Abstract
Cells infected with Sindbis virus (SV) make two positive-strand RNAs, a genomic-length RNA (G) RNA and a subgenomic (SG) RNA. In cells infected with SVstd, and in general in cells infected with wt alphaviruses, more SG RNA is made than G RNA. How the balance between synthesis of G RNA and SG RNA is regulated is not known. SVpzf and SVcpc are nsP4 mutants of SV which, in mosquito cells, make more G RNA than SG RNA. When low concentrations of pyrazofurin (inhibits the synthesis of UTP and CTP) were added to SVpzf-infected cells, the yield of virus was increased, and the ratio of SG/G RNA was changed from <1 to >1. These effects were reversed by uridine. In SVcpc-infected cells, but not in SVstd-infected cells, synthesis of viral RNA was inhibited by the addition of either uridine or cytidine, and viral yields were lowered. Our findings suggest that the activities of the viral RNA-synthesizing complexes in cells infected with SVpzf or SVcpc, in contrast to those in SVstd-infected cells, are sensitive to high concentrations of UTP or CTP. Using a cell-free system that synthesizes both SG and G RNA, we measured viral RNA synthesis as a function of the UTP/CTP concentrations. The results indicated that the presence of the SVpzf mutations in nsP4 and the SG promoter produced a pattern quite different from that seen with the SVstd nsP4 and SG promoter. As the UTP/CTP concentrations were increased, the SVpzf system, in contrast to the SVstd system, made more G RNA than SG RNA, reflecting the situation in cells infected with SVpzf.
Sindbis virus (SV), the prototype virus of the family Togaviridae, genus Alphavirus, is one of the simplest of the enveloped RNA viruses (20, 23). It has a positive-strand RNA genome about 11,700 nucleotides (nt) in length, which is capped at its 5′ end and polyadenylated at its 3′ end. Two positive-strand mRNAs are made in SV-infected cells, a genomic-length (G) RNA and a subgenomic (SG) RNA; the SG RNA is about 4,100 nt in length, and its sequence is exactly colinear with the 3′ sequence of the G RNA. The G RNA serves as the message for two polyproteins, P123 and P1234, from which are derived the four nonstructural (ns) proteins nsP1, nsP2, nsP3, and nsP4. The functions of the ns proteins are as follows. nsP1, by virtue of its guanylyltransferase and methyltransferase activities, is involved in the capping and methylation of the two mRNAs (1, 18). nsP2 has a protease domain which is responsible for the proteolytic processing of P123 and P1234, and it also has an RNA helicase domain (3). nsP4 is the viral RNA polymerase. The exact role of nsP3, the only one of these four proteins that is phosphorylated, is not known (see references 20 and 23). Besides serving as a message, newly synthesized G RNA, is also packaged into progeny virions. The SG RNA serves as the message for the three structural proteins of the virus, the capsid protein, and the two envelope proteins, E2 and E1; it is not packaged into virions.
Upon infection of a susceptible cell, the incoming G RNA is translated into two polyproteins, P123 and P1234. The first step in the processing of these polyproteins is the cleavage of nsP4 from P1234, giving rise to RNA-synthesizing complexes composed of P123/nsP4. These complexes can use the G RNA as a template for the synthesis of a genomic-length negative-strand RNA, but they synthesize the positive-strand viral RNAs very poorly (10, 19). Further processing of P123 gives rise to nsP1 and P23 and finally to nsP2 and nsP3. Multiprotein complexes in which the polyproteins have been processed to the four individual ns proteins are no longer capable of synthesizing negative-strand RNA. The shutdown of negative-strand RNA synthesis after the first few hours of viral infection can be explained by the increased concentration of nsP2 with time after infection, which thereby quickens the processing of P123 and P1234. Complexes containing the four processed ns proteins, however, become much more efficient at using the negative-strand RNA as a template to synthesize the positive-strand G and SG RNAs. In the switch from the synthesis of negative-strand RNA to the synthesis of positive-strand RNA, it is the cleavage between nsP2 and nsP3 that is especially critical (9, 25).
Thus, with respect to the synthesis of viral RNA, there are at least two important regulatory issues. One is the switch from the synthesis of negative-strand RNA to positive-strand RNA. As just described, this switch is dependent upon the processing of the viral polyproteins. The second regulatory issue relates to the relative amounts of the G RNA and SG RNA which are made in infected cells. Although both of these RNAs are made from the negative-strand template, the mechanisms of synthesis must be quite different. Synthesis of G RNA begins at or very close to the 3′ end of the negative-strand template, whereas synthesis of the SG RNA begins via internal initiation on the negative-strand template (11).
Generally, whether measured in absolute amounts or in molar terms, cells infected with wild-type (wt) alphaviruses make SG RNA in excess of G RNA. Furthermore, it seems reasonable that to generate maximal amounts of virus, there must be an optimal ratio of viral SG/G RNA that is made. If infected cells synthesized only G RNA or only SG RNA, no virus progeny would be made. What the optimal SG/G RNA ratio is and how the proper balance between synthesis of G RNA and SG RNA is maintained are not well understood.
Mutants of SV have been described that make decreased amounts of SG RNA relative to the amount of G RNA made, i.e., the SG/G RNA ratio is less than one. Such a phenotype can be caused by mutations in the nsP2 coding region, specifically in the region coding the protease domain of nsP2 (24). In some cases, this phenotype may be related to inefficient processing of P123, specifically between nsP2 and nsP3, or in other cases to the altered cellular localization of nsP2 (24). Other reports have demonstrated that mutations in the promoter for the synthesis of SG RNA also adversely affect synthesis of this RNA (4, 6, 16); furthermore, the effects of promoter mutations can vary depending on the host cell. Finally, a mutation in the nsP3 coding region that greatly reduced the synthesis of SG RNA but not G RNA has been described (8).
We have described two mutants of Sindbis virus, SVpzf and SVcpc, that are resistant to pyrazofurin (PZF) and cyclopentenylcytosine (CPC), respectively (12, 17). PZF is a nucleoside analog, the monophosphate form of which inhibits the enzyme orotate monophosphate (OMP) decarboxylase and thereby prevents the synthesis of both UTP and CTP. Therefore, cells treated with PZF have low levels of both UTP and CTP; SVpzf is able to replicate in such cells. The mutations responsible for the SVpzf phenotype are in the nsP4 coding region and result in the following changes in nsP4: M287L, K592I, and P609T (17). Complicating the analysis of SVpzf was the fact that the same mutation responsible for the P609T change also changed the nucleotide at position −5 of the promoter for SG RNA synthesis (where transcription of the SG RNA begins at the +1 position). A second phenotype of SVpzf is that it was restricted in BHK cells and made less SG RNA than G RNA. Both the restriction in BHK cells and the altered RNA pattern were reversible by the addition of adenosine to the infected cells (16).
The second mutant, SVcpc, is able to grow in cells treated with CPC, a nucleoside analog that inhibits the enzyme CTP synthase (12). Cells treated with CPC have low levels of CTP. The mutation responsible for the SVcpc phenotype was, as expected, also in the nsP4 coding sequence and changes Leu 585 to Phe. While recently reviewing our sequencing data for SVcpc, we became aware that we made an error in this paper in stating that the mutation responsible for the amino acid change at position 585 was A7523C. The mutation was, in fact, G7523U.
A surprising property of SVpzf is that when mosquito cells infected with SVpzf were treated with low concentrations of PZF, the yield of virus was actually increased (17). We began the experiments described in this report to understand this unexpected observation. We observed that in SVpzf-infected cells, the ratio of SG/G RNA was <1, that this ratio was reversed by the addition of PZF to the infected cells, and that the PZF effect was in turn abolished by the addition of uridine to the infected cells. The addition of either uridine or cytidine to SVcpc-infected cultures depressed viral RNA synthesis and reduced the virus yield; no such effect was seen with SVstd-infected cultures. Making use of a cell-free system that makes both G and SG RNA, we found that the SVpzf mutations in nsP4 and the SG promoter change how the concentrations of UTP and CTP affect the balance between the synthesis of G RNA and SG RNA. We conclude that the mutations in SVpzf and SVcpc render the activity of their RNA-synthesizing complexes unduly sensitive to changes in the concentrations of UTP and/or CTP.
MATERIALS AND METHODS
Cells and viruses.
The Aedes albopictus mosquito cells (C7-10) were described previously (2); they were grown in Eagle's minimal essential medium supplemented with nonessential amino acids, glutamine, and 5% fetal bovine serum. The chicken embryo fibroblast (CEF) line was obtained from ATCC (CRL-12203) and grown in Dulbecco's modified Eagle's medium, as modified by ATCC to contain 4 mM l-glutamine, 4.5 g glucose, and 1.5 g sodium bicarbonate/liter; the medium was supplemented with 10% fetal bovine serum.
Our standard Sindbis virus (SVstd) was derived from the HR strain of SV (21). SVpzf is a mutant of SV that is resistant to PZF, an inhibitor of pyrimidine biosynthesis (16, 17), and SVcpc is a mutant of SV that is resistant to CPC, an inhibitor of CTP synthesis only (12).
Mosquito cells were infected with Sindbis virus at the indicated multiplicities of infection (MOIs) (using the titers determined with CEFs). Virus adsorption was performed at 28°C or 34.5°C for 1 h, after which the infected cultures were maintained at 34.5°C in medium containing 0.2% bovine serum albumin in place of serum. Media were harvested from the infected cultures at the times indicated and titrated for infectious virus by plaque assay on CEFs as described earlier (17); neutral red was added 24 or 48 h after infection. Two to three hours later, the neutral red was removed, and the plaques were counted.
The recombinant vaccinia viruses expressing SV P123, SV nsP4, or T7 polymerase were kindly provided to us by Charles M. Rice and Richard Hardy (Rockefeller University and Indiana University, respectively) (9). The nsP2-coding sequence in P123 contained the N614D substitution that results in more rapid processing of P123 than is observed with the wt P123 (10, 23). The recombinant vaccinia virus expressing nsP4 with the three SVpzf mutations was constructed as described earlier (14).
Northern blot analysis.
To extract RNA, Trizol reagent was pipetted onto cell monolayers (1 ml/60-mm plate), and the cells were lysed by repeated pipetting. Chloroform was added to the Trizol lysate (200 μl/ml lysate); the mixture was shaken vigorously and then allowed to stand at room temperature for 1 to 2 min. After centrifugation at 12,000 × g for 15 min, the aqueous phase was removed, and the RNA was precipitated by the addition of isopropanol (500 μl/ml of Trizol lysate). The RNA was pelleted by centrifugation at 12,000 × g for 10 min at 4°C, washed with 70% ethanol, dried, and dissolved in RNase-free water. Twenty micrograms of total RNA (as measured by the optical density at 260 nm) from each sample was denatured by the addition of formaldehyde and formamide (final concentrations of 2.2 M and 50%, respectively) and resolved on a 0.8% agarose gel. The RNAs were then blotted onto a nylon membrane (GeneScreen Plus; Perkin Elmer). A minus-strand oligodeoxyribonucleotide probe with a sequence corresponding to the SV positive-strand sequence from nt 8206 to nt 8229 (this probe would recognize both SG and G RNA) was end labeled with [γ-32P]ATP and used to probe the RNAs on the nylon membrane. After UV cross-linking of the RNA to the membrane, the blots were prehybridized for 2 h at 42°C in Ultrahyb hybridization buffer (Ambion) followed by overnight hybridization with the 32P-labeled probe. The blots were washed twice, each time for 15 min at 25°C with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate, and then exposed for 24 h on a storage phosphor screen. The signals were read on a Molecular Dynamics Typhoon PhosphorImager scanner and analyzed by ImageQuant software.
In vitro synthesis of SV RNA.
The reaction mixture for the in vitro synthesis of SV RNA (25 μl) contained 5 μl of 5× reaction buffer (200 mM Tris-HCl [pH 7.9], 30 mM MgCl2, and 50 mM NaCl), 10 mM dithiothreitol, 40 units of RNase inhibitor (Promega), 2 μg negative-strand RNA which serves as the promoter/template, and 12.2 μl of P15 extract prepared from BSC40 cells infected with recombinant vaccinia viruses expressing the T7 RNA polymerase, the SV polyprotein, P123, and wt SV nsP4 or SVpzf nsP4; the protein concentrations of the P15 fractions were adjusted to contain 6.4 μg protein/μl. The nucleoside triphosphate (NTP) concentrations were 3 mM ATP and 0.5 mM GTP. [32P]GTP (800 Ci/mM; 10 μCi/ml) was included to label the transcripts. The concentrations of CTP and UTP were equal but were varied as indicated. Incubation was performed at 37°C for 1 h. RNA was then extracted using phenol-chloroform and electrophoresed on a 6% denaturing polyacrylamide gel containing 7 M urea. The gel was dried, exposed to a storage phosphor screen, scanned, and analyzed as described above.
The negative-strand RNA promoter/template was synthesized basically by the method of Li and Stollar (15), except that the deleted form of pToto, which was described there and which was used as the template for synthesis of the negative-strand RNA, was further digested and religated step by step with NdeI, NarI, BspEI, and BamHI; the end product was a deleted form of pToto with one deletion between nt 421 and 7334 and a second deletion between nt 7869 and 11311 (transcription of the SG RNA begins with an A at nt 7598). PCR was used to amplify the deleted form of pToto using upstream and downstream primers with added T7 and SP6 promoters, respectively. RNA was transcribed from the PCR product using the AmpliScribe SP6 high-yield transcription kit (Epicenter), as described previously (13).
RESULTS
Effects of PZF and uridine on viral yield and viral RNA synthesis in mosquito cells infected with SVstd or SVpzf.
Figure 1 makes two points. First, it contrasts the effect of PZF on SVstd and SVpzf replication in mosquito cells. Whereas PZF reduced the yield of SVstd by close to 1,000-fold (Fig. 1A), it increased the yield of SVpzf about 10-fold (Fig. 1B). In other experiments, this increase has ranged from 10- to 50-fold. Second, Fig. 1 demonstrates that whether the effect of PZF was to reduce the virus yield, as with SVstd, or to increase the virus yield, as was the case with SVpzf, the effect of PZF was reversed by the addition of uridine to the infected cells.
FIG. 1.
Effects of PZF and uridine on the replication of SVstd and SVpzf in mosquito cells. (A) Confluent monolayers of mosquito cells were infected with SVstd (MOI of about 4 PFU/cell). After 1 hour for adsorption of virus, cells were treated with PZF (1 μM) and the indicated concentrations of uridine (•). One culture was not treated with uridine (▪). Media were harvested 23 h after infection and assayed for infectious virus by plaque assay on CEFs. (B) The procedure was performed as in panel A except that the cells were infected with SVpzf and the MOI was 10 PFU/cell. Media were harvested at 18 h after infection.
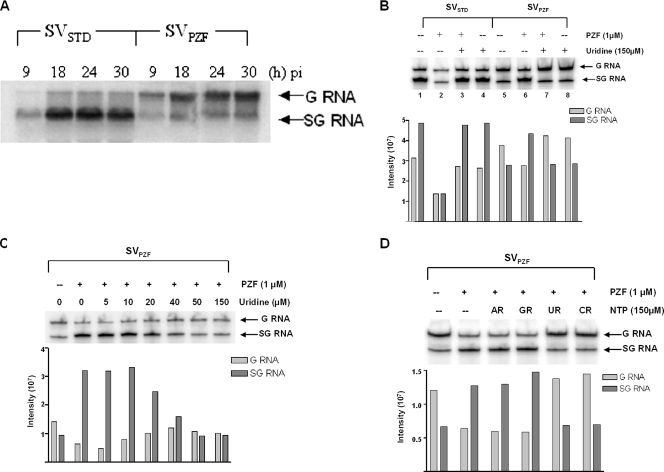
To better understand the effects of PZF and uridine on the replication of SVstd and SVpzf, we used Northern blot analysis to examine the synthesis of viral RNA in infected mosquito cells. Figure 2A shows that in contrast to SVstd-infected mosquito cells which made more SG RNA than G RNA, in SVpzf-infected cells, the reverse was true; G RNA was made in excess over SG RNA. (A similar pattern of viral RNA was seen in SVpzf-infected BHK cells [16].) Furthermore, in each case, the pattern of viral RNA synthesis was the same, whether examined at 9, 18, 24, or 30 h after infection.
FIG. 2.
Analysis of SV SG and G RNA in mosquito cells infected with SVstd or SVpzf. (A) Comparison of SG RNA and G RNA at different times after infection. Mosquito cells were infected at an approximate MOI of 4 PFU/cell with SVstd or SVpzf as indicated. At 9, 18, 24, and 30 h after infection, total RNA was extracted and analyzed by Northern blotting as described in Materials and Methods. pi, postinfection. (B) Effects of PZF and uridine on the levels of SG and G RNA. Mosquito cells were infected with SVstd or SVpzf (MOI of 1 to 2 PFU/cell) and treated (+) with PZF and uridine as indicated. RNA was extracted at 18 h after infection and analyzed by Northern blotting. In the graph below the gel, the intensity of the bands (in arbitrary units) as measured from the phosphorimager scan is plotted for each of the lanes. (C) Reversal of the PZF effect on the viral RNA pattern as a function of the uridine concentration. Mosquito cells were infected with SVpzf as described above for panel B and treated with PZF and various concentrations of uridine. RNA was extracted at 18 h after infection and analyzed by Northern blotting as described above for panel B. (D) Effects of the different nucleosides on the pattern of viral RNA synthesis in SVpzf-infected cells treated with PZF. Mosquito cells were infected with SVpzf as described above for panel B and treated with PZF, adenosine (AR), guanosine (GR), uridine (UR), or cytidine (CR) as indicated. At 20 h after infection, RNA was extracted and analyzed by Northern blotting as described above for panel B.
Figure 2B shows the effect of uridine on the pattern of viral RNA synthesis in mosquito cells that had also been treated with PZF. As already shown, in SVstd-infected cells, SG RNA was made in excess of G RNA (Fig. 2B, lane 1), whereas in SVpzf-infected cells, more G RNA was made than SG RNA (lane 5). As expected, PZF inhibited the synthesis of viral RNA in the SVstd-infected cells, reducing the synthesis of both G and SG RNA (lane 2); in contrast, in SVpzf-infected cells, the total amount of viral RNA made was not affected by the addition of PZF (lane 6). What PZF did do, however, was to modify the pattern of viral RNA synthesis so that it now resembled that in SVstd-infected cells. When uridine was added to the SVstd-infected cells along with PZF, it completely reversed the effect of PZF (lane 3), so that the pattern was similar to that seen in lane 1. Uridine also reversed the effect of PZF in SVpzf-infected cells (lane 7), so that the ratio of SG to G RNA which was made was similar to that in the untreated cells (lane 5). The addition of uridine alone had little effect on viral RNA synthesis in the SVstd-infected cells (lane 4) but shifted the balance slightly toward greater synthesis of G RNA in the SVpzf-infected cells (compare lanes 5 and 8).
In Fig. 2C, the uridine effect on cells infected with SVpzf and treated with PZF was titrated. As in Fig. 2B, untreated cells made more G than SG RNA. The addition of PZF to the infected cultures resulted in a marked stimulation of SG RNA synthesis. As increasing amounts of uridine were added, the PZF effect was reversed. The reversal by uridine began at a concentration of 20 μm, and at 50 μM, the reversal was complete.
To test whether the ability to reverse the effect of PZF on viral RNA synthesis was unique to uridine, mosquito cells were infected with SVpzf, treated with PZF, and then, as indicated, treated with uridine, cytidine, adenosine, or guanosine, each at a final concentration of 150 μM. As before, the addition of PZF to the infected cells altered the pattern of viral RNA synthesis so that the amount of SG RNA made exceeded that of G RNA (Fig. 2D). Neither adenosine nor guanosine altered this pattern. In contrast, both uridine and cytidine abolished the PZF effect so that the ratio of SG/G RNA made was similar to that observed in the untreated cells.
Table 1 shows that the viral yields correlated well with the RNA patterns shown in Fig. 2D. In this experiment, PZF increased the yield of SVpzf more than 40-fold. Neither adenosine nor guanosine had any effect on the stimulation by PZF. On the other hand, both uridine and cytidine completely reversed the PZF effect, reducing the virus yield to what it was in untreated cells.
TABLE 1.
Effects of PZF and different nucleosides on the replication of SVpzfa
| Addition | Virus yield (PFU/ml) |
|---|---|
| None | 4.0 × 106 |
| PZF | 1.8 × 108 |
| PZF + adenosine | 1.3 × 108 |
| PZF + guanosine | 1.7 × 108 |
| PZF + uridine | 3.2 × 106 |
| PZF + cytidine | 3.0 × 106 |
Mosquito cells were infected with SVpzf at an MOI of approximately 3 PFU/ml. After adsorption, cultures were refed with medium; PZF, adenosine, guanosine, uridine, and cytidine were then added to the cultures as indicated. PZF was added to a final concentration of 1 μM, and the nucleosides were added to a final concentration of 150 μM. After 24 h, media were harvested and assayed for infectious virus on CEFs.
Effects of PZF, uridine, and cytidine on the NTP pools in mosquito cells.
To clarify how PZF and uridine affect viral RNA synthesis in mosquito cells, we measured the effects of these compounds on the pool sizes of the different NTPs (Table 2). In untreated cells, ATP was present at the highest concentration, followed by UTP, CTP, and then GTP; the GTP concentration was only about one-third that of ATP. These results are similar to what we observed previously with mosquito cells, except that in our earlier work, the GTP concentration was slightly higher than the CTP concentration (22).
TABLE 2.
Effects of PZF, uridine, and cytidine on the NTP pools in mosquito cellsa
| Treatment | Size of NTP pool (pmol/106 cells)b
|
Concn (pmol/106 cells)c of:
|
|||||
|---|---|---|---|---|---|---|---|
| ATP | GTP | UTP | CTP | UDPGd | OMP | NAD | |
| None (control) | 1,583 ± 61 | 466 ± 11 | 1,348 ± 125 | 615 ± 44 | 1,033 ± 119 | 36 ± 2 | 182 ± 30 |
| PZF | 2,399 ± 32 | 775 ± 50 | 81 ± 12 | 56 ± 16 | 210 ± 28 | 614 ± 27 | 480 ± 22 |
| Uridine | 1,620 ± 36 | 558 ± 11 | 3,770 ± 111 | 1,326 ± 46 | 2,605 ± 69 | 0 | 445 ± 12 |
| Cytidine | 1,841 ± 55 | 596 ± 32 | 3,040 ± 179 | 1,255 ± 85 | 2,125 ± 105 | 21 ± 0 | 491 ± 32 |
| PZF + uridine | 1,550 ± 127 | 551 ± 48 | 4,243 ± 300 | 1,402 ± 100 | 2,605 ± 206 | 14 ± 12 | 449 ± 42 |
| PZF + cytidine | 1,827 ± 58 | 572 ± 24 | 1,231 ± 59 | 3,195 ± 125 | 828 ± 37 | 256 ± 16 | 484 ± 18 |
Mosquito cells in 60-mm plates were treated with 1 μM PZF, 100 μM uridine, or 100 μM cytidine as indicated, for 8 h. Extraction and analysis of NTPs were done as described earlier (1a, 12).
NTP concentrations are expressed as picomoles per 106 cells initially plated.
Concentrations are expressed as picomoles per 106 cells initially plated.
UDPG, uridine diphosphate glucose.
When cells were treated with PZF (1 μM), the levels of UTP and CTP were reduced to less than 1/10 the levels observed in untreated control cells. Since the monophosphate of PZF inhibits the enzyme OMP decarboxylase, the increase in OMP, which rose almost 20-fold, is not surprising. Treatment with PZF also increased the levels of both ATP and GTP by approximately 50%. The addition of uridine (100 μM) to mosquito cells increased the pool size of UTP by almost threefold and that of CTP by more than twofold. The level of ATP was not significantly changed, but the level of GTP was increased by about 20%. Cytidine (100 μM) had similar effects on the UTP and CTP levels and increased the levels of ATP and GTP by about 12% and 20%, respectively.
Figures 1 and 2 showed that PZF had quite different effects on SVstd- and SVpzf-infected mosquito cells; PZF inhibited the synthesis of SVstd RNA and reduced the virus yield, but in SVpzf-infected cells, it reversed the ratio of SG/G RNA and increased the yield of virus. Whereas the inhibition of SVstd can be explained by the precipitous decrease in the pool sizes of UTP and CTP, essential substrates for the synthesis of viral RNA, the explanation for how PZF increased the yield of SVpzf is less clear. Elsewhere we suggested that the marked stimulation of SVpzf replication in BHK cells by the addition of adenosine might be mediated by the resultant marked increase in ATP (16). The addition of adenosine to BHK cells, like the addition of PZF to mosquito cells, decreased the levels of UTP and CTP, although not nearly to the same extent (16). Thus, in BHK cells treated with adenosine and in mosquito cells treated with PZF, the increased yield of SVpzf was associated not only with an increased level of ATP, but also with decreased levels of UTP and CTP.
Table 2 shows that concentrations of uridine or cytidine that reversed the stimulatory effect of PZF on the yield of SVpzf (Fig. 1 and Table 1) also reversed the effects of PZF on the levels of UTP and CTP. Thus, in cells treated with PZF plus uridine, the UTP and CTP levels were similar to those in cells treated with uridine alone. In cells treated with PZF plus cytidine, the UTP level was only 40% of that in cells treated only with cytidine, whereas the CTP level was 2.5 times higher. In contrast to cells treated only with PZF, in cells treated with PZF plus uridine or PZF plus cytidine, the increases in ATP and GTP relative to the control cells were much more modest.
Marked changes in the level of UTP invariably correlated with changes in uridine diphosphate glucose. It is noteworthy that whereas the level of OMP was not increased in cells treated with PZF plus uridine, the level of OMP did increase in cells treated with PZF plus cytidine, although to a lesser degree than in cells treated only with PZF. This suggests that cytidine is not as effective as uridine in downregulating the pathway for the synthesis of pyrimidine nucleotides. The levels of NAD were increased between two- and threefold following the addition of PZF alone, PZF with uridine or cytidine, and uridine or cytidine alone. The significance of this result is not known, but it suggests that any major change in the NTP pools may lead to an increase in NAD.
Effects of uridine and cytidine on viral replication and on the RNA pattern in SVcpc-infected mosquito cells.
After observing that uridine and cytidine were able to reverse the effects of PZF on the replication of SVpzf, we wished to test the effects of different nucleosides on the replication of SVcpc. In contrast to SVpzf, SVcpc was selected for its ability to replicate in cells in which only CTP was reduced (12).
As shown in Table 3, neither adenosine nor guanosine had any effect on the yield of SVcpc from mosquito cells. Both uridine and cytidine, on the other hand, reduced the yield of SVcpc approximately 100-fold. None of the nucleosides affected the yield of SVstd at the concentrations used. A concentration of 100 μM uridine was required to lower the yield of SVcpc (Table 4).
TABLE 3.
Effects of the different nucleosides on the yields of SVstd and SVcpca
| Addition | Yield of virus (PFU/ml)
|
|
|---|---|---|
| SVstd | SVcpc | |
| None | 4.0 × 108 | 3.5 × 108 |
| Adenosine | 3.6 × 108 | 5.0 × 108 |
| Guanosine | 1.3 × 108 | 3.0 × 108 |
| Uridine | 6.1 × 108 | 2.7 × 106 |
| Cytidine | 6.4 × 108 | 3.9 × 106 |
Mosquito cells were infected with either SVstd or SVcpc at an MOI of approximately 2 PFU/cell. After adsorption of virus for 1 h, cultures were fed with 2 ml of medium; adenosine, guanosine, uridine, and cytidine were then added to a final concentration of 200 μM as indicated. Media were harvested 24 h after infection and assayed for infectious virus on CEFs.
TABLE 4.
Yield of SVcpc as a function of the concentration of uridine addeda
| Uridine concn (mM) | Yield of virus (PFU/ml)
|
|
|---|---|---|
| SVSTD | SVCPC | |
| 0 | 6.4 × 108 | 1.7 × 108 |
| 10 | 5.7 × 108 | 3.8 × 108 |
| 50 | 5.2 × 108 | 1.2 × 108 |
| 100 | 4.0 × 108 | 1.7 × 107 |
| 200 | 2.8 × 108 | 8.2 × 106 |
Mosquito cells were infected with either SVstd or SVcpc at an MOI of approximately 2 PFU/cell. After adsorption, the cultures were refed with medium and uridine was added to the indicated final concentrations. Media were harvested at 24 h after infection and assayed for infectious virus.
We then examined the pattern of viral RNA synthesis in SVcpc-infected cells (Fig. 3A) as we did for SVpzf-infected cells. As was the case with SVpzf-infected cells and in contrast to SVstd-infected cells (Fig. 3A, compare lanes 1 and 5), SVcpc-infected cells made mainly G RNA (Fig. 3A). Lanes 4 and 8 in Fig. 3A show, as expected, that viral RNA synthesis was sensitive to CPC in SVstd-infected cells but resistant in SVcpc-infected cells. Also of interest, whereas PZF reversed the ratio of SG/G RNA in SVpzf-infected cells, CPC did not change this ratio in SVcpc-infected cells (compare lanes 5 and 8). However, whereas cytidine had no effect on viral RNA synthesis in SVstd-infected cells, it significantly reduced viral RNA synthesis in SVcpc-infected cells (compare lanes 2 and 3 with lanes 6 and 7).
FIG. 3.
SG RNA and G RNA in mosquito cells infected with SVcpc. (A) Effects of CPC and cytidine. Mosquito cells were infected with SVstd or SVcpc at an approximate MOI of 0.1 PFU/cell and treated with CPC (5 μM), cytidine (250 μM), or cytidine (500 μM) as indicated. Twenty hours after infection, RNA was extracted from the various samples and analyzed by Northern blotting. (B) Effects of CPC and uridine. Mosquito cells were infected as described above for panel A with SVstd or SVcpc and were either untreated or treated with uridine (500 μM) or cytidine (500 μM). Twenty hours after infection, RNA was extracted from the different samples and analyzed by Northern blotting.
In this same experiment, CPC reduced the yield of SVstd from 8 × 107 to <5 × 104 PFU/ml; with SVcpc, the virus yields were 6 × 107 and 4 × 107 PFU/ml in the absence and presence of CPC, respectively.
Figure 3B demonstrates that like cytidine, uridine reduced viral RNA synthesis in SVcpc-infected cells, but not in SVstd-infected cells. These findings suggest that the high levels of UTP and CTP resulting from the addition of uridine or cytidine (Table 2) are deleterious for viral RNA synthesis directed by SVcpc.
In vitro synthesis of Sindbis virus G and SG RNA.
To further explore how the changes in the NTP pools brought about by the addition of PZF, uridine, or cytidine alter the pattern of viral RNA synthesis in SV-infected cells, we made use of an in vitro system that synthesizes both SG RNA and G RNA. Since the major effect of adding PZF to cells is to drastically lower the concentrations of UTP and CTP, we set up two series of reactions: the first series (SVstd system) contained wt or SVstd forms of nsP1, nsP2, nsP3, and nsP4; a negative-strand RNA that contained the SVstd promoter sequences for both the G and SG promoters; ATP and GTP at concentrations of 3.0 and 0.5 mM, respectively, and various concentrations of UTP and CTP. The second series of reactions differed from the first in that the nsP4 contained the three amino acid substitutions found in nsP4 encoded by SVpzf, and the negative-strand RNA contained the same mutation at the −5 position of the SG promoter as was found in SVpzf. Thus, the second series of reactions mimicked the situation that would be found with SVpzf-infected cells. Although in both series the concentrations of UTP and CTP were varied, in each reaction mixture their concentrations were equal.
The reaction mixtures were incubated as described in Materials and Methods, after which the labeled RNA was extracted, fractionated by PAGE, and measured by exposure to a phosphorimager. The results are shown in Fig. 4. With the SVstd system containing SVstd nsP4 and the negative-strand RNA with the SVstd SG promoter (Fig. 4, left, lanes 1 to 9), the results were quite straightforward. At all concentrations of UTP/CTP, SG RNA was made in excess of G RNA, and the synthesis of both SG RNA and G RNA was dependent on the concentrations of UTP/CTP. At concentrations of UTP/CTP from 0.2 to 1.0 mM, synthesis of both RNAs was low but increased markedly, almost doubling when the concentrations of UTP/CTP were increased to 1.5 mM. Further increases in the UTP/CTP concentrations up to 5 mM did not result in higher levels of either SG or G RNA synthesis.
FIG. 4.
In vitro synthesis of SV SG and G RNA as a function of the concentrations of UTP and CTP. Reaction mixtures were set up and incubated as described in Materials and Methods. The RNA was extracted and fractionated by denaturing polyacrylamide gel electrophoresis. After the gel was allowed to dry, it was exposed to a storage phosphor screen, scanned, and analyzed as described in Materials and Methods. The graphs below the gel show the intensity of the bands (in arbitrary units) as measured from the phosphorimager scans. C, control without promoter/template (P/T).
The results were quite different with the SVpzf system (Fig. 4, right, lanes 1 to 9). At the two lowest concentrations of UTP/CTP, 0.2 and 0.5 mM, more SG RNA was made than G RNA. However, when the concentrations of UTP/CTP were increased to 1.0 mM, synthesis of SG RNA decreased sharply so that SG RNA and G RNA were made in roughly equal amounts. With 1.5 mM UTP/CTP, synthesis of G RNA doubled but then showed no further increase at higher concentrations of these substrates. SG RNA synthesis remained low but steady from 1.0 to 5.0 mM UTP/CTP. Thus, with the SVpzf system, at the very lowest substrate concentrations, more SG RNA was made than G RNA; however, at concentrations from 1.5 mM to 5.0 mM, more G RNA was made than SG RNA.
Finally, we note that at 0.2 and 0.5 mM UTP/CTP, the lowest concentrations tested, more total RNA was made in the reaction mixtures containing the SVpzf components than in the reaction mixtures containing the wt virus components. This may explain the ability of SVpzf to replicate in the face of low concentrations of UTP/CTP in infected cells.
DISCUSSION
This report deals with two mutants of Sindbis virus, SVpzf and SVcpc. These mutants were selected for resistance to PZF and CPC, respectively, nucleoside analogs which interfere in the first case with the biosynthesis of both UTP and CTP, and in the second case only with the biosynthesis of CTP. Thus, SVpzf is able to replicate in mosquito cells with markedly decreased levels of UTP and CTP, and SVcpc is able to replicate in mosquito cells with abnormally low levels only of CTP. The effects of treating mosquito cells with PZF on the NTP pools were shown in Table 2; the effects of CPC on the NTP pools have been reported by Li et al. (12).
In general, wt alphaviruses make SG RNA in infected cells in excess of G RNA. This is true of SVstd (Fig. 2A). In contrast, both SVpzf and SVcpc make less SG RNA than G RNA. In the case of SVcpc, only one amino acid change in nsP4 is required for the phenotype; SVpzf, on the other hand, has three mutations in the nsP4-coding sequence, all of which result in amino acid changes, and because of the overlap between the nsP4-coding sequence and the SG promoter, the 3′ most of these mutations also results in a mutation at the −5 position of the SG promoter. All of these mutations are required for resistance to PZF in mosquito cells (17). In contrast, SVcpc is an example in which an nsP4 mutation alone results in the synthesis of less SG RNA than G RNA.
In experiments with BHK cells infected with two subgenomic viruses, we showed that the SVpzf mutation at the −5 position of the SG promoter by itself was sufficient to reduce synthesis of SG RNA (16). However, we did not rule out the possibility that the nsP4 mutations could also contribute to this phenotype. Other examples of mutations in the sequence of the SG promoter that decrease the synthesis of SG RNA have been reported (5-7). In addition to promoter mutations, mutations that are located in the region of nsP2 that encodes the protease domain of this protein can also result in decreased SG RNA synthesis (24). In many cases, cells infected with mutant viruses that make less SG than G RNA also make less virus than cells infected with wt viruses. However, this does not seem to be the case with SVcpc, which generates virus yields in mosquito cells similar to those of SVstd, in spite of the fact that it makes less SG RNA than G RNA.
The simplest explanation for how PZF and CPC inhibit the synthesis of SVstd RNA and thereby viral replication in mosquito cells is that they lower the concentrations of UTP and/or CTP to levels too low for efficient synthesis of viral RNA. It is more difficult to understand why low concentrations of PZF [or of N-(phosphonoacetyl)-L-aspartic acid, another inhibitor of pyrimidine biosynthesis], increase the yield of SVpzf from infected cells (17). As shown in Results, one of our first findings was that the addition of PZF to SVpzf-infected cells reversed the ratio of SG/G RNA, so that instead of making less SG than G RNA, these cells made more SG than G RNA, i.e., they showed the wt pattern of viral RNA synthesis. Whether or not this change in the pattern of viral RNA synthesis is sufficient to explain the increase in virus yield is not clear. A second point of interest is that the effects of PZF on SVpzf-infected cells, i.e., both the reversal of the ratio of SG/G RNA synthesis and the increase in virus yield, were completely abolished by the addition of uridine to infected cells. Thus, whether PZF inhibits virus replication, as is the case with SVstd, or whether it stimulates replication, as is the case with SVpzf, the effects of PZF are reversed by uridine.
The addition of uridine or cytidine to mosquito cells, as would be expected, caused major alterations of the NTP pools, mainly increasing the levels of UTP and CTP; these changes had no effect on the replication of SVstd. However, the addition of uridine to PZF-treated SVpzf-infected cells, as just noted, did have significant effects on both virus yield and viral RNA synthesis. Equally striking was the inhibition by uridine and cytidine of viral RNA synthesis in SVcpc-infected cells and the subsequent reduction in virus yield.
How can the effects of PZF, CPC, uridine, and cytidine on the synthesis of viral RNA in cells infected with SVpzf or SVcpc be explained? We showed earlier using cell-free systems prepared from SV-infected cells, that SVcpc generates an RNA-synthesizing complex which has a Km for CTP which is about fourfold lower than the complex made in SVstd-infected cells (12) (the Kms of the two complexes for UTP, however, were similar). This likely reflects a more efficient binding of CTP by the SVcpc complex relative to the SVstd complex. While this more efficient binding may be advantageous when the concentration of CTP is limiting, it may, for various reasons, prove deleterious when the concentration of CTP is elevated above normal. Thus, the increased level of UTP and CTP following the addition of uridine or cytidine might be one possible explanation for the inhibition of viral RNA synthesis by SVcpc following the additions of these nucleosides to infected cultures. Similar considerations could apply to SVpzf-infected cultures with respect to the levels of UTP and CTP.
We also made use in our study of an in vitro system which we recently described and which synthesizes both G RNA and SG SV RNA (15). The critical components in this system are a cytoplasmic extract of cells containing the four SV ns proteins expressed by recombinant vaccinia virus virions and a negative-strand RNA molecule, which serves as the template and contains promoter sequences for the synthesis of both G GNA and SG RNA. In the experiment shown in Fig. 4, using both the SVstd and the SVpzf systems, we examined the pattern of viral RNA synthesis as a function of the concentrations of UTP and CTP.
Several interesting points emerge from our results. First, whereas the pattern with the wt system was quite straightforward, and more SG RNA was made than G RNA at all UTP/CTP concentrations tested, the pattern with the SVpzf system was more complex. Second, at the lowest concentrations of UTP/CTP, the system with the SVpzf mutations made more RNA than did the system with the wt sequences. This could explain the resistance of SVpzf to PZF and its ability to replicate in cells with low levels of UTP/CTP. Third, although at the lowest concentrations of UTP/CTP, more SG RNA was made by the SVpzf system than G RNA, as the concentration of UTP/CTP was increased, more G RNA was made than SG RNA, reflecting the situation seen in SVpzf-infected cells. Thus, the pattern of viral RNA with the SVpzf-system was dependent on the concentrations of UTP/CTP.
Why the pattern with the SVpzf system changes as the UTP/CTP concentrations is increased is not understood, but it could be related to competition between the G and SG promoters for the RNA-synthesizing complex and a preference of the RNA-synthesizing complexes for the G promoter rather than the SG promoter as the UTP/CTP concentration is increased. Work is in progress with this cell-free system to further examine the effects of mutations in nsP4 and in the G and SG promoters on the pattern of SV RNA synthesis. Also of special interest will be the question of whether it will be possible in some way to reverse the pattern of viral RNA synthesis seen in SVcpc-infected cells, as was done with SVpzf-infected cells.
Acknowledgments
We thank Yan Hu for technical support.
This work was funded by U.S. Public Health Service grants AI-49273 and AI-070728.
Footnotes
Published ahead of print on 28 May 2008.
REFERENCES
- 1.Ahola, T., and L. Kaariainen. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. USA 92507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Bofill, M., L. D. Fairbanks, K. Ruckemann, M. Lipman, and H. A. Simmonds. 1995. T lymphocytes from AIDS patients are unable to synthesize ribonucleotides de novo in response to mitogenic stimulation. Impaired pyrimidine responses are already evident at early stages of HIV-1 infection. J. Biol. Chem. 27029690-29697. [PubMed] [Google Scholar]
- 2.Durbin, R. K., and V. Stollar. 1984. A mutant of Sindbis virus with a host-dependent defect in maturation associated with hyperglycosylation of E2. Virology 135331-344. [DOI] [PubMed] [Google Scholar]
- 3.Gomez de Cedrón, M., N. Ehsani, M. L. Mikkola, J. A. García, and L. Kääriäainen. 1999. RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett. 44819-22. [DOI] [PubMed] [Google Scholar]
- 4.Grakoui, A., R. Levis, R. Raju, H. V. Huang, and C. M. Rice. 1989. A cis-acting mutation in the Sindbis virus junction region which affects subgenomic RNA synthesis. J. Virol. 635216-5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hahn, Y. S., A. Grakoui, C. M. Rice, E. G. Strauss, and J. H. Strauss. 1989. Mapping of RNA− temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J. Virol. 631194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hertz, J. M., and H. V. Huang. 1995. Evolution of the Sindbis virus subgenomic mRNA promoter in cultured cells. J. Virol. 697768-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hertz, J. M., and H. V. Huang. 1995. Host-dependent evolution of the Sindbis virus promoter for subgenomic mRNA synthesis. J. Virol. 697775-7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lastarza, M. W., A. Grakoui, and C. M. Rice. 1994. Deletion and duplication mutations in the C-terminal nonconserved region of Sindbis virus nsP3: effects on phosphorylation and on virus replication in vertebrate and invertebrate cells. Virology 202224-232. [DOI] [PubMed] [Google Scholar]
- 9.Lemm, J. A., A. Bergqvist, C. M. Read, and C. M. Rice. 1998. Template-dependent initiation of Sindbis virus RNA replication in vitro. J. Virol. 726546-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemm, J. A., T. Rumenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 132925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levis, R., S. Schlesinger, and H. V. Huang. 1990. Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J. Virol. 641726-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, M. L., Y. H. Lin, H. A. Simmonds, and V. Stollar. 2004. A mutant of Sindbis virus which is able to replicate in cells with reduced CTP makes a replicase/transcriptase with a decreased Km for CTP. J. Virol. 789645-9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, M. L., Y. H. Lin, and V. Stollar. 2005. A cell-free system for the synthesis of Sindbis virus subgenomic RNA: importance of the concentration of the initiating NTP. Virology 34124-33. [DOI] [PubMed] [Google Scholar]
- 14.Li, M. L., and V. Stollar. 2004. Identification of the amino acid sequence in Sindbis virus nsP4 that binds to the promoter for the synthesis of the subgenomic RNA. Proc. Natl. Acad. Sci. USA 1019429-9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, M. L., and V. Stollar. 2007. Distinct sites on the Sindbis virus RNA-dependent RNA polymerase for binding to the promoters for the synthesis of genomic and subgenomic RNA. J. Virol. 814371-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, Y. H., H. A. Simmonds, and V. Stollar. 2002. Restriction of a Sindbis virus mutant in BHK cells and relief of the restriction by the addition of adenosine. Virology 29278-86. [DOI] [PubMed] [Google Scholar]
- 17.Lin, Y. H., P. Yadav, R. Ravatn, and V. Stollar. 2000. A mutant of Sindbis virus that is resistant to pyrazofurin encodes an altered RNA polymerase. Virology 27261-71. [DOI] [PubMed] [Google Scholar]
- 18.Mi, S., R. Durbin, H. V. Huang, C. M. Rice, and V. Stollar. 1989. Association of the Sindbis virus RNA methyltransferase activity with the nonstructural protein nsP1. Virology 170385-391. [DOI] [PubMed] [Google Scholar]
- 19.Sawicki, S. G., and D. L. Sawicki. 1986. The effect of overproduction of nonstructural proteins on alphavirus plus-strand and minus-strand RNA synthesis. Virology 152507-512. [DOI] [PubMed] [Google Scholar]
- 20.Schlesinger, S., and M. J. Schlesinger. 2001. Togaviridae, p. 567-588. In D. M. Knipe and P. M. Howley (ed.), Fundamental virology. Lippincott Williams & Wilkins Press, Philadelphia, PA.
- 21.Shenk, T. E., and V. Stollar. 1973. Defective-interfering particles of Sindbis virus. II. Homologous interference. Virology 55530-534. [DOI] [PubMed] [Google Scholar]
- 22.Stollar, V., and F. Malinoski. 1981. The effects of adenosine and guanosine on the replication of Sindbis and vesicular stomatitis viruses in Aedes albopictus cells. Virology 11557-66. [DOI] [PubMed] [Google Scholar]
- 23.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suopanki, J., D. L. Sawicki, S. G. Sawicki, and L. Kaariainen. 1998. Regulation of alphavirus 26S mRNA transcription by replicase component nsP2. J. Gen. Virol. 79309-319. [DOI] [PubMed] [Google Scholar]
- 25.Wang, Y. F., S. G. Sawicki, and D. L. Sawicki. 1994. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J. Virol. 686466-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]