreceptor inhibitors in their respective indicator cell lines. Thus, HI variants demonstrated the typical properties of a dual-tropic virus, while the later HR-bearing variants used almost exclusively the CXCR4 coreceptor. Variants with HI insertions in the V3 loop, therefore, are functional intermediates as well as evolutionary ones.
FIG. 3.
Coreceptor usage of predominant week 20 HI insertion-bearing viruses. (A) CCR5 and CXCR4 usage of viruses pseudotyped with Env of SHIVSF162P3N (P3N), SHIVBR24N (BR24N), and week 20 viruses bearing HI insertions (HI20) was determined by entry inhibition into TZM-bl CCR5+ CXCR4+ (TZM) cells with 1 μM CXCR4 inhibitor (AMD3100) or CCR5 inhibitors (TAK-779 and PSC-RANTES). Infection and blocking with cognate coreceptor inhibitors were also performed in the U87.CD4.CCR5 (B) and U87.CD4.CXCR4 (C) cells. Error bars indicate standard errors of the mean of data in triplicate wells. Results shown are representative of at least two independent experiments.
R5X4 intermediates have reduced entry efficiency into CCR5-expressing cells and use CCR5 and CXCR4 less efficiently.
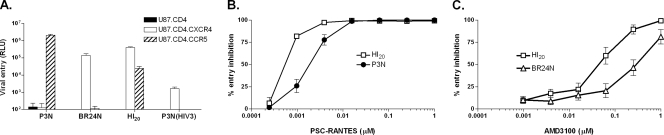
To assess the costs and benefits of coreceptor switching in rhesus macaques, we examined the entry and coreceptor usage efficiencies of the inoculating R5, R5X4 intermediate, and final X4 viruses by infectivity and dose titration entry-blocking experiments. Results of single-round infectivity assays showed that EnvHI20 mediated similar entry into CXCR4-expressing cells compared to EnvBR24N but showed a 1.5 log lower entry into CCR5 expressing cells than the parental R5 EnvP3N (Fig. 4A). Substitution of the V3 loop of EnvHI20 into EnvP3N generated a recombinant EnvP3N(HI-V3) that now mediated entry only in CXCR4-expressing cells, albeit much less efficiently than the parental dual-tropic HI20 and the final X4 Envs, indicating that mutations outside of the V3 loop of the R5X4 intermediate virus contribute to coreceptor usage and virus entry efficiencies. Increased sensitivity to antagonists of both coreceptors was also observed for viruses expressing EnvHI20. Concentrations of the CCR5 inhibitor PSC-RANTES needed to suppress infection of the R5X4 intermediate were fourfold less than those required to inhibit the parental R5 virus (IC50s of 0.6 nM and 2.4 nM, respectively) (Fig. 4B). Similarly, sensitivity to inhibition with the CXCR4 blocker AMD3100 was enhanced 4.5-fold compared to the final X4 virus (IC50s of 71 nM and 323 nM, respectively) (Fig. 4C). The findings of less efficient entry into CCR5-expressing cells and increased sensitivity to CCR5 and CXCR4 blockers for R5X4 viruses suggest a loss of competitive fitness of these dual-tropic intermediates compared to the parental R5 and the final X4 viruses.
FIG. 4.
Entry and coreceptor usage efficiencies of R5, R5X4, and X4 SHIV viruses. (A) Relative entry into U87.CD4 (closed bars), U87.CD4.CXCR4 (open bars), and U87.CD4.CCR5 (hatched bars) cells of luciferase reporter virus expressing Env of R5 SHIVSF162P3N (P3N), X4 SHIVBR24N (BR24N), R5X4 (HI20), and V3 loop recombinant EnvP3N(HI-V3). (B) Blocking of P3N (•) and R5X4 (□) virus entry into U87.CD4.CCR5 cells with increasing concentrations of the CCR5 inhibitor PSC-RANTES. (C) Blocking of BR24N (▵) and R5X4 (□) virus entry into U87.CD4.CXCR4 cells with increasing concentrations of the CXCR4 inhibitor AMD3100. Data shown are means ± standard errors of the mean of at least three independent experiments. RLU, relative light units.
R5X4 variants are neutralization-sensitive antigenic intermediates.
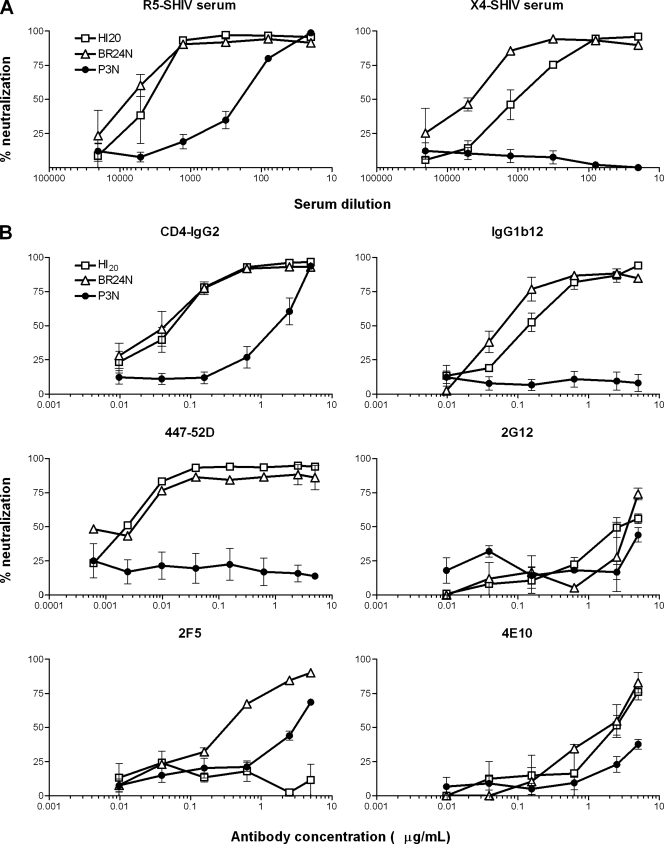
We showed previously that the final X4 SHIVBR24N virus was more sensitive to antibody-mediated neutralization than the parental R5 SHIVSF162P3N virus, particularly to CD4-IgG2 and MAb IgG1b12 that are directed against the CD4BS (7, 23, 77). To determine whether the R5X4 intermediates present at 20 wpi share this characteristic with the final virus that uses only CXCR4, as well as to assess the antigenic relatedness of the viral Envs, the neutralization profile of these viruses to serum from macaques infected with the parental R5 SHIVSF162P3N and the final X4 variant SHIVBR24N (23) was first determined. Results showed that EnvHI20 and EnvBR24 virus pseudotypes were equally sensitive to neutralization with antibodies present in the R5 SHIV serum and more so than viruses bearing the parental R5 EnvP3N. Ninety percent neutralization of the CXCR4-using viruses was achieved at a serum dilution of 1:1,500, but a similar degree of neutralization of the R5 virus required a 30-fold higher serum titer. Furthermore, while the parental R5 virus was resistant to neutralization with serum antibodies from an X4 SHIV-infected macaque, the R5X4 virus was efficiently (90%) neutralized at a dilution of 1:120, and the final X4 virus was the most sensitive of all, with 90% neutralization at a serum dilution of 1:750 (Fig. 5A). The finding that EnvHI20 can be differentiated from EnvBR24 in terms of neutralization sensitivity to the X4 SHIV serum implies differences in their antigenic structures.
FIG. 5.
Neutralization sensitivity of R5, R5X4, and X4 SHIVs. The neutralization susceptibility of the parental R5 SHIVSF162P3N (•), the final X4 SHIVBR24N (▵), and the R5X4 intermediate (□) virus to antibodies in serum from R5 SHIVSF162P3N-infected (R5-SHIV serum) and X4 SHIVBR24N-infected (X4-SHIV serum) macaques (A) and to soluble CD4 (CD4-IgG2) and MAbs IgG1b12, 447-52D, 2G12, 2F5, and 4E10 (B) was determined in TZM-bl cells as described in Materials and Methods. Data shown are the means and standard errors of at least three independent neutralization experiments.
Neutralization with several well-defined broadly neutralizing MAbs was performed to better understand the mechanism underlying enhanced sensitivity to neutralization of the R5X4 intermediate virus to SHIV-positive macaque serum. Soluble CD4 (CD4-IgG2) neutralized the two CXCR4-using viruses with equal potency, achieving 50% neutralization at concentrations that were 40-fold lower than those required to neutralize the parental R5 virus (IC50s of 0.05 and 0.07 μg/ml for the R5X4 and X4 viruses, respectively, compared to 2.2 μg/ml for the R5 virus). While the parental virus was resistant, the R5X4 virus was sensitive to neutralization with the CD4BS MAb IgG1b12 but with an IC50 that was twofold higher than the final X4 virus (IC50s of 0.15 and 0. 075 μg/ml, respectively) (Fig. 5B). Both CXCR4-using viruses but not the parental R5 virus were potently neutralized by the V3 loop MAb 447-52D (IC50 of 0.004 μg/ml). In contrast, no difference in sensitivity of the three viruses to 2G12 directed at sugar moieties of gp120 was observed. Of the two MAbs directed against the membrane-proximal external region of gp41 tested, 4E10 moderately neutralized virus pseudotypes bearing the HI20 and BR24N Envs (IC50 of >1 μg/ml) but not P3N, while 2F5 neutralized the final X4 (IC50 of 0.4 μg/ml) and to some extent the parental R5 virus (IC50 of 3 μg/ml), but the R5X4 virus was resistant. A K-to-N mutation in the 2F5 epitope of the dual-tropic virus accounted for its resistance (data not shown). Taken together, the results showed that the R5X4 intermediate shares a generalized increased sensitivity to neutralization with the final X4 virus but differs from the final X4 virus due to its reduced sensitivity to X4 SHIV serum and to IgG1b12 neutralization. The finding of enhanced neutralization susceptibility of the dual-tropic and the X4 viruses to agents directed at the CD4BSs and the V3 loop compared to the parental R5 virus suggests adoption of an increasingly open conformation of the Env gp120 of viruses in macaque BR24 over time, with exposure of both the CD4 and coreceptor binding sites. The data further support the hypothesis proposed by us (23) and others (6, 36) that lowered or absent antibody-mediated selective pressure is one of the factors favoring the emergence of neutralization-sensitive, CXCR4-using variants.
CXCR4 usage and sCD4 neutralization sensitivity are functionally linked.
CXCR4 usage and sCD4 neutralization sensitivity have been reported to be regulated by similar mechanisms via the V3 domain (27, 47), with the conserved 301N glycosylation site in the V3 base playing a particularly important role (35, 38, 51). To examine whether the increase in sCD4 neutralization sensitivity of the R5X4 and X4 SHIV viruses is functionally linked to their ability to use the CXCR4 coreceptor, the neutralization profile of R5 SHIVSF162P3N derivatives that contained the V3 loop of the R5X4 or X4 SHIV virus to CD4-IgG2 was examined. These derivatives, designated P3N(HI-V3) and P3N(HR-V3), respectively, altered the tropism of SHIVSF162P3N from CCR5 to CXCR4 use (Fig. 4A) (23). Results showed that the CXCR4-using V3 loop recombinant viruses displayed an sCD4 neutralization profile comparable to that observed with either the R5X4 or X4 SHIV virus (Table 1). Thus, mutations in V3 loop of the R5X4 intermediate and the final X4 SHIV variant that confer CXCR4 use are also sufficient to determine sCD4 neutralization sensitivity.
TABLE 1.
CXCR4 usage and sCD4 neutralization sensitivity are functionally linkeda
| ENV type | Replication of virus on:
|
sCD4 neutralization (IC50 [μg/ml] ± SEM)b | |
|---|---|---|---|
| U87.CD4.CCR5 cells | U87.CD4.CXCR4 cells | ||
| P3N | Yes | No | 2.170 ± 0.103 |
| HI20 | Yes | Yes | 0.021 ± 0.004 |
| P3N(HI-V3) | No | Yes | 0.043 ± 0.001 |
| BR24 | No | Yes | 0.038 ± 0.000 |
| P3N(HR-V3) | No | Yes | 0.025 ± 0.002 |
The coreceptor preference and CD4-IgG2 neutralization susceptibility (concentrations achieving IC50) of virus pseudotypes bearing the envelopes of the parental R5 SHIVSF162P3N (P3N), the week 20 R5X4 intermediate SHIV (HI20), the final X4 SHIVBR24N (BR24N), and SHIVSF162P3N derivatives containing the V3 loop of the R5X4 [P3N(HI-V3)] or the final X4 [P3N(HR-V3)] SHIV are shown.
Values represent three independent neutralization experiments.
DISCUSSION
The characterization of SHIV variants present in a macaque at the time of X4 virus emergence provides further insight into the process of coreceptor switching. We find that viruses capable of using both coreceptors predominated in plasma and lymphoid tissues of macaque BR24 at week 20 but represented only a minor population at the time of necropsy 8 weeks later. The dual-tropic viruses occupied an intermediate position between the parental R5 and final X4 SHIV virus in the phylogenetic tree, differing from these viruses in Env gp120 sequence regions known to modulate receptor and coreceptor binding. In vitro studies showed that R5X4 SHIV viruses were compromised in the efficiency of use of each of the coreceptors but shared Env conformations with both the parental R5 and X4 SHIV viruses, as probed by sensitivity to antibody neutralization. Thus, dual-tropic viruses represent temporal, evolutionary, functional, and antigenic intermediates in the pathway to coreceptor switch in rhesus macaques.
At necropsy, dual-tropic and X4 viruses predominated in peripheral LNs (axillary, iliac, and inguinal) but not in intestinal LNs (colonic and mesenteric) or LPL isolated from the jejunum or plasma. Memory CD4+ T lymphocytes are enriched in the gut-associated lymphoid tissue (GALT) and have much higher CCR5 expression than lymphocytes from blood and LNs (22, 69). Accordingly, the GALT is an early site of R5 virus replication and massive destruction of CD4+ T cells in humans and in macaques (4, 18, 19, 39, 40, 68). CCR5 expression levels have also been shown to be higher on CD4+ T cells in the mesenteric LN than in the axillary LN (69). Thus, intestinal LNs could provide optimal sites for R5 virus replication and pathogenesis following the depletion of susceptible target cells in the gut. In this regard, our finding that X4 viruses, which evolved late in infection, reside principally in the peripheral lymph nodes suggests that a significant proportion of CD4+ target T cells in the gut and intestinal LNs express both CCR5 and CXCR4, such that prior seeding and depletion of these CCR5/CXCR4-coexpressing target cells by R5 viruses limited the establishment of niches by the emerging X4 viruses at these sites. Alternatively, higher levels of stromal cell derived factor 1, the ligand of CXCR4, could be present in the GALT and intestinal LNs. Interestingly, a recent report in pediatric HIV infections indicates that the thymus or secondary lymphoid tissues may also play an important role in the evolution/amplification of coreceptor variants (55). Evolution and localization of X4 variants in peripheral lymphoid organs which are not frequently sampled could explain the observation of X4 emergence seen in only a subset of patients progressing to AIDS. Tissue data are very limiting for HIV-1 infection of humans, highlighting the usefulness of the SHIV model in providing a detailed picture of X4 evolution over time and in different tissue compartments of the host.
The dual-tropic SHIV intermediate virus entered CCR5-expressing cells less well and was more sensitive to entry inhibition with the CCR5 analog PSC-RANTES than the parental R5 virus. It was also more sensitive than the final X4 SHIV to inhibition with the CXCR4 inhibitor AMD3100. Correlation between diminished coreceptor usage efficiency and enhanced sensitivity to neutralization with coreceptor inhibitors of HIV-1 variants has been reported (48, 52) although in the case of AMD3100 differences in sensitivity could be due to differences in V3 loop sequence that influence the overall conformation of gp120 (15). Increased sensitivity to PSC-RANTES is also a robust indicator of better patient outcome (64), suggesting that increased sensitivity to CCR5 antagonists may also be related to reduced viral fitness. Accordingly, our findings indicate that the R5X4 intermediates in macaque BR24 have reduced replicative capacity and decreased CCR5 and CXCR4 binding efficiency. HIV-1 R5X4 intermediates have also been found to have reduced fitness (49, 50, 67) and display greater sensitivity to CCR5 antagonists and lower binding affinity to CCR5 than R5 viruses (1, 57, 59, 76). Decreasing sensitivity to CXCR4 antagonists of X4 HIV-1 variants that evolved during the natural course of infection has also been reported (62). Collectively, the data support a similar mechanistic basis for coreceptor switching in humans and macaques. Ongoing evolution of Env variants toward CXCR4 coreceptor usage in both rhesus macaques and humans is at the expense of CCR5 use, and R5X4 intermediate viruses are inefficient in engaging each of the chemokine receptors.
Why, then, does the switch from CCR5 to CXCR4 need to transition through an apparently weak and inefficient intermediate? It is conceivable that the intermediate arises by chance as a result of high virus replication and mutation rates. Indeed, the HI insertions seen in the V3 loop of the R5X4 intermediate SHIV could arise from copying error, resulting in gene duplication of the HI sequence immediately upstream of the GPGR crown in one of the major variants in the R5 SHIV inoculum (Fig. 1A, 162P3N-2). The mutations might become fixed because they confer the selective advantage of CXCR4 use and, hence, target cell expansion. Furthermore, at this stage of the infection within the host, R5X4 viruses may, in fact, be more fit than coexisting X4 viruses. The transient rise in viremia that accompanied the appearance of these intermediates at 20 wpi in macaque BR24 suggests that they are indeed fit, and preliminary data indicate that compared to the R5X4 intermediate, CXCR4-using variants that harbor HR insertions present at 20 wpi are poor in mediating viral entry (data not shown). Lastly, we found a small but consistent difference in neutralization susceptibility of the CXCR4-using SHIVs to serum antibodies and IgG1b12, with the R5X4 intermediate being more resistant than the X4 variant. Thus, the possibility exists that in the presence of even minimal residual humoral immune pressure, perhaps localized at tissue sites that were not sampled in this study, replication of a more neutralization-resistant virus like the HI insertions bearing intermediate and not the highly sensitive final X4 SHIV will be favored. Coreceptor switching was recently observed in a second R5 SHIVSF162P3N-infected macaque (24). It will be of interest to determine whether R5X4 intermediate viruses are also present in this second animal and whether similar costs and benefits are associated with switching via a dual-tropic intermediate.
Compared to the parental R5 virus, the dual-tropic and X4 viruses were highly susceptibility to neutralization with sCD4 and the anti-V3 loop antibody 447-52D, but, as noted above, the final X4 virus was twofold more sensitive than the R5X4 intermediate to neutralization with the CD4BS MAb IgG1b12. Structural studies showed that the CD4BS is recessed on the virion surface, shielded by the V1/V2 loop and associated carbohydrate structures (8, 33, 34, 37, 56, 71, 72, 75). The V1/V2 loop also masks the coreceptor binding site on gp120 that is composed of parts of the V3 loop and a surface formed by the bridging sheet (13, 25, 54, 73). Higher sensitivity of viruses to neutralization with sCD4, IgG1b12, and anti-V3 loop antibodies implies greater exposure of the CD4BS and chemokine receptor binding site that are usually sequestered away from the humoral immune response (32, 34), indicative of a more open Env conformation. Enhanced neutralization susceptibility of the CXCR4-using viruses to MAb directed against epitopes in gp41 lends further support to an open Env configuration of these viruses. The observation that the final X4 SHIV virus is more sensitive than the R5X4 intermediate to neutralization with SHIV serum and IgG1b12 suggests that there is greater exposure of the receptor binding site on the virus that uses only CXCR4 than on viruses that are capable of interacting with both coreceptors. We show that acquisition of CXCR4 use and sCD4 neutralization sensitivity of the R5X4 and X4 SHIV viruses are mediated by similar mechanisms. Furthermore, X4 emergence in BR24 (23) and in another SHIVSF162P3N-infected macaque (24) lags rather than precedes the precipitous drop in CD4+ T cells. In humans, CXCR4 use is associated with a lower CD4 cell count (5, 26, 41, 45, 65, 70), and X4 viruses are frequently recovered from rapid progressors (31, 42). Thus, it is tempting to speculate that adoption of a more open Env conformation, driven perhaps by the need to bind CD4 efficiently as the number of CD4+ target T cells and/or amount of the receptor molecule on target cells becomes limited, will also facilitate interaction with CXCR4 but will, at the same time, reduce protection from antibody-mediated neutralization.
In conclusion, as in HIV-1 infection, evolution from CCR5 to CXCR4 in SHIVSF162P3N-infected rhesus macaque BR24 transitions through an R5X4 intermediate phase characterized by reduced entry capacity and less efficient coreceptor usage in vitro. Similar to the final X4 virus, the dual-tropic intermediate SHIV is sensitive to sCD4 and anti-V3 loop antibody neutralization. This, coupled with the finding that sCD4 neutralization sensitivity and CXCR4 use are regulated by similar mechanisms, supports a link between open Env configuration, increased CD4 binding, better CXCR4 use, and enhanced humoral immune recognition that merits further investigation. R5 SHIVSF162P3N infection of rhesus macaques, in which the precise timing, tissue sites, and cause for change in Env glycoprotein structure and coreceptor preference can be studied, provides a valid and invaluable animal model to further elucidate the reasons for coreceptor switching.
Acknowledgments
We thank Lisa Chakrabarti for critical and insightful comments on the manuscript, Zhiwei Chen for tree analysis, and Matt LaRoche for help with p24 antigen assays. AMD3100, TAK-799, TZM-bl cells, and the U87.CD4 indicator cell lines were obtained through the NIH AIDS Research and Reference Reagent Program.
This work was supported by National Institutes of Health grants RO1 AI46980 and R37AI41945.
Footnotes
Published ahead of print on 14 May 2008.
REFERENCES
- 1.Baik, S. S., R. W. Doms, and B. J. Doranz. 1999. HIV and SIV gp120 binding does not predict coreceptor function. Virology 259267-273. [DOI] [PubMed] [Google Scholar]
- 2.Berger, E. A., R. W. Doms, E. M. Fenyo, B. T. Korber, D. R. Littman, J. P. Moore, Q. J. Sattentau, H. Schuitemaker, J. Sodroski, and R. A. Weiss. 1998. A new classification for HIV-1. Nature 391240. [DOI] [PubMed] [Google Scholar]
- 3.Bjorndal, A., H. Deng, M. Jansson, J. R. Fiore, C. Colognesi, A. Karlsson, J. Albert, G. Scarlatti, D. R. Littman, and E. M. Fenyo. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 717478-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenchley, J. M., T. W. Schacker, L. E. Ruff, D. A. Price, J. H. Taylor, G. J. Beilman, P. L. Nguyen, A. Khoruts, M. Larson, A. T. Haase, and D. C. Douek. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brumme, Z. L., J. Goodrich, H. B. Mayer, C. J. Brumme, B. M. Henrick, B. Wynhoven, J. J. Asselin, P. K. Cheung, R. S. Hogg, J. S. Montaner, and P. R. Harrigan. 2005. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J. Infect. Dis. 192466-474. [DOI] [PubMed] [Google Scholar]
- 6.Bunnik, E. M., E. D. Quakkelaar, A. C. van Nuenen, B. Boeser-Nunnink, and H. Schuitemaker. 2007. Increased neutralization sensitivity of recently emerged CXCR4-using human immunodeficiency virus type 1 strains compared to coexisting CCR5-using variants from the same patient. J. Virol. 81525-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 2661024-1027. [DOI] [PubMed] [Google Scholar]
- 8.Cao, J., N. Sullivan, E. Desjardin, C. Parolin, J. Robinson, R. Wyatt, and J. Sodroski. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 719808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrillo, A., and L. Ratner. 1996. Cooperative effects of the human immunodeficiency virus type 1 envelope variable loops V1 and V3 in mediating infectivity for T cells. J. Virol. 701310-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casper, C., L. Naver, P. Clevestig, E. Belfrage, T. Leitner, J. Albert, S. Lindgren, C. Ottenblad, A. B. Bohlin, E. M. Fenyo, and A. Ehrnst. 2002. Coreceptor change appears after immune deficiency is established in children infected with different HIV-1 subtypes. AIDS Res. Hum. Retrovir. 18343-352. [DOI] [PubMed] [Google Scholar]
- 11.Cheng-Mayer, C., D. Seto, M. Tateno, and J. A. Levy. 1988. Biologic features of HIV-1 that correlate with virulence in the host. Science 24080-82. [DOI] [PubMed] [Google Scholar]
- 12.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cormier, E. G., and T. Dragic. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 768953-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delwart, E. L., and C. J. Gordon. 1997. Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods 12348-354. [DOI] [PubMed] [Google Scholar]
- 15.De Vreese, K., I. Van Nerum, K. Vermeire, J. Anne, and E. De Clercq. 1997. Sensitivity of human immunodeficiency virus to bicyclam derivatives is influenced by the three-dimensional structure of gp120. Antimicrob. Agents Chemother. 412616-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fouchier, R. A., M. Groenink, N. A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 663183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groenink, M., R. A. Fouchier, S. Broersen, C. H. Baker, M. Koot, A. B. van't Wout, H. G. Huisman, F. Miedema, M. Tersmette, and H. Schuitemaker. 1993. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science 2601513-1516. [DOI] [PubMed] [Google Scholar]
- 18.Guadalupe, M., E. Reay, S. Sankaran, T. Prindiville, J. Flamm, A. McNeil, and S. Dandekar. 2003. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J. Virol. 7711708-11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harouse, J. M., A. Gettie, R. C. Tan, J. Blanchard, and C. Cheng-Mayer. 1999. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science 284816-819. [DOI] [PubMed] [Google Scholar]
- 20.Hartley, O., P. J. Klasse, Q. J. Sattentau, and J. P. Moore. 2005. V3: HIV's switch-hitter. AIDS Res. Hum. Retrovir. 21171-189. [DOI] [PubMed] [Google Scholar]
- 21.Heckman, K. L., and L. R. Pease. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2924-932. [DOI] [PubMed] [Google Scholar]
- 22.Ho, S. H., L. Shek, A. Gettie, J. Blanchard, and C. Cheng-Mayer. 2005. V3 loop-determined coreceptor preference dictates the dynamics of CD4+-T-cell loss in simian-human immunodeficiency virus-infected macaques. J. Virol. 7912296-12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho, S. H., S. Tasca, L. Shek, A. Li, A. Gettie, J. Blanchard, D. Boden, and C. Cheng-Mayer. 2007. Coreceptor switch in R5-tropic simian/human immunodeficiency virus-infected macaques. J. Virol. 818621-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho, S. H., N. Trunova, A. Gettie, J. Blanchard, and C. Cheng-Mayer. 2 April 2008. Different mutational pathways to CXCR4 coreceptor switch of CCR5-using SHIV. J. Virol. doi: 10.1128/JVI.00145-08. (Subsequently published, J. Virol. 82:5653-5656, 2008.) [DOI] [PMC free article] [PubMed]
- 25.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 3101025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, W., S. H. Eshleman, J. Toma, S. Fransen, E. Stawiski, E. E. Paxinos, J. M. Whitcomb, A. M. Young, D. Donnell, F. Mmiro, P. Musoke, L. A. Guay, J. B. Jackson, N. T. Parkin, and C. J. Petropoulos. 2007. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J. Virol. 817885-7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang, S. S., T. J. Boyle, H. K. Lyerly, and B. R. Cullen. 1992. Identification of envelope V3 loop as the major determinant of CD4 neutralization sensitivity of HIV-1. Science 257535-537. [DOI] [PubMed] [Google Scholar]
- 28.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23403-405. [DOI] [PubMed] [Google Scholar]
- 29.Koito, A., G. Harrowe, J. A. Levy, and C. Cheng-Mayer. 1994. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J. Virol. 682253-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuiken, C. L., J. J. de Jong, E. Baan, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Evolution of the V3 envelope domain in proviral sequences and isolates of human immunodeficiency virus type 1 during transition of the viral biological phenotype. J. Virol. 664622-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuipers, H., C. Workman, W. Dyer, A. Geczy, J. Sullivan, and R. Oelrichs. 1999. An HIV-1-infected individual homozygous for the CCR-5 Δ32 allele and the SDF-1 3′A allele. AIDS 13433-434. [DOI] [PubMed] [Google Scholar]
- 32.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420678-682. [DOI] [PubMed] [Google Scholar]
- 33.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 7710557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, Y., M. A. Rey-Cuille, and S. L. Hu. 2001. N-linked glycosylation in the V3 region of HIV type 1 surface antigen modulates coreceptor usage in viral infection. AIDS Res. Hum. Retrovir. 171473-1479. [DOI] [PubMed] [Google Scholar]
- 36.Lusso, P., P. L. Earl, F. Sironi, F. Santoro, C. Ripamonti, G. Scarlatti, R. Longhi, E. A. Berger, and S. E. Burastero. 2005. Cryptic nature of a conserved, CD4-inducible V3 loop neutralization epitope in the native envelope glycoprotein oligomer of CCR5-restricted, but not CXCR4-using, primary human immunodeficiency virus type 1 strains. J. Virol. 796957-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 746769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malenbaum, S. E., D. Yang, L. Cavacini, M. Posner, J. Robinson, and C. Cheng-Mayer. 2000. The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J. Virol. 7411008-11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattapallil, J. J., Z. Smit-McBride, M. McChesney, and S. Dandekar. 1998. Intestinal intraepithelial lymphocytes are primed for gamma interferon and MIP-1β expression and display antiviral cytotoxic activity despite severe CD4+ T-cell depletion in primary simian immunodeficiency virus infection. J. Virol. 726421-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehandru, S., M. A. Poles, K. Tenner-Racz, A. Horowitz, A. Hurley, C. Hogan, D. Boden, P. Racz, and M. Markowitz. 2004. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J. Exp. Med. 200761-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melby, T., M. Despirito, R. Demasi, G. Heilek-Snyder, M. L. Greenberg, and N. Graham. 2006. HIV-1 coreceptor use in triple-class treatment-experienced patients: baseline prevalence, correlates, and relationship to enfuvirtide response. J. Infect. Dis. 194238-246. [DOI] [PubMed] [Google Scholar]
- 42.Michael, N. L., J. A. Nelson, V. N. KewalRamani, G. Chang, S. J. O'Brien, J. R. Mascola, B. Volsky, M. Louder, G. C. White, 2nd, D. R. Littman, R. Swanstrom, and T. R. O'Brien. 1998. Exclusive and persistent use of the entry coreceptor CXCR4 by human immunodeficiency virus type 1 from a subject homozygous for CCR5 Δ32. J. Virol. 726040-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore, J. P., S. G. Kitchen, P. Pugach, and J. A. Zack. 2004. The CCR5 and CXCR4 coreceptors—central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retrovir. 20111-126. [DOI] [PubMed] [Google Scholar]
- 44.Moore, J. P., M. Thali, B. A. Jameson, F. Vignaux, G. K. Lewis, S. W. Poon, M. Charles, M. S. Fung, B. Sun, and P. J. Durda. 1993. Immunochemical analysis of the gp120 surface glycoprotein of human immunodeficiency virus type 1: probing the structure of the C4 and V4 domains and the interaction of the C4 domain with the V3 loop. J. Virol. 674785-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moyle, G. J., A. Wildfire, S. Mandalia, H. Mayer, J. Goodrich, J. Whitcomb, and B. G. Gazzard. 2005. Epidemiology and predictive factors for chemokine receptor use in HIV-1 infection. J. Infect. Dis. 191866-872. [DOI] [PubMed] [Google Scholar]
- 46.Nelson, J., T. Riddle, N. Shire, M. Sherman, K. Franco, and H. Sheppard. 2007. Sequential turnover of env variants and co-receptor switching during HIV-1 chronic infection, abstr 252. Abstr. 14th Conf. Retrovir. Opportunistic Infect., Los Angeles, CA. Foundation for Retrovirology and Human Health, Alexandria, VA.
- 47.O'Brien, W. A., I. S. Chen, D. D. Ho, and E. S. Daar. 1992. Mapping genetic determinants for human immunodeficiency virus type 1 resistance to soluble CD4. J. Virol. 663125-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pastore, C., R. Nedellec, A. Ramos, O. Hartley, J. L. Miamidian, J. D. Reeves, and D. E. Mosier. 2007. Conserved changes in envelope function during human immunodeficiency virus type 1 coreceptor switching. J. Virol. 818165-8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pastore, C., R. Nedellec, A. Ramos, S. Pontow, L. Ratner, and D. E. Mosier. 2006. Human immunodeficiency virus type 1 coreceptor switching: V1/V2 gain-of-fitness mutations compensate for V3 loss-of-fitness mutations. J. Virol. 80750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pastore, C., A. Ramos, and D. E. Mosier. 2004. Intrinsic obstacles to human immunodeficiency virus type 1 coreceptor switching. J. Virol. 787565-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pollakis, G., S. Kang, A. Kliphuis, M. I. Chalaby, J. Goudsmit, and W. A. Paxton. 2001. N-linked glycosylation of the HIV type-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 coreceptor utilization. J. Biol. Chem. 27613433-13441. [DOI] [PubMed] [Google Scholar]
- 52.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 9916249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Regoes, R. R., and S. Bonhoeffer. 2005. The HIV coreceptor switch: a population dynamical perspective. Trends Microbiol. 13269-277. [DOI] [PubMed] [Google Scholar]
- 54.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retrovir. 16741-749. [DOI] [PubMed] [Google Scholar]
- 55.Salemi, M., B. R. Burkhardt, R. R. Gray, G. Ghaffari, J. W. Sleasman, and M. M. Goodenow. 2007. Phylodynamics of HIV-1 in lymphoid and non-lymphoid tissues reveals a central role for the thymus in emergence of CXCR4-using quasispecies. PLoS ONE 2e950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saunders, C. J., R. A. McCaffrey, I. Zharkikh, Z. Kraft, S. E. Malenbaum, B. Burke, C. Cheng-Mayer, and L. Stamatatos. 2005. The V1, V2, and V3 regions of the human immunodeficiency virus type 1 envelope differentially affect the viral phenotype in an isolate-dependent manner. J. Virol. 799069-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scarlatti, G., E. Tresoldi, A. Bjorndal, R. Fredriksson, C. Colognesi, H. K. Deng, M. S. Malnati, A. Plebani, A. G. Siccardi, D. R. Littman, E. M. Fenyo, and P. Lusso. 1997. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat. Med. 31259-1265. [DOI] [PubMed] [Google Scholar]
- 58.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 661354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simmons, G., J. D. Reeves, S. Hibbitts, J. T. Stine, P. W. Gray, A. E. Proudfoot, and P. R. Clapham. 2000. Co-receptor use by HIV and inhibition of HIV infection by chemokine receptor ligands. Immunol. Rev. 177112-126. [DOI] [PubMed] [Google Scholar]
- 60.Simmons, G., D. Wilkinson, J. D. Reeves, M. T. Dittmar, S. Beddows, J. Weber, G. Carnegie, U. Desselberger, P. W. Gray, R. A. Weiss, and P. R. Clapham. 1996. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J. Virol. 708355-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh, A., and R. G. Collman. 2000. Heterogeneous spectrum of coreceptor usage among variants within a dualtropic human immunodeficiency virus type 1 primary-isolate quasispecies. J. Virol. 7410229-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stalmeijer, E. H., R. P. Van Rij, B. Boeser-Nunnink, J. A. Visser, M. A. Naarding, D. Schols, and H. Schuitemaker. 2004. In vivo evolution of X4 human immunodeficiency virus type 1 variants in the natural course of infection coincides with decreasing sensitivity to CXCR4 antagonists. J. Virol. 782722-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suphaphiphat, P., M. Essex, and T. H. Lee. 2007. Mutations in the V3 stem versus the V3 crown and C4 region have different effects on the binding and fusion steps of human immunodeficiency virus type 1 gp120 interaction with the CCR5 coreceptor. Virology 360182-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trkola, A., H. Kuster, C. Leemann, C. Ruprecht, B. Joos, A. Telenti, B. Hirschel, R. Weber, S. Bonhoeffer, and H. F. Gunthard. 2003. Human immunodeficiency virus type 1 fitness is a determining factor in viral rebound and set point in chronic infection. J. Virol. 7713146-13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Rij, R. P., M. D. Hazenberg, B. H. van Benthem, S. A. Otto, M. Prins, F. Miedema, and H. Schuitemaker. 2003. Early viral load and CD4+ T cell count, but not percentage of CCR5+ or CXCR4+ CD4+ T cells, are associated with R5-to-X4 HIV type 1 virus evolution. AIDS Res. Hum. Retroviruses 19389-398. [DOI] [PubMed] [Google Scholar]
- 66.van't Wout, A. B., N. A. Kootstra, G. A. Mulder-Kampinga, N. Albrecht-van Lent, H. J. Scherpbier, J. Veenstra, K. Boer, R. A. Coutinho, F. Miedema, and H. Schuitemaker. 1994. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J. Clin. Investig. 942060-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van't Wout, A. B., H. Blaak, L. J. Ran, M. Brouwer, C. Kuiken, and H. Schuitemaker. 1998. Evolution of syncytium-inducing and non-syncytium-inducing biological virus clones in relation to replication kinetics during the course of human immunodeficiency virus type 1 infection. J. Virol. 725099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Veazey, R. S., M. DeMaria, L. V. Chalifoux, D. E. Shvetz, D. R. Pauley, H. L. Knight, M. Rosenzweig, R. P. Johnson, R. C. Desrosiers, and A. A. Lackner. 1998. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280427-431. [DOI] [PubMed] [Google Scholar]
- 69.Veazey, R. S., K. G. Mansfield, I. C. Tham, A. C. Carville, D. E. Shvetz, A. E. Forand, and A. A. Lackner. 2000. Dynamics of CCR5 expression by CD4+ T cells in lymphoid tissues during simian immunodeficiency virus infection. J. Virol. 7411001-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilkin, T. J., Z. Su, D. R. Kuritzkes, M. Hughes, C. Flexner, R. Gross, E. Coakley, W. Greaves, C. Godfrey, P. R. Skolnik, J. Timpone, B. Rodriguez, and R. M. Gulick. 2007. HIV type 1 chemokine coreceptor use among antiretroviral-experienced patients screened for a clinical trial of a CCR5 inhibitor: AIDS Clinical Trial Group A5211. Clin. Infect. Dis. 44591-595. [DOI] [PubMed] [Google Scholar]
- 71.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393705-711. [DOI] [PubMed] [Google Scholar]
- 72.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 695723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 2801884-1888. [DOI] [PubMed] [Google Scholar]
- 74.Wyatt, R., M. Thali, S. Tilley, A. Pinter, M. Posner, D. Ho, J. Robinson, and J. Sodroski. 1992. Relationship of the human immunodeficiency virus type 1 gp120 third variable loop to a component of the CD4 binding site in the fourth conserved region. J. Virol. 666997-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye, Y., Z. H. Si, J. P. Moore, and J. Sodroski. 2000. Association of structural changes in the V2 and V3 loops of the gp120 envelope glycoprotein with acquisition of neutralization resistance in a simian-human immunodeficiency virus passaged in vivo. J. Virol. 7411955-11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yi, Y., F. Shaheen, and R. G. Collman. 2005. Preferential use of CXCR4 by R5X4 human immunodeficiency virus type 1 isolates for infection of primary lymphocytes. J. Virol. 791480-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou, T., L. Xu, B. Dey, A. J. Hessell, D. Van Ryk, S. H. Xiang, X. Yang, M. Y. Zhang, M. B. Zwick, J. Arthos, D. R. Burton, D. S. Dimitrov, J. Sodroski, R. Wyatt, G. J. Nabel, and P. D. Kwong. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]