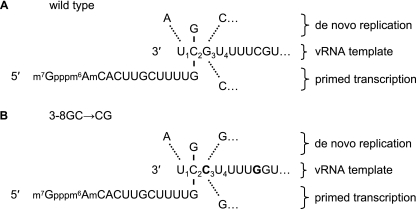Abstract
The mechanisms regulating the synthesis of mRNA, cRNA, and viral genomic RNA (vRNA) by the influenza A virus RNA-dependent RNA polymerase are not fully understood. Previous studies in our laboratory have shown that virion-derived viral ribonucleoprotein complexes synthesize both mRNA and cRNA in vitro and early in the infection cycle in vivo. Our continued studies showed that de novo synthesis of cRNA in vitro is more sensitive to the concentrations of ATP, CTP, and GTP than capped-primer-dependent synthesis of mRNA. Using rescued recombinant influenza A/WSN/33 viruses, we now demonstrate that the 3′-terminal sequence of the vRNA promoter dictates the requirement for a high nucleoside triphosphate (NTP) concentration during de novo-initiated replication to cRNA, whereas this is not the case for the extension of capped primers during transcription to mRNA. In contrast to some other viral polymerases, for which only the initiating NTP is required at high concentrations, influenza virus polymerase requires high concentrations of the first three NTPs. In addition, we show that base pair mutations in the vRNA promoter can lead to nontemplated dead-end mutations during replication to cRNA in vivo. Based on our observations, we propose a new model for the de novo initiation of influenza virus replication.
The negative-sense RNA genome of influenza A virus comprises eight segments. Each viral genomic RNA (vRNA) segment is packaged as a ribonucleoprotein (vRNP), with a trimeric RNA polymerase complex bound to the promoter, and one nucleoprotein (NP) molecule positioned along every 24 nucleotides of RNA (5, 24). The minimal vRNA promoter comprises the 13 5′-terminal nucleotides and the 12 3′-terminal nucleotides of each segment, which are base paired into a so-called “corkscrew-like” structure (8). The RNA segments are transcribed (vRNA → mRNA) and replicated (vRNA → cRNA → vRNA) by the polymerase complex, employing two distinct mechanisms. Replication is initiated de novo with a 5′-terminal triphosphate, whereas mRNA transcription is initiated by a capped primer that is endonucleolytically cleaved from cellular mRNAs by the polymerase complex. Transcription terminates by polyadenylation at a sequence of 5 to 7 U residues 16 to 17 nucleotides from the 5′ end of the vRNA template. The current model for polyadenylation suggests that premature termination and stuttering are caused by steric hindrance of the transcribing polymerase bound to the 5′ end of the vRNA template (reviewed in reference 9). On the other hand, replication is terminated by runoff transcription of the vRNA template to form full-length cRNA. The cRNA, the promoter of which possesses a similar “corkscrew-like” structure, is replicated to vRNA, also by de novo initiation. However, in this case, de novo initiation occurs internally on template positions 4 and 5, yielding a dinucleotide which is then realigned as a terminal primer (7).
It is not clear how transcription and replication are regulated. A previously widely accepted model suggested that replication requires free or soluble NP, i.e., NP that is not associated with RNPs, but the mechanism remained unclear (2, 21, 27). In this model, transcription occurs until there is sufficient soluble NP to switch the polymerase from transcription to replication. Therefore, transcription and replication would be in a dynamic balance controlled by the concentration of free NP. However, we have shown that both transcription (vRNA → mRNA) and replication (vRNA → cRNA) of virion-derived vRNPs occur in the absence of free NP. We proposed that differential accumulation of mRNA and cRNA in vivo is dependent on their stability (32, 33). Whereas mRNA is protected from degradation by the presence of the 5′ cap structure and the 3′-terminal poly(A) tail, newly transcribed cRNA is degraded by host cell nucleases, unless it is bound by newly synthesized polymerase and NP to form complementary RNPs (cRNPs). According to this model, transcription and replication, being dependent essentially on the availability of components for the formation of an active initiation complex, are regulated stochastically.
Extensive research has been carried out to determine the mechanism of elongation of the capped primer on the vRNA template. It has been found that host mRNAs are cleaved about 9 to 15 nucleotides from their 5′ caps, usually after a purine residue, with a preference for fragments terminating with CA (1, 28). In most cases, transcription is initiated by the addition of a G residue directed by the penultimate C at the 3′ end of the vRNA, although in some cases initiation occurs by the incorporation of a C residue directed by the G at position 3 of the vRNA template (12, 14).
In contrast, influenza virus replication occurs by de novo initiation, the details of which are not well understood. The synthesis of short RNA oligonucleotides by successive rounds of abortive de novo transcription by RNA-dependent RNA polymerases has been reported for a number of viruses, including brome mosaic virus (30), turnip crinkle carmovirus (22), reovirus (34), and rotavirus (4). We have previously shown that influenza virus RNA polymerase similarly synthesizes abortive transcripts in vitro, templated by the 3′-terminal residues of a model vRNA promoter (7).
The Kms of the replicases of a number of viruses are higher (weaker affinity) for the initiation nucleoside triphosphates (NTPi), which are usually either GTP or ATP, than they are for the NTPs used in elongation (16, 19, 31). Our previous data (32) confirmed that influenza virus polymerase requires a high concentration (>100 μM) of the NTPi (ATP) for de novo initiation of replication. We also showed that reduced concentrations of ATP, CTP, and GTP (the first three nucleotides to be incorporated during vRNA-templated polymerization), but not UTP, selectively inhibit replication to cRNA without affecting mRNA transcription. We speculated that this difference was related to the initiation mechanisms employed by the polymerase, namely, de novo initiation during replication as opposed to extension of a primer during transcription.
In order to test this hypothesis here, we rescued a recombinant virus with a mutant promoter sequence in the neuraminidase (NA) gene, such that the incorporation of CTP during vRNA-templated polymerization was delayed. We demonstrate that in vitro, replication of the mutant gene is insensitive, relative to that of the wild-type gene to the concentration of CTP. In addition, analysis of the genetic effects of the mutation during in vivo replication revealed that nontemplated “dead-end” mutations can arise during replication of the mutant promoter vRNA in vivo. Based on our observations, we propose a new mechanism for de novo initiation of influenza virus replication.
MATERIALS AND METHODS
Strains and culture conditions.
Influenza A/WSN/33 virus stocks were prepared and titrated in MDBK cell monolayers maintained in minimal essential medium with 10% fetal calf serum at 37°C and with 5% CO2. Human embryonic kidney (293T) cells were maintained under similar culture conditions.
Plasmids.
The protein expression plasmids pcDNA-PB1, pcDNA-PB1-D445A/D446A, pcDNA-PB2, pcDNA-PA, and pcDNA-NP have been described previously (10, 33). The pPOLI-PB1-RT, pPOLI-PB2-RT, pPOLI-PA-RT, pPOLI-HA-RT, pPOLI-NP-RT, pPOLI-NA-RT, pPOLI-M-RT, and pPOLI-NS-RT RNA transcription plasmids have also been described (11). The pPOLI-cNA-RT plasmid was generated by Ryu Yoshida by using PCR amplification and cloning. The GC→AU, GC→CG, and GC→UA base pair mutations at positions 3 and 8 of NA (referred to herein as the 3-8 promoter mutant NA vRNA) and the A→G mutation at position 1′ and the CG→GC base pair mutation at positions 3′ and 8′ of NA cRNA were engineered into pPOLI-NA-RT or pPOLI-cNA-RT by site-directed PCR mutagenesis using appropriate primers and were confirmed by sequencing.
In vivo vRNP reconstitution assays.
Human embryonic kidney (293T) cells in 35-mm dishes were transfected in suspension with 1 μg each of pcDNA-PB2, pcDNA-PA, pcDNA-NP, and pcDNA-PB1 or pcDNA-PB1-D445A/D446A (as a negative control), together with 1 μg of pPOLI-NA-RT (wild type or mutant) or pPOLI-cNA-RT (wild type or mutant) and 5 μl of Lipofectamine 2000 reagent (Invitrogen) in 1.14 ml of minimal essential medium containing 10% fetal calf serum. Total cell RNA was extracted 24 to 48 h posttransfection using Tri reagent (Ambion).
Primer extension analysis.
One twentieth of the RNA isolated from a 35-mm dish was analyzed by primer extension as described previously (32). A primer detecting host 5S rRNA was included as an internal control where required (15). The sequences of the primers are available on request.
Generation and amplification of recombinant viruses.
The plasmid-based rescue technique (11, 23) was used to generate recombinant influenza A/WSN/33 viruses, incorporating point mutations into the NA gene segment. Briefly, the pPOLI-NA-RT transcription plasmid encoding wild-type or mutant NA vRNAs, seven pPOLI transcription plasmids encoding the other seven viral gene segments, and four protein expression plasmids encoding PB1, PB2, PA, and NP (1 μg of each plasmid) were transfected into 293T cells in suspension in 35-mm dishes, using either 12 μl Lipofectamine 2000 or 18 μl Fugene HD (Roche) according to the manufacturers' instructions. Virus collected 48 h posttransfection was amplified for 2 to 3 days on MDBK cells. Following plaque purification on MDBK cells, stocks of passage-2 viruses were prepared and titrated. Plaques were visualized by staining with a Coomassie brilliant blue solution (0.25% Coomassie brilliant blue, 50% methanol, 10% acetic acid).
NA assay.
NA activity was determined essentially as described by Rowley et al. (26). Briefly, 293T cells in a 35-mm dish were infected with a recombinant virus containing either wild-type NA vRNA or the promoter mutant NA vRNA with mutations at positions 3-8 at a multiplicity of infection (MOI) of 2. The cells were harvested 4 h postinfection, resuspended in 100 μl lysis buffer (250 mM Tris-HCl [pH 7.4], 1 M KCl, 250 mM CaCl2, 2% Triton X-100), vortexed, and then sonicated. Ten microliters of the lysate was diluted in 0.1 M KPO4, pH 5.9, with 0.5 mM 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (4-MUNANA; Sigma) substrate in black, low-fluorescence, U-bottomed microtiter plates in a total volume of 100 μl. Plates were incubated in a water bath at 37°C for 1 h, and reactions were stopped by the addition of 150 μl 0.1 M glycine, pH 10.7, containing 25% ethanol. Fluorescence was read with 355-nm excitation and 460-nm emission wavelengths.
Preparation of virion-derived vRNPs.
Virion-derived vRNPs of recombinant viruses carrying 3-8 promoter mutant NA vRNA were prepared essentially as described previously (32), with an additional purification step for the virions prior to lysis. This involved centrifuging the virion pellet from the sucrose cushion through a step gradient of 2.5 ml each of 30%, 40%, 50%, and 60% sucrose in SW41 tubes at 35,000 rpm for 2.5 h at 4°C. A virion-enriched fraction (visible as the lower of two light-blue bands) was extracted with a 25-gauge needle, and the virions were pelleted by centrifugation at 30,000 rpm for 1 h at 4°C for lysis as described previously (32).
In vitro virion-derived vRNP polymerase reactions.
In vitro replication and transcription assays of 3-8 promoter mutant NA gene-specific virion-derived vRNPs were carried out and analyzed by primer extension as described previously (32).
Terminal sequencing of vRNAs.
The 5′ and 3′ termini of influenza virus RNA were sequenced as described previously (32).
RESULTS
Rescue of influenza viruses with alternative 3′-terminal promoter sequences.
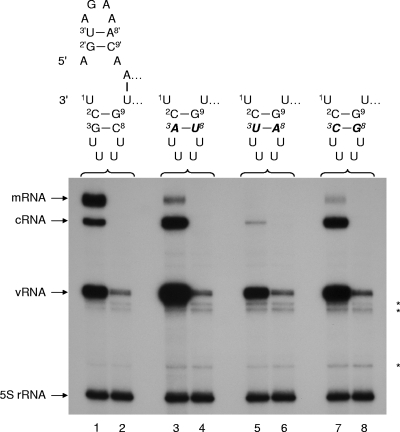
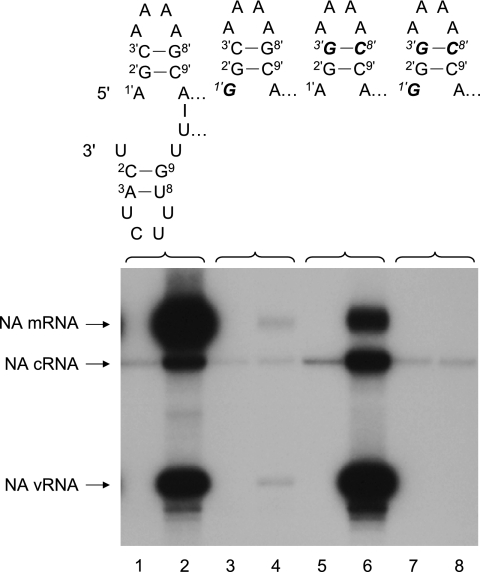
We have previously speculated that there is a correlation between 3′-terminal template sequences and a requirement for high NTP concentrations during in vitro, de novo-initiated replication with virion-derived vRNPs (32). To test this hypothesis, it was necessary to generate mutant vRNA templates with substitutions in the 3′-terminal position 1, 2, or 3 (Fig. 1) that are suitable for the rescue of recombinant viruses. Crow et al. (6) previously demonstrated with a chloramphenicol acetyltransferase reporter gene that single point mutations at positions 1′, 2′, or 3′ at the 5′ end of the cRNA promoter (which corresponds to the 3′ end of the vRNA promoter) renders the promoter inactive. However, promoters with base pair substitutions at positions 3′ and 8′ retained at least partial functionality. We therefore generated pPOLI-NA-RT mutants to direct in vivo synthesis of NA-specific vRNA with substitutions of the GC base pairs at positions 3 and 8 in the 3′ end of the promoter with AU, CG, or UA (referred to as 3-8GC→AU, 3-8GC→CG, and 3-8GC→UA, respectively) (Fig. 1). Primer extension assays were performed to analyze the activities of vRNPs reconstituted with either wild-type or mutant NA-specific vRNA in transiently transfected 293T cells (Fig. 1). Substitution of the GC base pair with AU or CG at positions 3 and 8 in the 3′ end of the vRNA promoter resulted in increased vRNA and cRNA levels but substantially inhibited mRNA levels compared to those of the wild type (Fig. 1, compare lanes 1, 3, and 7). On the other hand, both replication and transcription from the vRNA promoter were considerably inhibited by the 3-8GC→UA substitution (Fig. 1, compare lanes 1 and 5). Therefore, the results obtained for base pair substitutions at positions 3 and 8 in the 3′ end of the NA vRNA promoter confirmed previous results for the corresponding base pair substitution at positions 3′ and 8′ in the 5′ end of a chloramphenicol acetyltransferase reporter cRNA promoter (6).
FIG. 1.
Effects of base pair substitutions at positions 3 and 8 in the 3′-terminal hairpin loop of the vRNA promoter on polymerase activity. vRNPs were reconstituted in vivo by transfection of a plasmid transcribing wild-type or mutant NA vRNA together with plasmids expressing PA, PB2, NP, and either wild-type PB1 (odd-numbered lanes) or PB1 with a DD→AA double mutation in the active site (even-numbered lanes show plasmid-derived input vRNA). RNAs were analyzed at 31 h posttransfection by NA gene-specific primer extension (see Materials and Methods). The sequences and proposed corkscrew structures (8) of the wild-type and mutant vRNA promoters are shown above the gel. The bold, italic letters indicate the mutant residues. The positions of the vRNA, cRNA, and mRNA signals are shown to the left of the gel. The 5S rRNA signal was used as an internal control. Asterisks indicate background products synthesized from the 5S rRNA-specific primer.
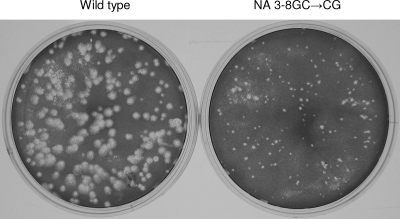
Next, we attempted to rescue recombinant viruses containing seven genes with wild-type promoters and an NA gene with either the wild-type promoter or a mutant (3-8GC→AU, 3-8GC→CG, or 3-8GC→UA) promoter. Following five attempts during which viruses with the wild-type NA gene were rescued every time, only a recombinant virus with the 3-8GC→CG base pair substitution in the NA gene promoter could be rescued (at the first attempt). Following plaque purification, stocks of the rescued wild-type and NA 3-8GC→CG mutant viruses were prepared in MDBK cells and titrated. Titers of NA 3-8GC→CG mutant viral stocks were 1 × 107 PFU/ml, while wild-type viral stock titers were 5 × 107 PFU/ml. As can be seen in Fig. 2, NA 3-8GC→CG mutant viruses produced smaller plaques than the wild-type virus, presumably due to reduced mRNA transcription of the mutant gene (Fig. 1) and, hence, reduced NA expression and activity. The finding of reduced activity was confirmed by determining the NA activities of infected cell lysates harvested at 4 h postinfection (see Materials and Methods). NA 3-8GC→CG mutant virus-infected cell lysates yielded approximately 8.5-fold less NA activity (average of two measurements) than lysates of cells infected with the wild-type virus at the same MOI.
FIG. 2.
Visualization of plaques generated by the infection of MDBK cells with influenza A/WSN/33 viruses carrying either eight wild-type genome segments (left) or seven wild-type genome segments and an NA genome segment with a 3-8GC→CG promoter mutation (right). Infected cell monolayers were stained with Coomassie blue.
Effect of the NA 3-8GC→CG mutations on the NTP concentration requirement for replication and transcription.
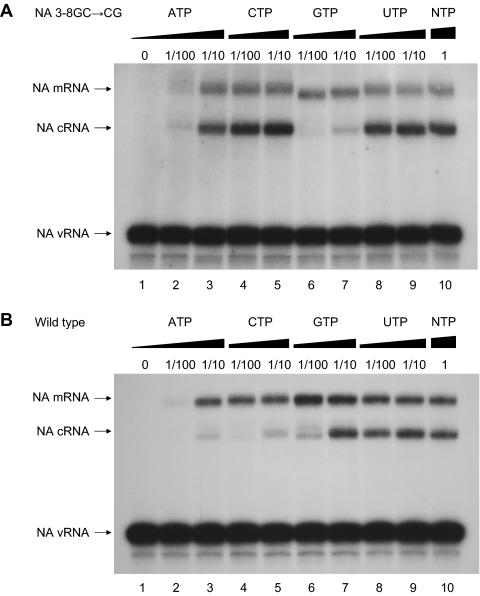
We have previously suggested that the viral polymerase requires a higher concentration of the initial NTPs (ATP, CTP, and GTP) incorporated during de novo replication than extension of a capped primer during mRNA transcription in order to overcome abortive initiation (32). This mechanism predicts that replication of an NA gene containing a 3-8GC→CG base pair substitution in the promoter should be insensitive to the concentration of CTP relative to replication of the wild-type NA gene, as the first C residue incorporated during replication of the mutant NA gene is at position 8′ (AGGAAAACC) instead of position 3′ in wild-type NA cRNA (AGCAAAAGC). The NTP concentration requirements for replication and transcription of the mutant NA gene were determined by in vitro polymerase activity assays of vRNPs derived from the rescued NA 3-8GC→CG virus, followed by primer extension (see Materials and Methods) (32). It should be noted that this assay specifically analyzes the synthesis of mRNA and cRNA from the input vRNP-associated vRNA template and not the replication of cRNA to vRNA (32). Figure 3 shows that the cRNA replication product of the NA gene with a 3-8GC→CG base pair promoter mutation was not reduced even with a 100-fold dilution of CTP (Fig. 3A, compare lanes 4 and 5 with lane 10), in contrast with a marked reduction in the cRNA replication product of the wild-type NA gene (Fig. 3B, compare lanes 4 and 5 with lane 10). In contrast, replication of the 3-8GC→CG base pair promoter mutant NA gene to cRNA was substantially inhibited by reduced concentrations of GTP (Fig. 3A, compare lanes 7 and 10) compared to the replication of the wild-type gene (Fig. 3B, compare lanes 7 and 10), consistent with the early requirement for GTP at positions 2′ and 3′ in the mutant cRNA (AGGAAAACC) instead of at positions 2′ and 8′ in the wild-type cRNA (AGCAAAAGC). Intriguingly, replication of the 3-8GC→CG promoter mutant NA gene to cRNA was less severely inhibited by a 10-fold dilution of ATP (Fig. 3A, compare lanes 3 and 10) than was replication of the wild-type gene (Fig. 3B, compare lanes 3 and 10) (see Discussion).
FIG. 3.
Titration of NTP concentration for in vitro transcription and replication by vRNPs derived from virions carrying either eight wild-type genome segments (A) (reproduced from reference 32 for comparison) or seven wild-type genome segments and an NA genome segment with a 3-8GC→CG promoter mutation (B). vRNPs were incubated in the presence of 18 ng/μl globin mRNA with various concentrations of NTPs. RNAs were analyzed by NA gene-specific primer extension. Lane 10 in each gel represents in vitro transcription under standard conditions (1 mM ATP and 0.5 mM of each CTP, GTP, and UTP). The dilutions of individual NTPs relative to the standard conditions are indicated above each gel. The positions of the vRNA, cRNA, and mRNA signals are shown to the left of each gel.
As expected (Fig. 1), in vitro mRNA transcription of the promoter mutant NA gene was inhibited compared to that of the wild type in the presence of the same concentrations of globin mRNA (compare Fig. 3A and B, lanes 10) but required NTP concentrations similar to those required by the wild type (compare Fig. 3A and B, lanes 2 to 10). Surprisingly, however, limiting concentrations of GTP resulted in the synthesis of shorter mRNA transcripts from the NA 3-8GC→CG promoter mutant template when globin mRNA was used as a source of capped primers (Fig. 3A, compare lanes 6 and 7 with lane 10). The transcripts were estimated to be, on average, 1 or 2 residues shorter at their 5′ ends based on the mobility shift of the primer extension products. α- and β-globin mRNA have been shown to be endonucleolytically cleaved predominantly after G10 and G13, respectively, by influenza A virus RNA polymerase and the resultant capped primer to be extended by incorporation of a C residue directed by the G at position 3 from the 3′ end of a wild-type vRNA template (25). We speculate that limiting concentrations of GTP promote alternative downstream positioning of globin mRNA-derived capped primers terminating with a G residue on the C residue at position 3 of NA 3-8GC→CG promoter mutant templates. This alternative would allow the capped primer to be extended by the incorporation of an A residue (templated by U at position 4) instead of a G residue (templated by C at position 3) and would yield α- and β-globin mRNA-primed transcripts which are 1 residue shorter (Fig. 3A, lanes 6 and 7).
Overall, it appears that, in vitro, the polymerase is sensitive to the concentrations of the first three NTPs incorporated into cRNA during de novo replication, but less so during globin mRNA-primed transcription (32).
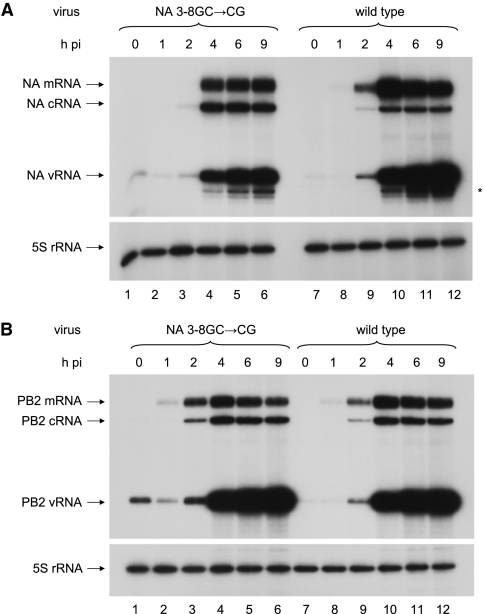
Effect of the NA 3-8GC→CG promoter mutation on the accumulation levels of vRNA, cRNA, and mRNA in virus-infected cells.
In order to study the effects of the NA 3-8GC→CG promoter mutation in the context of a viral infection, we carried out time course analyses of 293T cells infected, at equal MOIs, with viruses containing NA genes with either wild-type or NA 3-8GC→CG mutant promoters. The accumulation of vRNA, cRNA, and mRNA in infected cells at different times postinfection was analyzed by NA gene-specific primer extension assays (Fig. 4A), using PB2 and NP gene-specific analyses as controls (Fig. 4B; NP data not shown). The accumulation of 3-8GC→CG promoter mutant NA gene-specific vRNA, cRNA, and mRNA differed markedly from that of wild-type NA gene-specific RNA (Fig. 4A, compare lanes 1 to 6 with lanes 7 to 12). The 3-8GC→CG promoter mutation in the NA gene inhibited mRNA transcription relative to that of the wild type, consistent with the reduced NA activity seen in cell lysates infected with the mutant virus compared with those infected with the wild-type virus, as described above. Intriguingly, the kinetics of mutant mRNA accumulation, which reached a plateau at 4 h postinfection, differed from that of wild-type mRNA accumulation, which reached a peak at 4 h postinfection. In addition, the level of cRNA derived from the NA 3-8GC→CG promoter mutant vRNA was increased relative to that of the wild type. On the other hand, NA 3-8GC→CG promoter mutant vRNA accumulation was reduced, probably contributing to the reduced NA 3-8GC→CG mutant promoter viral titers relative to the titers of the wild-type viruses (see above). In contrast, as expected, the accumulation levels of PB2 and NP gene-specific RNA were similar in cells infected with a virus containing the wild-type or 3-8GC→CG promoter mutant NA gene (Fig. 4B, compare lanes 1 to 6 with lanes 7 to 12; also NP data not shown).
FIG. 4.
Analysis of RNA accumulation during a time course of infection with influenza A/WSN/33 viruses carrying either eight wild-type genome segments (lanes 7 to 12) or seven wild-type genome segments and an NA genome segment with a 3-8GC→CG promoter mutation (lanes 1 to 6). RNA harvested at different hours postinfection (h pi), as indicated, were analyzed by primer extension by using either NA (A) or PB2 (B) gene-specific primers. The positions of the vRNA, cRNA and mRNA signals are shown at the left of both panels. The 5S rRNA signal was used as an internal control. The asterisk indicates minor products synthesized by alternative NA vRNA-specific priming.
It was noted that, despite infection at equal MOIs, increased levels of input vRNA (at 0 h postinfection) of all three genes analyzed were present for the NA 3-8GC→CG promoter mutant virus, relative to the wild-type virus (Fig. 4A and B, compare lanes 1 and 7), indicating that an increased amount of the NA 3-8GC→CG promoter mutant virus relative to the wild-type virus was required to achieve the same plaque-forming titer. Therefore, either the titer of the NA 3-8GC→CG promoter mutant virus was underestimated due to the formation of undetectable plaques or the ratio of infectious to noninfectious particles for the NA 3-8GC→CG promoter mutant virus may be lower than it is for wild-type virus. A reduced infectivity of the NA 3-8GC→CG promoter mutant virus may be due to the reduced NA expression and activity inhibiting virus entry into the cell (20) or to impaired packaging of the 3-8GC→CG promoter mutant NA gene segments.
Overall, the 3-8GC→CG promoter mutation affects the regulation of vRNA, cRNA, and mRNA accumulation in infected cells.
Nontemplated mutagenesis of cRNA during replication of NA 3-8GC→CG promoter mutant vRNA in infected 293T cells.
Due to the possibility of the selection of reversions or compensatory mutations during viral rescue, we sequenced the termini of NA gene-specific RNAs harvested from cells infected with the wild-type or NA 3-8GC→CG promoter mutant virus. Reverse transcription-PCR products of RNA harvested 6 h postinfection (Fig. 4A, lanes 4 and 5), obtained by RLM-RACE, were cloned and sequenced (32). The presence of the base pair mutation was confirmed in all NA gene-specific vRNA, cRNA, and mRNA obtained from NA 3-8GC→CG promoter mutant virus-infected cells (Table 1). 5′-Terminal sequences of 9 to 12 nucleotides which are derived from host cellular mRNA were identified in both wild-type and mutant NA gene-specific mRNA. However, two unexpected differences between the mutant and wild-type NA gene-specific RNA were observed. First, in contrast to the 3′-terminal degradation of the wild-type vRNA that we previously observed (32) and have confirmed here (Table 1; 0 out of 9 clones were full length), the 3′ end of the 3-8GC→CG promoter mutant vRNA was almost always entirely intact (9 out of 10 clones were full length) (Table 1). Second, 9 out of 16 of the 3′-8′CG→GC promoter mutant cRNA-specific clones possessed an additional nontemplated 5′-terminal 1′A→G mutation (Table 1). As no vRNA with a 3′-terminal 1U→C mutation was detected (Table 1) and on the basis of previous work by Crow et al. (6), this suggested that the 1′A→G mutant cRNA may be inactive as a template for further vRNA synthesis.
TABLE 1.
Terminal sequence analysis of RNAs synthesized during infection with influenza virus carrying either eight wild-type genome segments or seven wild-type genome segments and a mutant promoter NA genome segment
| RNA speciesa | Genotypeb | 5′- and 3′-terminal sequencec | No. of clonesd | ||
|---|---|---|---|---|---|
| vRNA | Wild type | AGTAGAAACAAGGAGTTT | ........................ | TAAACTCCTGCTTTTGC- | 4 |
| AGTAGAAACAAGGAGTTT | ........................ | TAAACTCCTGCTTTTG-- | 4 | ||
| AGTAGAAACAAGGAGTTT | ........................ | TAAACTCCTGCTTTT--- | 1 | ||
| 3-8GC→CG | AGTAGAAACAAGGAGTTT | ........................ | TAAACTCCTGGTTTTCCT | 9 | |
| AGTAGAAACAAGGAGTTT | ........................ | TAAACTCCTGGTTTTCC- | 1 | ||
| cRNA | Wild type | AGCAAAAGCAGGAGTTTA | ........................ | AAACTCCTTGTTTCTACT | 7 |
| AGCAAAAGCAGGAGTTTA | ........................ | AAACTCCTTGTTTCTAC- | 2 | ||
| AGCAAAAGCAGGAGTTTA | ........................ | AAACTCCTTGTTTCTA-- | 1 | ||
| 3-8GC→CG | AGGAAAACCAGGAGTTTA | ........................ | AAACTCCTTGTTTCTACT | 6 | |
| AGGAAAACCAGGAGTTTA | ........................ | AAACTCCTTGTTTCTA-- | 1 | ||
| GGGAAAACCAGGAGTTTA | ........................ | AAACTCCTTGTTTCTACT | 3 | ||
| GGGAAAACCAGGAGTTTA | ........................ | AAACTCCTTGTTTCTAC- | 4 | ||
| GGGGAAACCAGGAGTTTA | ........................ | AAACTCCTTGTTTC---- | 1 | ||
| GGGAAAACCAGGAGTTTA | ........................ | AAACCCCTTGTTT----- | 1 | ||
| mRNA | Wild type | GAAGCGCTTCCGCAAAAGCAGGAGT | ........................ | TCACCATTGACAAGTAGTTTGTTCA30 | 1 |
| ACCAGCAGAAGCAAAAGCAGGAGT | ........................ | TCACCATTGACAGGTAGTTTGT---- | 1 | ||
| CAACAAGATGGCAAAAGCAGGAGT | ........................ | TCACCAT------------------- | 1 | ||
| ATTTACAGCTCAGCAAAAGCAGGAGT | ........................ | TCA----------------------- | 1 | ||
| 3-8GC→CG | TTTTGCTGTGGAAAACCAGGAGT | ........................ | TCACCATTGACAAGTAGTTTGTTCA26 | 1 | |
| GAAAAGATGGGGAAAACCAGGAGT | ........................ | TCACCATTGACAAGTAGTTTGTTCA25 | 1 | ||
| CCAGCCGAGCGGGAAAACCAGGAGT | ........................ | TCACCATTGACAAGTAGTTTGT---- | 1 | ||
| AGATGGCGGGAGGAAAACCAGGAGT | ........................ | TCA----------------------- | 1 | ||
Viral RNA species analyzed.
Genotype of NA gene in infecting virus.
Terminal sequences of NA gene-specific RNAs isolated from infected cells. Sequences are shown 5′ to 3′. Templated base-pair substitutions are underlined. Nontemplated substitutions or deletions are in boldface type. Sequences derived from host cellular mRNA are italicized. mRNA transcripts were prematurely terminated before the poly(A) site or polyadenylated.
The number of clones identified with the corresponding sequences.
To test this hypothesis, pPOLI-cNA-RT mutant plasmids, which direct in vivo synthesis of NA-specific cRNA with substitutions of a G residue for the 5′-terminal A residue and/or of GC for the CG base pair at positions 3′ and 8′ in the 5′ end of the promoter, were generated (see Materials and Methods). Following in vivo reconstitution of cRNPs by transient-transfection and primer extension analyses of NA gene-specific RNA (Fig. 5), we found that a 1′A→G substitution alone considerably inhibits cRNA from acting as a template for vRNA synthesis (Fig. 5, compare lanes 2 and 4). In combination with a 3′-8′CG→GC base pair substitution, a 1′→G substitution prevents cRNA from acting as a template for vRNA synthesis (Fig 5, compare lanes 2 and 8). As a control, the 3′-8′CG→GC base pair substitution in cRNA compared to the wild type yields a phenotype similar to that produced by the 3-8GC→CG substitution in vRNA compared to the wild type (compare Fig. 5, lanes 2 and 6, with Fig. 1, lanes 1 and 7). Therefore, the introduction of a nontemplated 5′-terminal 1′A→G mutation in cRNA replicated from 3-8GC→CG promoter mutant vRNA renders the cRNA inactive for further cycling to vRNA.
FIG. 5.
Effects of 1′A→G and 3′-8′CG→GC substitutions in the 5′-terminal hairpin loop of the cRNA promoter on polymerase activity. cRNPs were reconstituted in vivo by transfection of a plasmid transcribing wild-type or mutant NA cRNA together with plasmids expressing PA, PB2, NP, and either wild-type PB1 (even-numbered lanes) or PB1 with a DD→AA double mutation in the active site (odd-numbered lanes show plasmid-derived input cRNA). RNAs were analyzed at 42 h posttransfection by primer extension (see Materials and Methods). The sequences and proposed corkscrew structures (8) of the wild-type and mutant 5′ cRNA promoters are shown above the gels. The bold, italic letters indicate the mutant residues. The positions of the vRNA, cRNA and mRNA signals are shown to the left.
DISCUSSION
There has been significant progress in understanding the mechanisms regulating endonucleolytic cleavage and templated elongation of the capped primer during mRNA transcription by the influenza virus polymerase on the vRNA promoter (reviewed in reference 9). However, the mechanisms regulating de novo replication to cRNA are less well understood. We have previously shown that different concentrations of NTPs are required for in vitro replication and transcription of virion-derived vRNPs, through the development of an authentic in vitro transcription and replication assay (32). Specifically, we compared the nucleotide requirements for the elongation of a capped primer, endonucleolytically cleaved from globin mRNA, with those for de novo initiation during full-length transcription and replication. The aim of this study was to test the hypothesis that the requirement for different concentrations of NTPs is related to the identities of the nucleotides required to initiate de novo replication. This was achieved through the rescue of a recombinant virus containing a gene with a base pair substitution at positions 3 and 8 in the 3′ strand of the vRNA promoter (Fig. 2) and through analysis of the NTP concentration requirement for in vitro replication (Fig. 3). While the wild-type vRNA template sequence (3′-UCGUUUCGU…) requires high concentrations of ATP, CTP, and GTP for full-length de novo replication to cRNA, the promoter mutant vRNA template sequence (3′-UCCUUUUGGU…) requires high concentrations of only ATP and GTP for replication. Therefore, by delaying the incorporation of the first C residue from position 3 to position 8, the minimum CTP concentration required for efficient full-length replication to cRNA was decreased approximately 100-fold. In addition, the concomitant shift in the incorporation of a second G residue from position 8 to position 3 increased 10-fold the minimum GTP concentration required. Therefore, it appears that de novo replication of a vRNA template requires high concentrations of at least the first three nucleotides incorporated into cRNA.
Substitution of the base pair at positions 3′ and 8′ in the 5′-terminal hairpin loop of the cRNA promoter has previously been shown to inhibit mRNA transcription, in particular, and to also affect cRNA and vRNA accumulation (6). We have confirmed these findings for the corresponding base pair substitution at positions 3 and 8 in the 3′ end of the vRNA promoter of an authentic viral gene, both with in vivo reconstituted vRNPs (Fig. 1) and with recombinant mutant virus infections (Fig. 4). Leahy et al. (18) have shown that mutagenesis of this base pair specifically inhibits endonuclease cleavage for mRNA transcription, although the mechanism of inhibition is not known in any further detail. Similarly, the mechanism for how the identity of the base pair at positions 3 and 8 affects the relative accumulation of vRNA and cRNA was not known (6). Here, we have studied the phenotype in more detail, by sequencing the termini of in vivo-synthesized promoter mutant vRNA and cRNA, revealing an interesting additional mutation, the 5′-terminal A→G, in approximately 50% of the cRNAs (Table 1). Furthermore, we found that 3′-8′CG→GC cRNA with the additional 1′A→G mutation was inactive for replication to vRNA (Fig. 5), thus forming a separate population of inactive cRNAs. We propose that this may account for the increased levels of cRNA and potentially for the decreased levels of vRNA during mutant virus infection (Fig. 4).
The sequence of a promoter determines not only the efficiency with which it forms a complex with RNA polymerase but also the concentration of NTP required for initiating transcription (NTPi). In fact, rRNA transcription in bacteria is known to be regulated by the concentration of NTPi (13). We have previously confirmed that influenza virus polymerase requires a high concentration (>100 μM) (Fig. 3B, lane 3) of the NTPi for de novo initiation of replication (32). We have now shown that influenza virus polymerase also requires a high concentration (>50 μM) (Fig. 3A, lane 7, and B, lane 5) of the NTP templated by the third residue from the 3′ end (NTP3) and an intermediate concentration (>5 μM) (Fig. 3B, lane 6) of the NTP templated by the penultimate residue from the 3′ end (NTP2) during de novo initiation of replication. These data are similar to the bacteriophage T7 RNA polymerase transcription data, where the Kd of NTPi has been found to be 10 times weaker than the Kd of NTP2 (29). In addition, however, NTP2 was found to bind before NTPi and to direct the binding of NTPi, consistent with a model proposed by Butcher et al. (3) for de novo initiation and chain elongation by bacteriophage φ6 RNA polymerase complexes. According to this model, the template strand initially directs the binding of NTP2. Realignment of the template is subsequently required to allow binding and phosphodiester bond formation with NTPi.
In the case of influenza virus, in the presence of limiting concentrations of radiolabeled GTP, the polymerase synthesizes large quantities of pppApG and pppGpC from a vRNA template (7). This suggests that GTP (NTP2), which is required at intermediate concentrations, may be bound to the polymerase first and can form phosphodiester bonds with either ATP (NTPi) or CTP (NTP3), both of which are required at higher concentrations (lower Kd). In the case of the cRNA promoter, we have also previously shown that the viral polymerase initiates de novo replication by synthesis of a pppApG dinucleotide templated by residues 4 and 5 from the 3′ end (7). Realignment of the dinucleotide onto the complementary 3′-terminal residues 1 and 2 of the template allows the addition of residue 3 (U) and elongation to full-length vRNA. Although this process is distinct from vRNA-templated replication, it demonstrates that the influenza virus RNA polymerase indeed allows the realignment of a bound template and the subsequent elongation of de novo-synthesized dinucleotide. In addition, we have found that the influenza virus RNA polymerase requires high concentrations of ATP (NTPi), GTP (NTP2), and UTP (NTP3) for de novo initiation on a cRNA promoter (7; unpublished data). Overall, these data suggest that the first three NTPs incorporated during de novo initiation of replication are all required by the influenza virus RNA polymerase, which is bound to the promoter to form an active complex.
This model (Fig. 6), proposing the initial binding of GTP, offers an explanation for some of our observations in this study and in previous studies (32). First, it proposes a mechanism for the introduction of the additional 1′A→G mutation in cRNA derived from vRNA with an NA 3-8GC→CG promoter mutation (Table 1). If the viral polymerase initially binds GTP, directed by the 3′-terminal penultimate C residue in vRNA, we would argue that phosphodiester bond formation with the NTP directed by either template positions 1 or 3 may plausibly occur. In the case of the wild-type template, this would give rise to dinucleotide pppApG or pppGpC, both of which have previously been shown to be synthesized in vitro (7). While pppApG would extend to full-length wild-type cRNA, transcription of pppGpC would be expected either to abort or to yield cRNA lacking a 5′-terminal A residue (32). However, in the case of a vRNA template with the 3-8GC→CG promoter mutation, the mechanism proposed in the model (Fig. 6) would yield pppApG or pppGpG. pppApG would be extended to full-length cRNA (5′-pppAGG…). However, we propose that the pppGpG dinucleotide could be realigned to template positions 1 and 2, with an A-G mismatch at position 1 and G-C base pairing at position 2. This would allow an addition of a third G residue templated by position 3 and an extension to full-length cRNA (5′-pppGGG…). This is consistent with our data describing the detection of a mixed population of 3′-8′CG→GC cRNAs with different 5′-terminal nucleotides (Table 1). Similarly, at limiting concentrations of ATP in vitro, we suggest that the 3-8GC→CG promoter mutation in the template vRNA promotes the synthesis of pppGpG over pppApG for realignment and extension to full-length 3′-8′CG→GC mutant cRNA with an additional nontemplated 1′A→G substitution. This may explain the apparent insensitivity of the polymerase to reduced ATP concentrations for the de novo replication of the 3-8GC→CG promoter mutant template relative to that for the de novo replication of the wild-type promoter template (Fig. 3A and B, compare lanes 3 and 10). Finally, in vitro transcription of a wild-type vRNA template yields increased mRNA at limiting concentrations of GTP (Fig. 3B, lane 6). This may be explained by competitive binding of GTP for de novo initiation of replication and the capped fragment terminating with a G derived from globin mRNA by the polymerase (Fig. 6). Limiting the concentration of GTP would favor binding of the capped primer for transcription, leading to an increase in the yield of mRNA.
FIG. 6.
Model for vRNA-templated de novo initiation of replication to cRNA and extension of a capped primer during transcription to mRNA. The 3′-terminal sequences of the wild-type (A) and 3-8GC→CG promoter mutant (B) NA genes are shown. The sequence of the capped primer is derived from endonucleolytically cleaved β-globin mRNA (25). According to this model, replication or transcription is initiated by competitive stochastic binding of GTP or endonucleolytically cleaved capped primer, directed by the penultimate residue of the vRNA template. During replication, bound GTP can form phosphodiester bonds with ATP and either CTP on the wild-type template (A) or GTP on the 3-8GC→CG promoter mutant template (B). During transcription, the capped primer is extended by incorporation of CTP on the wild-type template (A) or GTP on the 3-8GC→CG promoter mutant template (B). The bold letters indicate the mutant residues.
Recently, a new regulatory mechanism for de novo replication of the influenza virus genome was proposed (17); in this mechanism, a DNA replicative helicase, MCM, stabilizes the abortive de novo-initiated transcription complex by preventing the release of nascent cRNA from the RNA polymerase and thereby promotes its transition to an elongating complex. In contrast to that report, we have not found it necessary to add MCM to our preparations of virion-derived vRNPs to achieve efficient in vitro synthesis of full-length cRNA and mRNA (32; this study).
In summary, we have shown here that the concentration of the first three nucleotides to be incorporated during replication by the influenza virus polymerase is critical for the synthesis of full-length transcripts. In addition, we present a model which shows that de novo initiation of influenza virus cRNA synthesis occurs at the penultimate residue of the vRNA template by binding of the cognate NTP, which then forms the scaffold for polymerization of the first and third residues prior to elongation.
Acknowledgments
We thank Ryu Yoshida for the construction of pPOLI-cNA-RT, Nicole Robb for designing the PB2 and NP gene-specific primers, Julian Robinson for sequencing, and Tao Deng and Ervin Fodor for helpful discussions.
This work was supported by MRC grants (G9523972 and G9901312) to G.G.B.
Footnotes
Published ahead of print on 7 May 2008.
REFERENCES
- 1.Beaton, A. R., and R. M. Krug. 1981. Selected host cell capped RNA fragments prime influenza viral RNA transcription in vivo. Nucleic Acids Res. 94423-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaton, A. R., and R. M. Krug. 1986. Transcription antitermination during influenza viral template RNA-synthesis requires the nucleocapsid protein and the absence of a 5′ capped end. Proc. Natl. Acad. Sci. USA 836282-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410235-240. [DOI] [PubMed] [Google Scholar]
- 4.Chen, D., and J. T. Patton. 2000. De novo synthesis of minus strand RNA by the rotavirus RNA polymerase in a cell-free system involves a novel mechanism of initiation. RNA 61455-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Compans, R. W., J. Content, and P. H. Duesberg. 1972. Structure of the ribonucleoprotein of influenza virus. J. Virol. 10795-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crow, M., T. Deng, M. Addley, and G. G. Brownlee. 2004. Mutational analysis of the influenza virus cRNA promoter and identification of nucleotides critical for replication. J. Virol. 786263-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, T., F. T. Vreede, and G. G. Brownlee. 2006. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J. Virol. 802337-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flick, R., G. Neumann, E. Hoffmann, E. Neumeier, and G. Hobom. 1996. Promoter elements in the influenza vRNA terminal structure. RNA 21046-1057. [PMC free article] [PubMed] [Google Scholar]
- 9.Fodor, E., and G. G. Brownlee. 2002. Influenza virus replication, p. 1-29. In C. W. Potter (ed.), Influenza. Elsevier Science, New York, NY.
- 10.Fodor, E., M. Crow, L. J. Mingay, T. Deng, J. Sharps, P. Fechter, and G. G. Brownlee. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 768989-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. Garcia-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 739679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1995. Characterization of the RNA-fork model of virion RNA in the initiation of transcription in influenza A virus. J. Virol. 694012-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaal, T., M. S. Bartlett, W. Ross, C. L. Turnbough, Jr., and R. L. Gourse. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 2782092-2097. [DOI] [PubMed] [Google Scholar]
- 14.Hagen, M., L. Tiley, T. D. Y. Chung, and M. Krystal. 1995. The role of template-primer interactions in cleavage and initiation by the influenza virus polymerase. J. Gen. Virol. 76603-611. [DOI] [PubMed] [Google Scholar]
- 15.Hara, K., F. I. Schmidt, M. Crow, and G. G. Brownlee. 2006. Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J. Virol. 807789-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao, C. C., and J.-H. Sun. 1996. Initiation of minus-strand RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: use of oligoribonucleotide primers. J. Virol. 706826-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawaguchi, A., and K. Nagata. 2007. De novo replication of the influenza virus RNA genome is regulated by DNA replicative helicase, MCM. EMBO J. 264566-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leahy, M. B., H. C. Dobbyn, and G. G. Brownlee. 2001. Hairpin loop structure in the 3′ arm of the influenza A virus virion RNA promoter is required for endonuclease activity. J. Virol. 757042-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matrosovich, M. N., T. Y. Matrosovich, T. Gray, N. A. Roberts, and H. D. Klenk. 2004. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J. Virol. 7812665-12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medcalf, L., E. Poole, D. Elton, and P. Digard. 1999. Temperature-sensitive lesions in two influenza A viruses defective for replicative transcription disrupt RNA binding by the nucleoprotein. J. Virol. 737349-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagy, P. D., C. D. Carpenter, and A. E. Simon. 1997. A novel 3′-end repair mechanism in an RNA virus. Proc. Natl. Acad. Sci. USA 941113-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 969345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ortega, J., J. Martin-Benito, T. Zurcher, J. M. Valpuesta, J. L. Carrascosa, and J. Ortin. 2000. Ultrastructural and functional analyses of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J. Virol. 74156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plotch, S. J., M. Bouloy, I. Ulmanen, and R. M. Krug. 1981. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23847-858. [DOI] [PubMed] [Google Scholar]
- 26.Rowley, K. V., R. Harvey, and W. S. Barclay. 1999. Isolation and characterization of a transfectant influenza B virus altered in RNA segment 6. J. Gen. Virol. 802353-2359. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro, G. I., and R. M. Krug. 1988. Influenza virus RNA replication in vitro: synthesis of viral template RNAs and virion RNAs in the absence of an added primer. J. Virol. 622285-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaw, M. W., and R. A. Lamb. 1984. A specific sub-set of host-cell mRNAs prime influenza virus mRNA synthesis. Virus Res. 1455-467. [DOI] [PubMed] [Google Scholar]
- 29.Stano, N. M., M. K. Levin, and S. S. Patel. 2002. The +2 NTP binding drives open complex formation in T7 RNA polymerase. J. Biol. Chem. 27737292-37300. [DOI] [PubMed] [Google Scholar]
- 30.Sun, J.-H., S. Adkins, G. Faurote, and C. C. Kao. 1996. Initiation of (-)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: synthesis of oligonucleotides. Virology 2261-12. [DOI] [PubMed] [Google Scholar]
- 31.Testa, D., and A. K. Banerjee. 1979. Initiation of RNA synthesis in vitro by vesicular stomatitis virus. Role of ATP. J. Biol. Chem. 2542053-2058. [PubMed] [Google Scholar]
- 32.Vreede, F. T., and G. G. Brownlee. 2007. Influenza virion-derived viral ribonucleoproteins synthesize both mRNA and cRNA in vitro. J. Virol. 812196-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vreede, F. T., T. E. Jung, and G. G. Brownlee. 2004. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J. Virol. 789568-9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamakawa, M., Y. Furuichi, K. Nakashima, A. J. LaFiandra, and A. J. Shatkin. 1981. Excess synthesis of viral mRNA 5′-terminal oligonucleotides by reovirus transcriptase. J. Biol. Chem. 2566507-6514. [PubMed] [Google Scholar]