Abstract
CD4 T cells are required for the maintenance and recall of antiviral CD8 T cells and for antibody responses. Little is known concerning the overall architecture of the CD4 response to complex microbial pathogens. In a whole-proteome approach, 180 predicted open reading frames (ORFs) in the vaccinia virus genome were expressed and tested using responder cells from 20 blood samples from 11 vaccinees. Validation assays established the sensitivity and specificity of the system. Overall, CD4 responses were detected for 122 ORFs (68%). A mean of 39 ORFs were recognized per person (range, 13 to 63). The most frequently recognized ORFS were present in virions, including A3L and A10L (core proteins), WR148 (a fragmented homolog of an orthopoxvirus protein that forms inclusions in cells), H3L (a membrane protein), D13L (a membrane scaffold protein), and L4R (a nucleic acid binding protein). Serum immunoglobulin G profiling by proteome microarray detected responses to 45 (25%) of the ORFs and confirmed recent studies showing a diverse response directed to membrane and nonmembrane antigens. Our results provide the first empirical whole-proteome data set regarding the global CD4 response to full-length proteins in a complex virus and are consistent with the theory that abundant structural proteins are immunodominant.
The eradiation of smallpox transmission by preventative infection with replication-competent vaccinia virus is a remarkable success. Unfortunately, zoonotic orthopoxviruses can periodically emerge (61), there remains a potential for use of smallpox as a bioweapon, and vaccination with live vaccinia virus has complications (33). These issues, together with the use of vaccinia virus as a vector for vaccines and immunotherapy (31), have raised interest in the immune response to wild-type and replication-incompetent vaccinia virus (1).
Vaccinia virus vaccination induces persistent cellular and humoral responses (2, 15, 18, 34, 41, 42, 56, 58, 73). Several lines of evidence indicate that CD4 T-cell help is required for both CD8 cytotoxic T-lymphocyte and antibody responses. CD8 cells clear primary vaccinia virus, and neutralizing antibodies mediate protection from heterologous challenge (8, 22). The priming, maturation, and survival of poxvirus-specific, functional memory CD8 T cells requires CD4 T-cell help (4, 9, 10, 12, 13, 25, 39, 40, 64). In animals that are CD4 depleted, and in CD4−/− or major histocompatibility complex class II-deficient mice, vaccinia virus clearance is delayed and CD8 T cells have proliferative and maturation defects (77, 78). Antibodies fail to develop in CD4-deficient animals (21, 23).
In the last several years, the breadth of CD8 T-cell responses to vaccinia virus has been studied in several labs using a variety approaches in humans and in wild-type and HLA-transgenic mice (20, 41-43, 51, 53, 57-59, 66, 69, 70, 72). Epitopes in a total of 103 vaccinia virus open reading frames (ORFs) have been found to induce CD8 T-cell responses restricted by human HLA class I molecules, with some data limited to transgenic mice (43). Much less is known about the specificity of CD4 responses. We reported on the dominance hierarchy of CD4 responses in three primary human vaccinees analyzed using a genomic expression library approach (41). The apparent diversity of the responses ranged from 8 to 20 antigenic ORFs per subject. Single DRB1*0101-restricted epitopes in ORFs A24R and D1R, enzymes involved in RNA metabolism, were defined using T-cell clones and bioinformatics-based peptide binding predictions (54). Most recently, Calvo-Calle et al. combined predictive algorithms to focus on DRB1*0101-restricted responses and obtained a relatively high hit rate to define 25 epitopes in the strain modified vaccinia virus Ankara in diverse ORFs (11). In mice, a similar predictive method was used to explore CD4 specificity, and responses to virion structural proteins were predominant (56).
The goal of the current study was to define the breadth and population-dominant antigens of the vaccinia virus-specific CD4 response using a nonpredictive approach. We used recombinant antigens covering the entire predicted vaccinia virus proteome. Quality control and titration experiments confirmed that proteins expressed by in vitro transcription/translation allowed sensitive and specific detection of CD4 responses. To cross-correlate CD4 T-cell and B-cell responses, we probed the same protein set, arrayed as spots on nitrocellulose-coated slides, for binding immunoglobulin G (IgG). Overall, we identified 122 vaccinia virus ORFs that stimulated CD4 responses, and 45 ORFs that stimulated antibody responses, in at least one T-cell line. The within-subject diversity was generally quite broad and far higher than has been presented for other infectious agents to date. The ORFs most frequently recognized by CD4 T cells were predominately virion structural proteins and tended to have high molecular weights and to be expressed with late expression kinetics during virus replication. Most ORFs that were immunodominant for antibody responses also frequently elicited CD4 responses, while many other antigens were recognized by CD4 cells only. This is the first proteome-wide screen of CD4 responses to a complex pathogen using full-length protein antigens.
MATERIALS AND METHODS
Subjects and specimens.
Eleven adults (Table 1) receiving immunization by scarification with Dryvax smallpox vaccine for occupational health purposes gave Institutional Review Board-approved consent. Numbering for subjects 1, 2, and 5 to 10 corresponds to that used in our previous reports (42, 43), while subjects 11 to 13 were newly enrolled. HLA class II typing was done on some subjects and is available upon request. Control subject 14 was herpes simplex virus type 2 (HSV-2) seropositive (3) and never vaccinated with vaccinia virus. No subject had lived in a region endemic for malaria or had a known malaria or Francisella tularensis infection. Blood for T-cell testing was drawn 2 weeks to several years after vaccination. Peripheral blood mononuclear cells (PBMC) were cryopreserved after Ficoll enrichment. Sera were stored at −80°C.
TABLE 1.
Subjects, vaccination history, time points studied, and responses to whole-cell-associated vaccinia virus
| Subject no.a | No. of vaccinations (timing)b | First blood sample
|
Second blood sample
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Mos. between vaccination and samplec | cpmd
|
CD4 IFN-γe (%) | Mos. between vaccinatio and samplec | cpmd
|
CD4 IFN-γe (%) | ||||
| Vacc | Mock | Vacc | Mock | ||||||
| 1 | 2 (43) | 18 | 69,658 | 5,945 | 8.8 | 25 | 45,622 | 2,025 | 7.7 |
| 2 | 1 | 1 | 16,223 | 1,701 | 8.3 | 43 | 23,459 | 1,575 | 8.7 |
| 5 | 3 (52, 28) | 0.5 | 20,288 | 351 | 15.3 | 38 | 70,887 | 322 | 5.9 |
| 6 | 2 (41) | 0.5 | 27,247 | 341 | 3.3 | 28 | 35,480 | 727 | 6.3 |
| 7 | 1 | 1 | 17,274 | 327 | 6.1 | 10 | 29,820 | 786 | 6.6 |
| 8 | 2 (36) | 1 | 33,627 | 247 | 15.1 | 25 | 38,418 | 157 | 8.3 |
| 9 | 2 (35) | 44 | 22,674 | 212 | 11.6 | 48 | 44,134 | 441 | 3.7 |
| 10 | 1 | 2 | 25,984 | 1,372 | 14.3 | 30 | 36,961 | 1,118 | 9.8 |
| 11 | 1 | 6 | 64,832 | 766 | 22.2 | 17 | 49,748 | 377 | 20.0 |
| 12 | 1 | 60 | 65,089 | 146 | 27.5 | NA | NA | NA | NA |
| 13 | 4 (0.1, 29, 39) | 1 | 31,242 | 169 | 3.9 | NA | NA | NA | NA |
| 14 | 0 | NA | 811 | 545 | 0 | NA | NA | NA | NA |
Numbering for subjects 1 to 10 corresponds with the numbering used in our prior publications (41, 42).
The number of total lifetime vaccinations is followed in parentheses by the interval(s) in years between the current and past vaccinations. All vaccinations were clinical “takes” (63) except for subject 13, who had a poor take just prior to a typical clinical take.
Months between most recent vaccination and first blood sample for T-cell studies. NA, not applicable.
All cpm values are means of duplicate [3H]thymidine incorporation determinations. Vacc, vaccinated group; NA, not applicable.
The net percentage of CD4+ CD8− cells accumulating intracellular IFN-γ minus the percentage of similar cells that were IFN-γ positive in response to mock antigen. NA, not applicable.
Cell and viral culture.
Enrichment and expansion of vaccinia virus-specific T cells have been described (41, 42). Briefly, PBMC were seeded at 106/ml in 2 ml of T-cell medium (TCM) in 24-well plates. Live or UV-inactivated vaccinia virus strain New York City Board of Health (NYCBH) was added at a multiplicity of infection of 10 (or equivalent). Interleukin-2 (IL-2; Hemagen, Columbia, MD) was started on day 5 (32 to 50 U/ml), and cultures were split and fed periodically with IL-2-TCM. On day 12 to 14, gamma interferon (IFN-γ) intracellular cytokine cytometry (ICC) was done to confirm enrichment of vaccinia virus-specific CD4 T cells (below). Cells were expanded one additional cycle in vitro using anti-CD3 monoclonal antibody and IL-2 (45). The resultant cells were cryopreserved in aliquots for functional assays. Vaccinia virus strain NYCBH was raised and titers were determined in BSC-40 cells (42). Cell-associated virus stocks were the supernatants of sonicated cell monolayers that had 90 to 100% of the surface area of the cell monolayer showing obvious cytopathic effect, infected at a multiplicity of infection of 1, 72 h previously, after clarification by centrifugation at 400 × g for 10 min. UV inactivation (30-min exposure to GT038 bulbs at 10 cm; General Electric, Cleveland, OH) eliminated detectable infectious virus. For vaccinia virus-naïve control subject 14, a similar polyclonal virus-reactive bulk culture was raised using UV-inactivated HSV-2 strain 333 (44) as described elsewhere (48).
Antigens.
The vaccinia virus strain WR proteome was expressed as previously described (17, 18). In brief, genomic viral DNA was used as template for PCR. PCR products corresponding to individual full-length ORFs were cloned into a T7 expression vector by homologous recombination. Purified plasmids were then expressed in the Escherichia coli-based rapid translation system (RTS-100) for in vitro transcription/translation (Roche, Pleasanton, CA). The reaction products are termed RTS proteins in this report. All reactions were verified for protein expression on dot blots or arrays by probing with antibodies to the N- and C-terminal epitope tags, followed by alkaline phosphatase-conjugated secondary antibodies and nitrotetrazolium blue substrate as described elsewhere (18). RTS proteins from 20-μl reaction volumes was used for CD4 assays without purification (17). The vaccinia virus antigen set contained 180 potential unique ORFs. Nomenclature for vaccinia virus ORFs is from vaccinia virus strain Copenhagen, or from strain WR for orthologs absent from the Copenhagen strain. A cross-correlation of strain/ORF nomenclature and GenBank accessions numbers is available (50). Negative control antigens included 5 ORFs from Plasmodium falciparum strain 3D7 (PF11_0284, PFE0225W, PFE0795C, PFE159W, and PF08-0054; available via http://www.plasmodb.org/plasmo/home.jsp) (30, 65) and 28 ORFs (FT292, 306, 345, 394, 425, 468, 701, 745, 790, 994, 1108, 1127, 1208, 1254, 1305, 1306, 1344, 1396, 1695, 1729, 1835, 1844, 1952, 1961, 1973, 2036, and 2045; from GenBank AJ749949 from F. tularensis strain Schu S4) (49), also made in RTS reactions, from our published proteome sets (26, 67). Some experiments used vaccinia virus strain NYCBH ORFs or ORF fragments expressed previously and described as in-frame fusions with β-galactosidase (41). ORF fragments from our published molecular library approach to antigen discovery (41) were expressed as fusions with β-galactosidase and used for comparison to RTS reaction products. Selected ORFs and truncated ORF fragments were newly expressed for this report. In brief, vaccinia virus strain NYCBH genomic DNA was PCR amplified (primer sequences available on request). Amplicons were ligated in frame into pBAD/Myc-His/lacZ (Invitrogen, Carlsbad, CA). E. coli cells harboring the desired plasmids were induced to express β-galactosidase fusion proteins as intracellular inclusions, which were partially purified and used at a final dilution of 1:1,000. For ORFs A3L and A7L (full length) and L4R (selected regions), peptides (typically 13-mers overlapping by 9 amino acids) from the predicted strain Copenhagen amino acid sequences (50) were synthesized (Mimotopes, San Diego, CA, or Sigma, St. Louis, MO) and resuspended at 10 mg/ml in dimethyl sulfoxide (DMSO). Pool-level enzyme-linked immunospot (ELISPOT) assays used mixtures of 20 or 21 peptides at final concentrations of 1 to 2 μg/ml for each peptide and a DMSO concentration of 0.2% or less. Single-peptide ELISPOT and ICC assays used a final concentrations of 1 μg/ml.
Lymphocyte functional assays.
Duplicate proliferation assays were used to probe the microbial ORF panel. U-bottom, 96-well plates contained 2 × 104 cultured bulk responders, 5 × 104 to 1 × 105 autologous PBMC treated with 3,300 rads gamma irradiation as antigen-presenting cells, and RTS protein in 200 μl TCM. Proliferation was assessed by [3H]thymidine incorporation on day 3 (41, 46). In addition to the P. falciparum and F. tularensis ORFs, negative controls included 5 RTS reaction mixtures containing empty vector pXi (identical to pXT7 (17) used for expressing microbial ORFs, 8 RTS reaction mixtures without added DNA, 14 wells with medium, and a UV-treated mock virus BSC-40 cell preparation diluted 1:1,000. The positive control was UV vaccinia virus NYCBH (titer, 2 × 109 PFU/ml) that was UV inactivated and diluted 1:1,000.
ICC was performed as described previously (41). In brief, bulk-cultured vaccinia virus-reactive T-cell lines (2.5 × 105/tube for peptides, 5 × 105 to 1 × 106/tube for whole vaccinia virus) were stimulated with UV-treated vaccinia virus diluted 1:1,000 or 1 μg/ml peptide. Autologous PBMC (obtained at a late postvaccination time point and cryopreserved and thawed prior to use) were added in an equal number as antigen-presenting cells, and stimulation lasted 6 h in medium containing brefeldin A (for the last 5 h), anti-CD49d, and anti-CD28. Cells were surface stained with anti-CD4, permeabilized, and stained with anti-IFN-γ. Data collected on a FACScan or FacsCanto (Becton Dickinson) were analyzed with FlowJo 8 (Treestar, Ashford, OR). Negative and positive controls were UV-treated mock virus, medium, DMSO, and phorbol myristate acetate/ionomycin, respectively (41). Gating selected cells in the lymphocyte forward/side scatter region. Selected cultures were stimulated with UV-inactivated cell-associated HSV-2 strain 333 as described elsewhere (32). For the IFN-γ ELISPOT we used our published procedures (47). For bulk-cultured polyclonal responder cells, 1 × 105 cultured lymphocytes and 1 × 105 autologous PBMC were plated per well. Results are reported as spot-forming units/106 cultured lymphocytes.
Statistical analysis.
For proliferative responses, the positivity cutoff was separately calculated for each 3H proliferation assay data set containing the complete set of responses for each polyclonal lymphocyte line. The raw counts per minute from each well was natural log transformed. For each set, the distribution of the natural log-transformed negative control group (54 wells containing 27 no DNA or empty vector RTS product antigens in duplicate) was tested for normality using the Kolmogorov-Smirnov goodness-of-fit test (SPSS, Chicago, IL). The means and standard deviations of the set of natural log-transformed counts per minute values for all of the negative control wells were calculated. Normality was satisfied in each case (see Results), and therefore the anti-natural log (e raised to the [mean + 3.09·standard deviation]) was set as the threshold for positive calls, corresponding to a 0.1% false positive rate by one-tailed analysis for a normal distribution (NORMDIST function, Excel 2003; Microsoft, Redmond, WA). For a viral ORF to be considered positive, both of the individual [3H]thymidine incorporation duplicate data points were required to be greater than the cutoff. The same normality test was applied to the negative control wells (n = 66, duplicates of 33 antigens) containing P. falciparum or F. tularensis antigens (see Results) and also to the combined control sets containing both the empty vector/no DNA and parasitic/bacterial proteins. The same method was used to calculate a threshold for antibody-positive calls, for each serum specimen, using the IgG microarray raw data (signal intensity). McNemar's test, two-tailed, with continuity correction (GraphPad, San Diego, CA) was used to evaluate the consistency of responses between paired specimens from some subjects by comparing the number of positive responses at the ORF level converting to a negative response with the number converting from negative to positive. Fisher's exact test, two-tailed (GraphPad), was used to evaluate differences between specimens or ORF characteristics. Linear regression was used to analyze relationships between the population prevalence of CD4 responses, grouped into five categories as described in Results, and mean predicted ORF length in amino acids, or percentage of ORFs detected in virions, using the linear regression function in Excel (Microsoft, Redmond, WA). To analyze the relationship between the presence of CD4 responses and virion abundance reported as the weight percent (14), a generalized estimating equation was used with response as the outcome and weight as the predictor. This regression technique accounts for the repeated measures done on individual subjects. Cluster analysis and tree visualization used Cluster 3.1 and TreeView (24).
Antibody responses.
Protein microarrays were printed from RTS proteins (singly) containing each vaccinia virus ORF, according to published procedures (17, 18). All proteins were expressed for 4 to 5 h in vitro (RTS-100 kits; Roche), except for the membrane protein L1R, which was expressed overnight in RTS disulfide kits (Roche) according to the manufacturer's instructions. Expression reactions were printed without purification onto nitrocellulose FAST slides (Whatman, Sanford, ME) within 6 h of the end of the reaction. Arrays were allowed to dry and stored at 18°C in a desiccator. Sera were diluted to 1:50 in protein array blocking buffer (Whatman) containing E. coli lysate at a final concentration of 4 to 5 mg/ml for 1 h to absorb antibodies to E. coli and then used to probe arrays for 18 h at 4°C with constant agitation. Binding of IgG was detected using biotinylated anti-human IgG γ chain (Jackson Immuno, West Grove, PA), followed by strepatividin-PBXL3 fluorophore (Martek, Columbia, MD). Slides were examined on a confocal glass slide scanner (Perkin-Elmer, Waltham, MA). Arrays showed no significant reactivity to the biotinylated secondary antibody and/or streptavidin-PBXL3. Fluorescence intensities were quantified using ScanArrayExpress software (Perkin-Elmer).
RESULTS
Responder cells.
The 21 responder cell lines included matched pairs from nine vaccinated individuals (five with vaccinations 1 month or less before sampling), single samples from two persons, and a negative control HSV-2-reactive cell line from a nonvaccinated subject (Table 1). Cell lines from vaccinia virus-immune donors had brisk responses to whole cell-associated vaccinia virus. The mean (± standard deviation) net cpm [3H]thymidine incorporation for vaccinia virus antigen was 37,478 ± 17,174. Responses to mock antigen were low (<6,000 cpm, and generally less than 1,000 cpm) (Table 1). A mean of 10.7% ± 6.6% (range, 3.2% to 27.5%) of responder CD4 T cells accumulated IFN-γ in response to whole vaccinia virus antigen (after subtraction of background IFN-γ, generally less than 1%) (data not shown). In contrast, a polyclonal HSV-2-reactive T-cell line (48) created from vaccinia virus-naïve, HSV-2-infected subject 14 (Table 1) had net [3H]thymidine incorporation of 266 cpm and 16,309 cpm for whole vaccinia virus and HSV-2 antigens. The CD4 IFN-γ responses measured by ICC were 0% and 9.7% to these viral antigens, respectively.
Validation of vaccinia virus ORFs transcribed and translated in vitro as T-cell antigens.
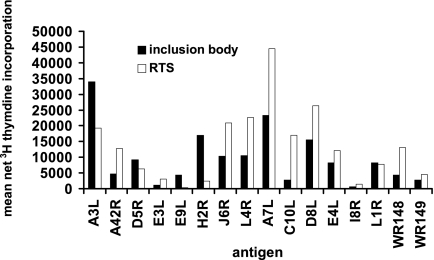
We studied the identity, diversity, and stability of CD4 responses to vaccinia virus by using a protein set covering the vaccinia virus proteome. The RTS set was derived from 180 unique, cloned ORFs that were judged to encode bona fide vaccinia virus proteins in strain WR. Validation experiments were performed to characterize the RTS reaction products and the T cells that react with them. Vaccinia virus ORF RTS reaction products typically have a single predominant band by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining (17). We expected, however, that E. coli proteins and other impurities might stimulate leukocytes. To test this possibility, cryopreserved aliquots of CD4+ polyclonal responder cells from previous research (41) containing T cells with known fine specificities were retested against a subset of the RTS reaction products. For titration, we used a cell line from the early time point from subject 2 (Table 1) and 15 RTS reactions, corresponding to the full-length equivalents of ORF fragments previously shown (41) to be antigenic for this cell line (not shown). Dilutions at 1:100 had toxicity, while 1:1,000 (chosen for subsequent studies) and 1:10,000 dilutions elicited strong [3H]thymidine proliferation for each ORF (data not shown). This cell line was reactive to both 1:1,000 RTS reaction products and to bacterial inclusion bodies containing fragments of the same ORFs known to be antigenic for this donor (Fig. 1).
FIG. 1.
Comparison of proliferative responses of polyclonal vaccinia virus-reactive cell lines to RTS reaction products from full-length vaccinia virus strain WR ORFs, diluted 1:1,000, and corresponding ORF fragments from strain NYCBH expressed as inclusion bodies and diluted 1:100. Proliferative responses are shown for subject 2 (Table 1), a primary vaccinee, at 1 month after vaccination. The mean values for inclusion bodies containing LacZ only or for RTS reactions containing only empty vector were less then 600 cpm and were subtracted to yield mean net [3H]thymidine incorporation.
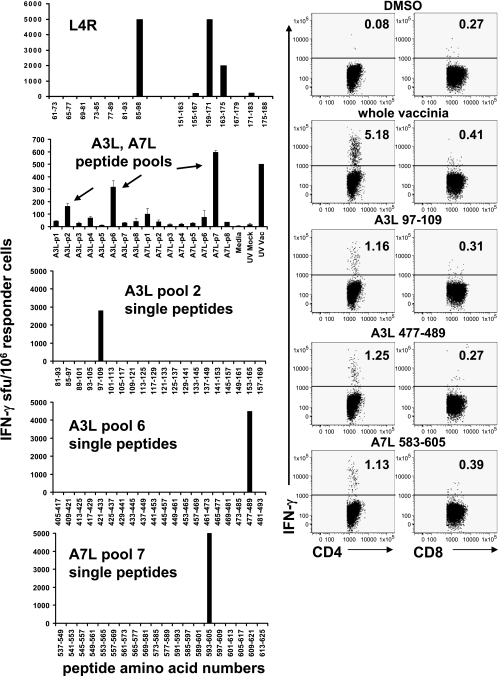
To further characterize the nature of the proliferative responses to RTS proteins, we first confirmed selected T-cell reactivities to the peptide level and determined that the responding lymphocytes had the CD4+ phenotype. For example, ORFs A3L, A7L, and L4R were positive (Fig. 2, left) for subject 9 (Table 1) when polyclonal cells from a blood sample from 48 months after vaccination were tested against the RTS panel. Truncation analyses for ORF L4R using truncated proteins expressed in E. coli and proliferation assays established the presence of at least two antigenic regions as evidenced by step-downs in lymphocyte proliferation with serial truncation (Fig. 2, right). Each predicted reactivity was confirmed with synthetic peptides and IFN-γ ELISPOT (Fig. 3). L4R, peptide 85-98 (DKLTIEAIENYFL), corresponds to the drop-off in reactivity between amino acids 61 to 197 and 91 to 197; likely, the minimal epitope extends N-terminal to amino acid 91. Overlapping peptides 159-171 and 163-175 (INIYKENMESAST and KENMESASTEYTP) were each positive, accounting for the second step-down in between the 151-197 and 181-251 fragments. It is not known if a single epitope is present in the overlap between these peptides. For ORFs A3L and A7L, peptide pools showed that A3L pools 2 and 6 and A7L pool 7 were antigenic. Deconvolution showed that A3L 97-109 (DWSTRLRNDGNAI), A3L 477-489 (ECYTGFRSLIDDT), and A7L 593-605 (FFPPNYKLLKDLF) stimulated strong IFN-γ release.
FIG. 2.
Proliferative responses of polyclonal PBMC-derived vaccinia virus-reactive cells to selected full-length and truncated vaccinia virus proteins. Responder cells are from subject 9, 48 months after vaccination (Table 1). Left: Proliferative responses, recorded as means ± standard deviations of duplicate [3H]thymidine incorporation, to pooled negative control RTS reactions (neg RTS), indicated full-length vaccinia virus RTS reactions, or mock or whole vaccinia virus. Right: Similar proliferative responses to full-length and truncated vaccinia virus ORF L4R expressed as fusions with β-galactosidase within heat-killed E. coli. L4R is predicted to contain 251 amino acids; numbers indicate amino acid coordinates of fragments. Negative controls were medium, β-galactosidase in killed E. coli, and mock virus; the positive control was whole vaccinia virus.
FIG. 3.
Synthetic peptides confirm reactivities assigned using RTS reactions for subject 9, 48 months after vaccination (Table 1). Top left: single peptides correspond to predicted antigenic regions of L4R. Data are means ± standard deviations of duplicate IFN-γ ELISPOT assay results. Second left panel: reactivity to pools of peptides covering ORFs A3L and A7L. Lower left panels: deconvolution of positive pools in A3L and A7L. Right: Reactivity IFN-γ ICC responses to control antigens and to A3L and A7L peptides reactive in the IFN-γ ELISPOT. Cells were gated on CD4+ (left column) or CD8+ (right column) events.
The phenotype of responding cells was then determined with ICC. The ICC protocol involved coincubating PBMC and T-cell lines in equal numbers. A portion of 5.18% of CD4+ cells from this donor reacted to positive control vaccinia virus antigen, while responses to DMSO (negative control) were near zero (Fig. 3, right). Each A3L peptide elicited specific IFN-γ synthesis among CD4+ T cells but not CD4-negative cells (Fig. 3). IFN-γ ICC data for A7L 593-605 and L4R 85-98, 159-171, and 163-175 also showed reactivity by CD4+ but not CD4− cells (not shown).
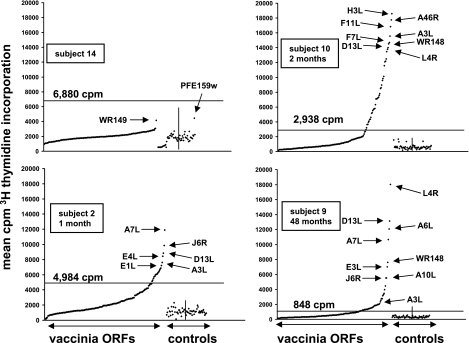
An additional example of CD4 reactivity with an antigen expressed in the RTS format came from subject 10, 2 months after primary vaccination (Table 1), for ORF F11L. We observed strong proliferative reactivity to the full-length ORF (Fig. 4, upper right) and CD4 IFN-γ responses to several F11L peptides (not shown). Both contiguous regions of overlapping F11L peptides (29-41 and 33-45, sequences PKKMNIVTDLENR and NIVTDLENRLKKN, and also peptides 41-53, 45-57, 49-61, and 53-65, sequences RLKKNSYIENTNQ, NSYIENTNQGNIL, ENTNQGNILMDSI, and QGNILMDSIFVST, respectively) and the isolated peptide 21-33 (SILARRPTPKKMN) were positive for this subject.
FIG. 4.
Proliferative responses to RTS reaction products covering a set of 180 PCR-amplified vaccinia virus strain WR ORFs and control antigens (33 P. falciparum and F. tularensis ORFs and 27 additional control wells). Responder cells are polyclonal virus-reactive PBMC-derived cell lines. The left portion of each plot ranks the vaccinia virus ORFs from left to right from lowest to highest mean [3H]thymidine incorporation (of duplicate experiments), indicated by dots, while the right portion shows the control antigens in arbitrary order. The short vertical line separates the no-DNA and empty vector control antigens (n = 27) (left) and the microbial control ORFs (n = 33) (right). The threshold for positive calls, determined as described in Materials and Methods, is indicated by a solid horizontal line. Top left: HSV-2-reactive T-cell line from an HSV-2-seropositive subject who never received vaccinia virus (negative control specimen). The most stimulatory vaccinia virus and negative control proteins are labeled. Top right: responder cells from subject 10 (Table 1), 2 months after primary vaccination. Bottom left: responder cells from subject 2, 1 month after primary vaccination. Bottom right: responder cells from subject 9, 48 months after vaccination. Selected reactive ORFs are labeled for each specimen.
For the subject used for the F11L studies, there was higher background among CD4-negative cells for IFN-γ secretion, due to residual activation from in vitro expansion. However, clear differentiation for the CD4+ population between the mock virus and DMSO negative control stimuli and the vaccinia virus peptides was readily apparent. Isotype control antibody staining showed no difference in staining between negatives control and active peptides. Several other reactivities have been verified by synthetic peptide ELISPOT and/or CD4 IFN-γ ICC (L. Jing et al., unpublished data). The large number of positive human subject/ORF combinations (sample data in Fig. 4) has limited this process from being carried out to completion to date.
To further validate that the proliferative responses to the vaccinia virus proteins made in the RTS format were specific, we tested the proteome-spanning RTS vaccinia virus ORF set with an HSV-2-specific polyclonal T-cell line from a vaccinia virus-naïve subject (Table 1). The mean ± standard deviation [3H]thymidine incorporation for the negative control wells was 1,636 ± 717 cpm. Inspection of the raw data (Fig. 4, upper left) showed that the overall pattern was very different from that obtained for representative vaccinia virus-reactive CD4 T-cell lines (Fig. 4, other panels). A single control ORF (P. falciparum ORF PFE159w; mean, 4,441 cpm) and a single vaccinia virus ORF (WR149; mean, 4,173 cpm) had higher proliferative responses than the other ORFs, but all vaccinia virus ORFs including WR149 were well under the cutoff value (6,880 cpm for this cell line).
Assessment of CD4 responses to vaccinia virus ORFs with a whole-proteome panel.
Taken together, the validation studies support the use of the RTS reaction product panel to screen the vaccinia virus proteome for CD4 T-cell responses. Strong proliferative responses were noted to many individual vaccinia virus proteins (sample data in Fig. 4, upper right and lower panels). The primary goal was to assign each vaccinia virus ORF as positive or negative for each responder cell population. Analyses of the magnitude of proliferative responses are considered less informative due to use of in vitro restimulation prior to the readout assay. An objective cutoff for positive calls was therefore established for each cell line. The negative control data (from wells containing medium or RTS reaction products with no DNA or empty vector substrate, or P. falciparum or F. tularensis ORF substrates; n = 60) were found, for each responder cell line, to be normally distributed. To determine whether the P. falciparum or F. tularensis proteins were driving T-cell responses, we examined data from subsets of the media/no substrate/empty vector substrate wells (n = 27) and of the parasitic/bacterial ORFs (n = 33). Again, each subset had a normal distribution for every responder cell line. Using the media/no substrate/empty vector subset to define a cutoff and a 0.1% false positive tolerance, no specific reactivity with P. falciparum or F. tularensis ORFs was noted for any subject or T-cell line. Given the absence of reactivity to irrelevant microbial proteins, we used the no-DNA and empty vector control sets (54 reactions per cell line) to calculate cutoffs for evaluating vaccinia virus ORFs.
Graphics representing the spectrum of data sets (Fig. 4) show that the cutoff levels appear reasonable when applied to continuously distributed responses (Fig. 4, lower left). Many data sets had “inflection points” in their distributions. At times, the mathematically calculated cutoff levels appeared to overshoot (Fig. 4, upper right) or undershoot (Fig. 4, lower right) these areas of changing slope, while for other specimens (not shown), the cutoff value corresponded very closely to this intuitive breakpoint. Use of the objective thresholds, based on the negative control antigens and individually calculated for each responder cell line, was maintained as a constant throughout our analyses.
Overall, 20 responder cell lines from 11 vaccinees were tested (Table 1). Among the 180 vaccinia virus ORFs studied, proliferative responses were detected in one or more polyclonal responder cell populations to 122 (68%) ORFs. A very large range of diversities was observed, from 2 to 60 ORFs per responder cell line (Table 2). A mean (± standard deviation) of 22.6 ± 15.6 ORFs were positive per cell line. The cell lines were derived at heterogeneous times after the most recent vaccination and from those who received either single or multiple vaccines. Regression analysis showed no overall relationship between the time after most recent vaccination and the number of ORFs recognized. Fisher's test showed no relationship between the number of lifetime vaccinations and the number of ORFs recognized. As work progressed, the format of vaccinia virus used for restimulation was changed from live (5 T-cell cultures) to UV-inactivated (15 cultures) virus (Table 1). This was done for reasons of safety but had the possibility of influencing the diversity or repertoire of the resultant T-cell lines. The number of antigenic ORFs detected in the live virus-stimulated cultures was 41.6 ± 16.3, while for the UV virus-stimulated cultures, 20.3 ± 15.2 ORFs were scored as positive (P = 0.03 by two-tailed Mann-Whitney test).
TABLE 2.
Number of vaccinia virus ORFs recognized by proliferations responses in PBMC from vaccinated subjects, virus stimulation, and relationship between proliferative responses in serial specimens
| Subject no. and stimulusa | Mos. after last vaccination
|
No. of positive ORFs
|
|||
|---|---|---|---|---|---|
| Sample 1 | Sample 2 | Sample 1 | Sample 2 | Sample 1 or 2 | |
| 1: L, UV | 18 | 25 | 53 | 2 | 53 |
| 2: L, UV | 1 | 43 | 18 | 6 | 18 |
| 5: L, UV | 0.5 | 38 | 60 | 17 | 63 |
| 6: UV, UV | 0.5 | 28 | 42 | 8 | 43 |
| 7: L, UV | 1 | 10 | 41 | 6 | 43 |
| 8: UV, UV | 1 | 25 | 23 | 13 | 26 |
| 9: UV, UV | 44 | 48 | 28 | 45 | 58 |
| 10: L, UV | 2 | 30 | 36 | 28 | 42 |
| 11: UV, UV | 6 | 17 | 3 | 23 | 23 |
| 12: UV | 60 | NA | 47 | NA | |
| 13: UV | 1 | NA | 13 | NA | |
Form(s) of vaccinia virus used to generate vaccinia virus-specific T-cell lines: L, live vaccinia virus; UV, UV-inactivated vaccinia virus. For subjects with cell lines generated on two different blood draw dates, the relevant stimuli are listed in sequence. NA, not applicable. See Materials and Methods for details.
Nine of the subjects were sampled twice each, providing an opportunity to examine temporal trends. It was considered biologically plausible that some responses might fall below the detection threshold over time but less likely that new responses would arise late after an infection that has no known persistent or latent state. Addition of a second blood specimen did increase the number of ORFs detected as positive (Table 2). For ORFs considered positive if either (or both) time point was above threshold, a mean ± standard deviation of 39 ± 16.7 (range, 13 to 63) vaccinia virus proteins were recognized per person. The assignments of individual ORFs as positive or negative, comparing the paired cell lines derived from blood specimens from the same subject, were 82.3% concordant. On a per-subject basis, concordance ranged from 71.7% to 93.3%. However, because of the change in the format of vaccinia virus used to enrich vaccinia virus-specific CD4 T cells from live to UV-inactivated viruses for five subjects, we did not attempt detailed analysis of the concordance between the two time points.
Characteristics of antigenic vaccinia virus ORFs.
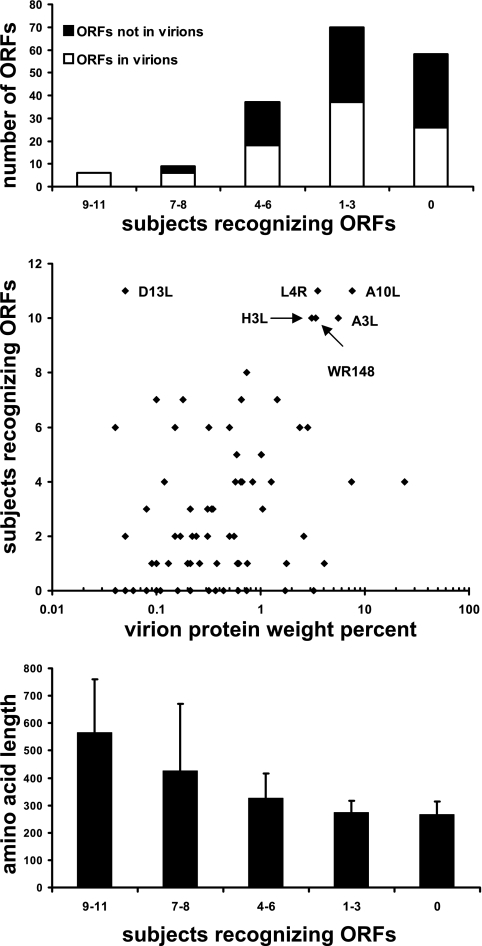
The published literature indicates that CD8 responses may emphasize transcription factors and other proteins expressed with early kinetics (42, 58, 79). The data set from this study was analyzed to determine if CD4 responses were biased toward vaccinia virus ORFs with certain structural or functional characteristics (Table 3; Fig. 5). Vaccinia virus proteins detected in purified virions were abstracted from three proteomics studies (14, 62, 80). Proteins observed in virions in one or more of these studies (93 of 180) were considered to be virion structural proteins for our analysis (Table 4). Among the virion proteins, 67 (72%) were CD4 antigens, compared to 55 of 87 (63.2%) of the ORFs not detected in virions (not significant by Fisher's exact test, two-tailed). To permit a semiquantitative estimate of immunodominance at the population level, vaccinia virus ORFs were placed into five groups. Group 5 ORFs (n = 6) were recognized by 9 to 11 subjects, and group 4 ORFs (n = 9) were recognized by 7 or 8 persons. Among the less frequently reactive ORFs, 37 were in group 3 (four to six persons), and 70 were in group 2 (one to three persons). Group 1 ORFs (n = 58) were recognized by none of the 11 subjects. There was a borderline association between dominance group and presence in virions (Fig. 5, top) by linear regression (r2 = 0.76; P = 0.052). There was no difference between the cultures derived using UV-treated or live vaccinia virus (Table 2) with regards to the number or proportion of structural or nonstructural ORFs with which these cultures showed T-cell reactivity.
TABLE 3.
Vaccinia virus ORFs recognized by CD4 T cells from 82 to 100% subjects (9 to 11 of 11)
| ORFa | Lengthb | Virionc | Promoterd | Human CD8e | Variola identityf | IgG prevalenceg | Comment(s)h |
|---|---|---|---|---|---|---|---|
| A3L | 644 | Yes | L | Yes | 99 | 0 | Capsid core |
| A10L | 891 | Yes | L | Yes | 97 | 10 | Capsid core |
| D13L | 551 | Yes | L | No | 99 | 8 | Membrane scaffold |
| H3L | 324 | Yes | E/L | Yes | 97 | 9 | IMV envelope, heparin binding |
| L4R | 251 | Yes | L | No | 100 | 4 | ssDNA/ssRNA binding; microtubule associated |
| WR148 | 725 | Yes | L | No | 92 | 10 | Fragment of homolog of cowpoxvirus A type inclusion |
Nomenclature from strain Copenhagen, except for WR148, which is from strain Western Reserve; actual PCR amplicons for protein expression were from strain WR (see text). WR148 is related to vaccinia virus Copenhagen gene A26R (see reference 50 for details).
Predicted length in amino acids from genomic information (50).
Yes indicates detection by mass spectroscopy in one or more studies of purified virions (14, 62, 80).
Based on personal communication with E. Lefkowitz, C. Hendrickson, and C. Wang, University of Alabama at Birmingham.
Based on a recent review (43).
Analysis for vaccinia virus Copenhagen (or WR for WR148) and variola Bangladesh 1975 was performed using the methods described in reference 50.
Percentage of the 10 subjects in this report with IgG reactivity to the listed ORF. Subject 11 had no serum samples available for analysis.
From reference 50 and a survey of the literature. IMV, intracellular mature virion; ssDNA, single-stranded DNA; ssRNA, single-stranded RNA.
FIG. 5.
Characteristics of vaccinia virus ORFs, expressed as RTS reaction products, grouped by population prevalence of T-cell recognition. ORFs were considered positive in subjects sampled twice if either time point was positive. Top: presence or absence of vaccinia virus ORFs in virions, determined as described in the text, versus the prevalence group for CD4 responses. Middle: relationship between prevalence of CD4 responses and weight percent of vaccinia virion proteins as reported by Chung et al. (14). The six ORFs with the highest population prevalence of CD4 responses are indicated with arrows. Bottom: length in amino acids (mean ± 1.95 times the standard deviation) of vaccinia virus ORFs versus the prevalence group for CD4 responses.
TABLE 4.
Vaccinia virus ORFs expressed in vitro by RTS reactions in CD4 T-cell and IgG immunologic assays
| ORFa | Virion componentb | No. of subjects (of 11) with proliferative responses |
|---|---|---|
| A1L | 0 | 1 |
| A2L | 0 | 4 |
| A3L | 1 | 10 |
| A4L | 1 | 4 |
| A5R | 1 | 1 |
| A6L | 1 | 6 |
| A7L | 1 | 7 |
| A8R | 0 | 1 |
| A9L | 1 | 0 |
| A10L | 1 | 11 |
| A11R | 1 | 0 |
| A12L | 1 | 3 |
| A13L | 1 | 0 |
| A14.5L | 1 | 0 |
| A14L (2) | 1 | 0 |
| A15L | 1 | 0 |
| A16L | 1 | 1 |
| A17L | 1 | 1 |
| A18R | 1 | 7 |
| A19L | 1 | 5 |
| A20R | 0 | 0 |
| A21L | 1 | 0 |
| A22R | 1 | 0 |
| A23R | 0 | 1 |
| A24R (1) | 1 | 4 |
| A26L/WR149 | 1 | 6 |
| A27L | 1 | 1 |
| A28L | 1 | 0 |
| A29L | 1 | 3 |
| A30L | 1 | 3 |
| A31R | 1 | 2 |
| A32L (1) | 1 | 0 |
| A33R | 1 | 0 |
| A34R | 1 | 3 |
| A35R | 0 | 2 |
| A36R | 0 | 1 |
| A37R | 0 | 0 |
| A38L | 0 | 1 |
| A39R (5) | 0 | 7 |
| A40R | 0 | 0 |
| A41L | 0 | 4 |
| A42R | 1 | 4 |
| A43R | 0 | 0 |
| A44L | 0 | 4 |
| A45R | 1 | 0 |
| A46R | 1 | 6 |
| A47L | 0 | 0 |
| A48R | 0 | 4 |
| A49R | 0 | 1 |
| A50R | 1 | 3 |
| A51R | 0 | 0 |
| A52R | 0 | 1 |
| A53R | 0 | 0 |
| A55R | 0 | 0 |
| A56R | 1 | 2 |
| B1R | 1 | 4 |
| B2R | 0 | 8 |
| B4R | 0 | 2 |
| B5R | 1 | 0 |
| B6R | 0 | 3 |
| B7R (1) | 0 | 2 |
| B8R | 0 | 1 |
| B9R | 0 | 1 |
| B11R (1) | 0 | 0 |
| B12R | 0 | 0 |
| B13R (2) | 0 | 2 |
| B14R | 0 | 0 |
| B15R | 0 | 4 |
| B17R | 0 | 0 |
| B18R | 0 | 0 |
| B19R | 0 | 4 |
| C1L | 0 | 0 |
| C2L | 0 | 0 |
| C3L | 0 | 3 |
| C4L | 0 | 1 |
| C5L | 0 | 3 |
| C6L | 1 | 0 |
| C7L | 0 | 1 |
| C8L | 0 | 3 |
| C9L | 0 | 0 |
| C10L | 0 | 4 |
| C11R | 0 | 0 |
| C12L | 0 | 4 |
| C14L (2) | 0 | 4 |
| C19L | 0 | 1 |
| C22L | 1 | 0 |
| C23L | 0 | 0 |
| D1R | 1 | 3 |
| D2L | 1 | 3 |
| D3R | 1 | 0 |
| D4R | 0 | 0 |
| D5R | 0 | 4 |
| D6R | 1 | 3 |
| D7R | 1 | 0 |
| D8L | 1 | 6 |
| D9R | 0 | 0 |
| D10R | 0 | 0 |
| D11L | 1 | 2 |
| D12L | 1 | 4 |
| D13L | 1 | 11 |
| E1L | 1 | 2 |
| E2L | 0 | 0 |
| E3L | 1 | 6 |
| E4L | 1 | 2 |
| E5R | 0 | 0 |
| E6R | 1 | 1 |
| E7R | 0 | 2 |
| E8R | 1 | 0 |
| E9L | 1 | 0 |
| E10R | 1 | 0 |
| E11L | 1 | 2 |
| F1L | 0 | 5 |
| F2L | 0 | 1 |
| F3L | 0 | 1 |
| F5L | 0 | 3 |
| F6L | 0 | 1 |
| F7L | 0 | 3 |
| F8L | 1 | 1 |
| F9L | 1 | 0 |
| F10L | 1 | 2 |
| F11L | 0 | 5 |
| F12L | 0 | 0 |
| F13L | 1 | 7 |
| F14L | 0 | 0 |
| F15L | 0 | 0 |
| F16L | 0 | 6 |
| F17R | 1 | 4 |
| G1L | 1 | 1 |
| G2R | 0 | 0 |
| G3L | 1 | 1 |
| G4L | 1 | 4 |
| G5R | 1 | 0 |
| G6R | 0 | 5 |
| G7L | 1 | 5 |
| G8R | 0 | 0 |
| G9R | 1 | 0 |
| G5.5R/WR083 | 0 | 6 |
| H1L | 1 | 1 |
| H2R | 1 | 3 |
| H3L (2) | 1 | 10 |
| H4L | 1 | 2 |
| H5R | 1 | 6 |
| H6R | 1 | 0 |
| H7R | 0 | 8 |
| I1L (1) | 1 | 2 |
| I2L | 1 | 8 |
| I3L (1) | 1 | 7 |
| I4L | 0 | 2 |
| I5L | 1 | 1 |
| I6L | 1 | 1 |
| I7L | 1 | 2 |
| I8R | 1 | 4 |
| J1R | 1 | 4 |
| J2R | 0 | 1 |
| J3R (2) | 1 | 5 |
| J4R | 1 | 1 |
| J5L | 1 | 2 |
| J6R | 1 | 7 |
| K1L | 0 | 1 |
| K2L | 0 | 6 |
| K3L | 0 | 6 |
| K4L | 1 | 0 |
| K6L | 0 | 4 |
| K7R | 0 | 2 |
| L1R-ss | 1 | 2 |
| L2R | 0 | 3 |
| L3L | 1 | 1 |
| L4R | 1 | 11 |
| L5R | 1 | 0 |
| M1L | 1 | 3 |
| M2L | 0 | 4 |
| N1L | 1 | 2 |
| N2L | 0 | 3 |
| O1L | 0 | 0 |
| O2L | 1 | 1 |
| WR011 | 0 | 0 |
| WR018 | 0 | 0 |
| WR148 | 1 | 10 |
| WR169 | 0 | 1 |
| WR208 | 0 | 0 |
ORFs were PCR amplified from strain WR. ORF nomenclature is based on that for vaccinia virus strain Copenhagen. GenBank accession numbers, gene sequences, and concordance are available elsewhere (50). Numbers in parentheses following the ORF name indicate the molecular clone used as the template in the RTS reaction.
The relative amounts of vaccinia virus proteins present in purified virions was measured by Chung et al. (14) and expressed as the weight percent or moles percent. We correlated the population prevalence of CD4 responses with these measures of abundance. In general, the frequently recognized ORFs were found to be abundant virion proteins (Fig. 5, middle). Statistical analysis showed a positive relationship between the population prevalence of CD4 responses and virion protein abundance, whether expressed as moles percent (P = 0.0006) or weight percent (P < 0.0001). The exception was D13L, measured as 0.05% (by weight) by Chung et al. (14, 50) yet driving CD4 responses in all 11 subjects. ORF WR148, frequently recognized in our study, was referred to as Copenhagen A25L in the virion proteome study of Chung et al. (14, 50, 74) and is a major component of virions and inclusions in vaccinia virus-infected cells (28). Given the association between virion proteins and late expression kinetics (55), we expected to find that ORFs driving prevalent CD4 responses would be expressed from promoters active later during viral replication. Because not all ORFs have experimentally derived promoter kinetic data, we analyzed predicted promoter activity (Table 3). Five of the six group 5 immunodominant ORFs had late predicted kinetics, while the sixth, H3L, has been shown experimentally to have late expression kinetics (16) despite its predicted early/late combined expression. The relationship between ORF length and CD4 reactivity was also analyzed, because longer proteins have more potential T-cell epitopes and might therefore have a higher prevalence of CD4 responses (Fig. 5, bottom). A direct relationship was noted, with the longer ORFs showing a higher population prevalence of T-cell proliferative responses (r2 = 0.89; P = 0.016 by linear regression).
The vaccinia virus ORFs most frequently recognized by CD4 T cells (Table 3), eliciting proliferative responses for 9 to 11 of the 11 subjects, were A3L, A10L, D13L, H3L, L4R, and WR148. These are all structural virion proteins with late expression kinetics. Integration with a recent tabulation of vaccinia virus ORF T-cell data (43) and a poxvirus bioinformatics resource (50) showed that A3L, A10L, and H3L are also known to contain at least one human CD8 T-cell epitope. Most are highly conserved with variola. A variety of protein functions are represented, including capsid, membrane, scaffold, and nucleic acid binding proteins. H3L is a transmembrane protein that is a known target of neutralizing antibodies (below). I2L, predicted to encode an intracellular mature virion membrane protein, and present and highly conserved in other orthopoxviruses (50), was atypical in being short (73 amino acids) but nonetheless was recognized by 6 of the 11 subjects.
Antibody responses.
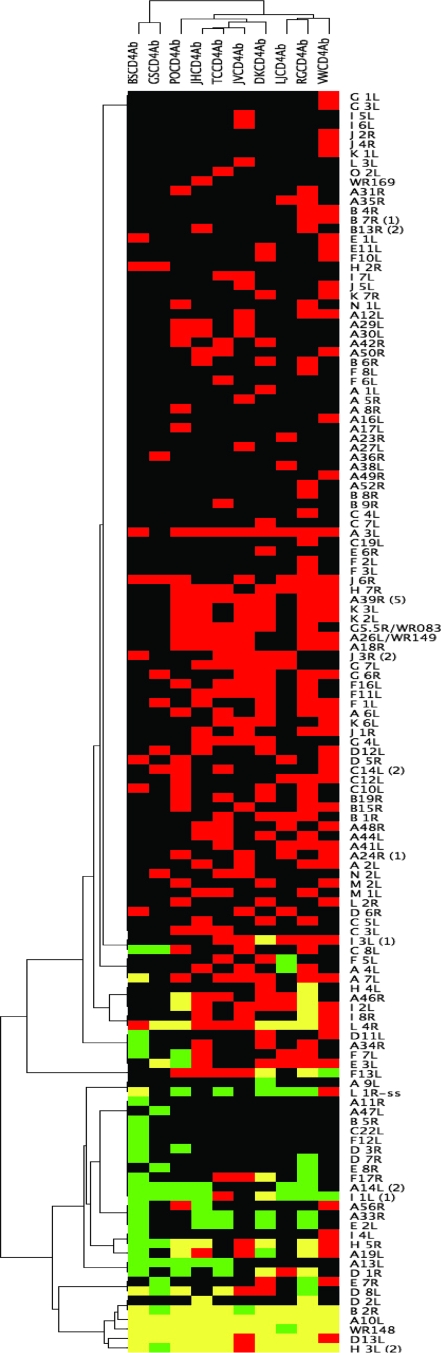
While the mechanisms of protection of smallpox by vaccination with vaccinia virus are unclear, antibodies are detected in humans for decades after vaccination (1, 15, 29, 34, 37, 38, 60, 75). In macaques, vaccinia virus-induced antibodies are necessary and efficient to protect against monkeypox virus infection (22). To investigate the correlation between the CD4 specificity and antibody development, we used protein microarrays spotted with vaccinia virus strain WR proteins, expressed as RTS reaction products, to detect IgG. Prevalent responses (present in 6 or more of the 10 subjects evaluated) were noted for 8 of 180 (4.4%) ORFs: A10L, WR148, B2R, H3L, D13L, I1L, A14L, and H5R (Fig. 6). These dominant antigens are similar to those reported earlier using this approach (17, 18, 66).
FIG. 6.
Cluster analysis of reactivity to vaccinia virus ORFs, expressed as RTS reaction products, for IgG (green) and proliferative T-cell (red) responses for 10 subjects. Yellow indicates the presence of both responses. Subjects are listed in columns, and ORFs are shown in the rows. Cutoffs for positive calls were set as described in Materials and Methods. An ORF was considered positive, for subjects sampled twice, if either or both time points were positive. Annotations for the vaccinia virus ORFs are available elsewhere (50).
In one theory of antigen-specific B-cell development, cognate effector CD4 T-helper-B-cell interactions are important in activating naive B cells in germinal centers. The latter undergo expansion, followed by diversification via somatic mutation and selection of cells secreting high-affinity variants into the long-lived memory B-cell compartment (35). The CD4 and B cells may recognize determinants on the same polypeptide or on physically associated molecules. ORFs that generate antibody responses may therefore also stimulate specific CD4 T-cell responses within the same individual. To analyze this, we merged antibody and CD4 responses for 10 vaccinees (subject 13 [Table 1] did not have serum available). For persons with two samples (Table 1), a response was scored positive if either time point was over the computed threshold. The ORFs with one or more positive calls for IgG, CD4 T cells, or both were grouped using pairwise average-linkage cluster analysis and TreeView (24) (Fig. 6). The antigenic ORFS clustered into several categories. A group of ORFs including A10L, H3L, WR148, D13L, and B2R elicited prevalent (6 or more of 10 subjects) IgG and CD4 responses. Of note, four of these ORFs (A10L, D13L, WR148, and H3L) were among the most frequently recognized CD4 antigens. Overlap between IgG and CD4 responses in four or more subjects was also noted for L4R and H5R. For I1L and A14L, antibody responses were prevalent, but CD4 responses were less frequently detected (Fig. 6). There was a relatively large group of ORFs with moderately prevalent CD4 responses, but mostly undetectable IgG detection (Fig. 6, middle region). A final group of ORFs (Fig. 6, upper part) had sporadic recognition by CD4 T cells and few or no detectable antibody responses.
DISCUSSION
CD4+ T cells play a central role in orchestrating acquired, effector immune responses such as CD8+ cytotoxic T lymphocytes and specific antibody synthesis. In addition, CD4+ T cells can have significant independent effector functions, including granule- and surface-mediated cytotoxicity and antiviral cytokine secretion. In the context of orthopoxvirus infections, CD4+ T cells have been shown to be essential for several aspects of specific CD8 T-cell function, to be required for virus-specific antibodies (5, 6, 64, 77, 78), and to be cytotoxic and secrete IFN-γ (41, 54). CD4 responses to vaccinia virus are remarkably durable in humans, in the absence of any known persistent or latent infection, with a half-life estimated at 8 to 15 years (34).
Relatively little is known concerning the breadth or fine specificity of vaccinia virus-specific CD4 T cells. Mitra-Kaushik et al. found HLA DR1-restricted CD4 epitopes in ORFs A24R and D1R from the predicted proteome of the Ankara modified vaccinia virus strain (54). In our study, these ORFs were recognized by 4 and 3 of 11 persons, respectively. Calvo-Calle et al. (11) found the majority of individuals had responses to ORFs A17L, F16L, A10L, and I6L; we detected proliferative responses in 1, 6, 11, and 6 persons, respectively, out of 11 subjects. Our previous study used a vaccinia virus expression library to detect responses in five vaccinated subjects. CD4 responses were documented to 35 vaccinia virus ORFs, with a diversity of 8 to 20 proteins per person (41). Among the six ORFs with the highest prevalence of responses to the whole ORF set (Table 3), all six were also detected by expression cloning (41). The whole-proteome method in this report captures a broader within-subject CD4 response than that detected with motif-driven peptide or expression library approaches.
The system reported herein is unique in that integrated responses restricted by HLA DRB1, DRB3/4/5, DQB1, and DPB1 loci are interrogated in a single screen, and it bypasses the need for comprehensive proteome-wide peptide synthesis to capture all potential responses. The proliferative responses at the core of our antigenic reactivity assignments (examples in Fig. 4) are highly likely to be CD4 mediated. Each example pursued to the peptide epitope level had the CD4+ phenotype (Fig. 3 and data not shown), and in vitro proliferative responses by memory T cells to exogenous protein antigens were far more efficient for CD4 than for CD8 T cells. The volume of antigen-human subject combinations uncovered with this technology (Fig. 4 and 6) limits the pace of the epitope deconvolution and responder CD4 phenotype confirmation processes. Future studies could deplete CD8 T cells from the responder populations to ensure isolation of CD4 T cells.
The observed within-specimen diversity was high, with a mean of 22, and up to 60 ORFs recognized per blood sample. Most individuals were studied twice, and while some variation was noted between serially collected blood specimens, concordance was much greater than would be expected by chance. By pooling the responses from these paired samples, the mean within-person diversity increased to 39 ORFs with a range of 13 to 63. To our knowledge, this is the highest CD4 diversity measured for the immune response to any infectious agent. We set our cutoff for antigenicity quite conservatively, at a false positive rate of 0.1%. In perhaps the only directly comparable study, Sylwester et al. surveyed the CD4 response to human cytomegalovirus (CMV) using overlapping peptides covering the entire proteome and a direct PBMC IFN-γ ICC readout (68). A median of 12, and up to 39, different CMV ORFs were recognized per subject by CD4 T cells in this format. CMV has a similar genome size and number of ORFs compared to vaccinia virus but differs dramatically in that it causes life-long, chronic, intermittently active infections.
The use of whole vaccinia virus IFN-γ ICC or ELISPOT has generally shown that the global CD4 response is of lower magnitude than the CD8 response in immune humans and mice (36, 78). This conclusion is tempered by the fact that not all pathogen-specific T cells make IFN-γ. Our lab has detected human CD4 clones that proliferate in response to vaccinia virus but do not secrete IFN-γ (41) and has confirmed the finding of Zaunders et al. (81), using ex vivo PBMC ICC, of vaccinia virus-reactive IL-2+ IFN-γ− CD4 T cells. With this caveat, and given the diversity we observe at the ORF level and multiple CD4 antigenic peptides per ORF (Fig. 2) (41), it is rational to conclude that IFN-γ+ CD4 T-cell populations reactive with single vaccinia virus peptides are of low magnitude in the blood. In contrast, CD8 T-cell populations may exceed the threshold required for tetramer detection ex vivo (71). A similar conclusion was reached in C57BL/6 mouse splenocytes using a proteome-wide ORF set and very large peptide sets predicted to bind haplotype-specific major histocompatibility complex class I or II. (56, 57, 73). To address the magnitude hierarchy in humans, the ORF-level “hits” from this study are being decoded to antigenic peptides and HLA-restricting alleles. When a large set is assembled, direct PBMC measurements will be feasible. Peptide-specific responses have been observed in preliminary direct PBMC IFN-γ ELISPOT assays.
Each proteome screen required only four microtiter plates, with about 4 × 107 each responder cells and PBMC and 400 μCi [3H]thymidine. The 20-μl RTS reaction mixtures provided antigen for up to 1,000 screens. The process of enriching microbe-specific CD4 T cells from PBMC or other lymphocyte sources is pathogen specific and becomes more complex if phase variation is present or whole-pathogen preparations are difficult to concentrate or detoxify. We used cell-associated virus, an antigen mixture we have previously shown restimulates memory CD4 T cells specific for both structural and nonstructural vaccinia virus proteins (41). After initial enrichment, the subsequent cycle of polyclonal expansion using anti-CD3 monoclonal antibody and cytokines is typically robust (∼1,000-fold increase in cell number) and is thought to be unbiased with regard to T-cell clonotypes (7). The most resource-intensive step is creation of the initial ORF set, a process requiring sequence information, and potentially the use of cDNA rather than genomic DNA. Efficiency is gained by in vivo ligation and bulk culture selection (26, 67).
With regards to assay sensitivity, CD4 T cells typically react to unmodified linear epitopes. Examples of CD4 epitopes requiring posttranslation modifications, expected to be absent in the RTS antigens, have been published (52, 76) but are rare. In contrast, antibodies that require glycosylation or other adducts, proper disulfide bonds, and tertiary and quaternary structures may fail to bind to the proteins expressed in our system. Recently, we have documented the absolute requirement for disulfide bonding for serological detection of the intracellular mature virion membrane protein using our array technology (19), and further improvements may be made from the use of eukaryotic cell-free expression systems. Ideally, natively folded proteins with full posttranslational processing that are also pure enough for direct PBMC studies would be available for every ORF for T-and B-cell studies, but the technical challenges of this grade of protein expression currently limit throughput. Given these caveats, we have de-emphasized analysis of CD4-B cell response correlation pending availability of an improved reagent set.
The majority (15) of the 20 vaccinia virus-reactive cell lines were made using virus treated with UV light, rendering it replication incompetent in BSC-40 cells. The remaining five cultures were expanded using live virus as part of our earlier CD8 work (42). While even replication-incompetent UV-treated vaccinia virus can express some protein in specific cell lines (27), it was expected that most antigen presentation in the UV-treated vaccinia virus cultures would be via the exogenous/phagocytic pathway. Endogenous presentation might contribute in the cultures stimulated with live virus. Side-by-side comparisons and additional samples will be required to determine the importance of this variable for diversity of fine specificity. We used whole-cell lysates as the source of virus for UV treatment in order to obtain a mixture of proteins including both virion structural and nonstructural proteins. At least some nonstructural proteins were present, as evidenced by detection of reactivity to nonstructural proteins in this and our previous report (41). Each positive call that we have pursued to date has been confirmed at the peptide level. High-throughput methodologies are of necessity a compromise, and future experiments may incorporate antigen standardization based on fusion tag detection to try to reduce variability.
In summary, the CD4 response to vaccinia virus is highly diverse within individuals, with a mean of 39 ORFs being recognized per subject. This is the highest measured diversity for CD4 responses to a viral pathogen. Within our small sample population, the majority of ORFs (68%) were recognized by at least one person. The ORFs recognized most frequently within the population are longer polypeptides that are structural virion antigens with late viral promoters. The relationship between ORF length and population prevalence response is expected if overall epitope density is relatively constant on a per-length basis. ORF I2L was an exception in having a very short predicted length while being quite immunogenic for T cells. Most of the ORFs recognized by 9 to 11 of our 11 subjects were quite abundant in virions, with the apparent exception of D13L. We also found considerable overlap between CD4 antigens and the known set of CD8 antigens (Table 3). The current CD8 and CD4 T cell antigen data sets (56-57, 72) are almost certainly incomplete. The tools studied in this report should speed full definition of the CD4 response to vaccinia virus and related, large-genome viral pathogens.
Acknowledgments
This work was supported by NIH AI061636 and AI067496 (D.M.K.) and U01AI056464, AI058365, and 1U01AI061363 (P.L.F.). The work was also partly supported by the Poxvirus T Cell Vaccine Discovery Consortium (PTVDC) under the Collaboration for AIDS Vaccine Discovery with support from the Bill and Melinda Gates Foundation.
D. M. Koelle and L. Jing are coinventors on a U.S. patent application that has not been issued and has no licensing activity, sought by the University of Washington on vaccinia virus antigens as vaccines. D. M. Molina is employed by ImmPORT Therapeutics, a company that uses RTS technology.
Footnotes
Published ahead of print on 14 May 2008.
REFERENCES
- 1.Amanna, I. J., M. K. Slifka, and S. Crotty. 2006. Immunity and immunological memory following smallpox vaccination. Immunol. Rev. 211320-337. [DOI] [PubMed] [Google Scholar]
- 2.Amara, R. R., P. Nigam, S. Sharma, J. Liu, and V. Bostik. 2004. Long-lived poxvirus immunity, robust CD4 help, and better persistence of CD4 than CD8 T cells. J. Virol. 783811-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashley, R. A., J. Militoni, F. Lee, A. Nahmias, and L. Corey. 1988. Comparison of Western blot (immunoblot) and glycoprotein G-specific immunoblot for detecting antibodies to herpes simplex types 1 and 2 in human sera. J. Clin. Microbiol. 26662-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashton-Rickardt, P. G. 2004. A license to remember. Nat. Immunol. 51097-1098. [DOI] [PubMed] [Google Scholar]
- 5.Bachmann, M. F., K. Schwarz, P. Wolint, E. Meijerink, S. Martin, V. Manolova, and A. Oxenius. 2004. Cutting edge: distinct roles for T help and CD40/CD40 ligand in regulating differentiation of proliferation-competent memory CD8+ T cells. J. Immunol. 1732217-2221. [DOI] [PubMed] [Google Scholar]
- 6.Bachmann, M. F., P. Wolint, K. Schwarz, and A. Oxenius. 2005. Recall proliferation potential of memory CD8+ T cells and antiviral protection. J. Immunol. 1754677-4685. [DOI] [PubMed] [Google Scholar]
- 7.Barcy, S., M. L. Huang, L. Corey, and D. M. Koelle. 2005. Longitudinal analysis of herpes simplex virus-specific CD4+ cell clonotypes in infected tissues and blood. J. Infect. Dis. 1912012-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belyakov, I. M., P. Earl, A. Dzutsev, V. A. Kuznetsov, M. Lemon, L. S. Wyatt, J. T. Snyder, J. D. Ahlers, G. Franchini, B. Moss, and J. A. Berzofsky. 2003. Shared modes of protection against poxvirus infection by attenuated and conventional smallpox vaccine viruses. Proc. Natl. Acad. Sci. USA 1009458-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett, S. R., F. R. Carbone, F. Karamalis, R. A. Flavell, J. F. Miller, and W. R. Heath. 1998. Help for cytotoxic-T-cell responses is mediated by CD40 signalling. Nature 393478-480. [DOI] [PubMed] [Google Scholar]
- 10.Bevan, M. J. 2004. Helping the CD8+ T-cell response. Nat. Rev. Immunol. 4595-602. [DOI] [PubMed] [Google Scholar]
- 11.Calvo-Calle, J. M., I. Strug, M. D. Nastke, S. P. Baker, and L. J. Stern. 2007. Human CD4+ T cell epitopes from vaccinia virus induced by vaccination or infection. PLoS Pathog. 3e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castellino, F., and R. N. Germain. 2006. Cooperation between CD4+ and CD8+ T cells: when, where, and how. Annu. Rev. Immunol. 24519-540. [DOI] [PubMed] [Google Scholar]
- 13.Castellino, F., A. Y. Huang, G. Altan-Bonnet, S. Stoll, C. Scheinecker, and R. N. Germain. 2006. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature 440890-895. [DOI] [PubMed] [Google Scholar]
- 14.Chung, C. S., C. H. Chen, M. Y. Ho, C. Y. Huang, C. L. Liao, and W. Chang. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 802127-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crotty, S., P. Felgner, H. Davies, J. Glidewell, L. Villarreal, and R. Ahmed. 2003. Cutting edge: long-term B cell memory in humans after smallpox vaccination. J. Immunol. 1714969-4973. [DOI] [PubMed] [Google Scholar]
- 16.da Fonseca, F. G., E. J. Wolffe, A. Weisberg, and B. Moss. 2000. Characterization of the vaccinia virus H3L envelope protein: topology and posttranslational membrane insertion via the C-terminal hydrophobic tail. J. Virol. 747508-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies, D. H., X. Liang, J. E. Hernandez, A. Randall, S. Hirst, Y. Mu, K. M. Romero, T. T. Nguyen, M. Kalantari-Dehaghi, S. Crotty, P. Baldi, L. P. Villarreal, and P. L. Felgner. 2005. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl. Acad. Sci. USA 102547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies, D. H., D. M. Molina, J. Wrammert, J. Miller, S. Hirst, Y. Mu, J. Pablo, B. Unal, R. Nakajima-Sasaki, X. Liang, S. Crotty, K. L. Karem, I. K. Damon, R. Ahmed, L. Villarreal, and P. L. Felgner. 2007. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics 71678-1686. [DOI] [PubMed] [Google Scholar]
- 19.Davies, D. H., L. S. Wyatt, F. K. Newman, P. L. Earl, S. Chun, J. E. Hernandez, D. M. Molina, S. Hirst, B. Moss, S. E. Frey, and P. L. Felgner. 2008. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus Ankara is comparable to that of Dryvax. J. Virol. 82652-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong, Y., and T. N. Denny. 2006. HLA-A2-restricted human CD8+ cytotoxic T lymphocyte responses to a novel epitope in vaccinia virus that is conserved among orthopox viruses. J. Infect. Dis. 194168-175. [DOI] [PubMed] [Google Scholar]
- 21.Edghill-Smith, Y., M. Bray, C. A. Whitehouse, D. Miller, E. Mucker, J. Manischewitz, L. R. King, M. Robert-Guroff, A. Hryniewicz, D. Venzon, C. Meseda, J. Weir, A. Nalca, V. Livingston, J. Wells, M. G. Lewis, J. Huggins, S. H. Zwiers, H. Golding, and G. Franchini. 2005. Smallpox vaccine does not protect macaques with AIDS from a lethal monkeypox virus challenge. J. Infect. Dis. 191372-381. [DOI] [PubMed] [Google Scholar]
- 22.Edghill-Smith, Y., H. Golding, J. Manischewitz, L. R. King, D. Scott, M. Bray, A. Nalca, J. W. Hooper, C. A. Whitehouse, J. E. Schmitz, K. A. Reimann, and G. Franchini. 2005. Smallpox vaccine-induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat. Med. 11740-747. [DOI] [PubMed] [Google Scholar]
- 23.Edghill-Smith, Y., D. Venzon, T. Karpova, J. McNally, J. Nacsa, W. P. Tsai, E. Tryniszewska, M. Moniuszko, J. Manischewitz, L. R. King, S. J. Snodgrass, J. Parrish, P. Markham, M. Sowers, D. Martin, M. G. Lewis, J. A. Berzofsky, I. M. Belyakov, B. Moss, J. Tartaglia, M. Bray, V. Hirsch, H. Golding, and G. Franchini. 2003. Modeling a safer smallpox vaccination regimen, for human immunodeficiency virus type 1-infected patients, in immunocompromised macaques. J. Infect. Dis. 1881181-1191. [DOI] [PubMed] [Google Scholar]
- 24.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 9514863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ekkens, M. J., D. J. Shedlock, E. Jung, A. Troy, E. L. Pearce, H. Shen, and E. J. Pearce. 2007. Th1 and Th2 cells help CD8 T-cell responses. Infect. Immun. 752291-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eyles, J. E., B. Unal, M. G. Hartley, S. L. Newstead, H. Flick-Smith, J. L. Prior, P. C. Oyston, A. Randall, Y. Mu, S. Hirst, D. M. Molina, D. H. Davies, T. Milne, K. F. Griffin, P. Baldi, R. W. Titball, and P. L. Felgner. 2007. Immunodominant Francisella tularensis antigens identified using proteome microarray. Proteomics 72172-2183. [DOI] [PubMed] [Google Scholar]
- 27.Fischer, M. A., D. C. Tscharke, K. B. Donohue, M. E. Truckenmiller, and C. C. Norbury. 2007. Reduction of vector gene expression increases foreign antigen-specific CD8+ T-cell priming. J. Gen. Virol. 882378-2386. [DOI] [PubMed] [Google Scholar]
- 28.Funahashi, S., T. Sato, and H. Shida. 1988. Cloning and characterization of the gene encoding the major protein of the A-type inclusion body of cowpox virus. J. Gen. Virol. 6935-47. [DOI] [PubMed] [Google Scholar]
- 29.Gallwitz, S., T. Schutzbank, R. L. Heberling, S. S. Kalter, and J. E. Galpin. 2003. Smallpox: residual antibody after vaccination. J. Clin. Microbiol. 414068-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, A. Pain, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. L. Salzberg, A. Craig, S. Kyes, M. S. Chan, V. Nene, S. J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davis, C. M. Fraser, and B. Barrell. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomez, C. E., J. L. Najera, V. Jimenez, K. Bieler, J. Wild, L. Kostic, S. Heidari, M. Chen, M. J. Frachette, G. Pantaleo, H. Wolf, P. Liljestrom, R. Wagner, and M. Esteban. 2007. Generation and immunogenicity of novel HIV/AIDS vaccine candidates targeting HIV-1 Env/Gag-Pol-Nef antigens of clade C. Vaccine 251969-1992. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez, J. C., W. W. Kwok, A. Wald, C. L. McClurkan, and D. M. Koelle. 2005. Programmed expression of cutaneous lymphocyte-associated antigen amongst circulating memory T-cells specific for HSV-2. J. Infect. Dis. 191243-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halsell, J. S., J. R. Riddle, J. E. Atwood, P. Gardner, R. Shope, G. A. Poland, G. C. Gray, S. Ostroff, R. E. Eckart, D. R. Hospenthal, R. L. Gibson, J. D. Grabenstein, M. K. Arness, and D. N. Tornberg. 2003. Myopericarditis following smallpox vaccination among vaccinia-naive US military personnel. JAMA 2893283-3289. [DOI] [PubMed] [Google Scholar]
- 34.Hammarlund, E., M. W. Lewis, S. G. Hansen, L. I. Strelow, J. A. Nelson, G. J. Sexton, J. M. Hanifin, and M. K. Slifka. 2003. Duration of antiviral immunity after smallpox vaccination. Nat. Med. 91131-1137. [DOI] [PubMed] [Google Scholar]
- 35.Hardtke, S., L. Ohl, and R. Forster. 2005. Balanced expression of CXCR5 and CCR7 on follicular T helper cells determines their transient positioning to lymph node follicles and is essential for efficient B-cell help. Blood 1061924-1931. [DOI] [PubMed] [Google Scholar]
- 36.Harrington, L. E., R. van der Most, J. L. Whitton, and R. Ahmed. 2002. Recombinant vaccinia virus-induced T-cell immunity: quantitation of the response to the virus vector and the foreign epitope. J. Virol. 763329-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrop, R., M. G. Ryan, H. Golding, I. Redchenko, and M. W. Carroll. 2004. Monitoring of human immunological responses to vaccinia virus. Methods Mol. Biol. 269243-266. [DOI] [PubMed] [Google Scholar]
- 38.Hooper, J. W., D. M. Custer, C. S. Schmaljohn, and A. L. Schmaljohn. 2000. DNA vaccination with vaccinia virus L1R and A33R genes protects mice against a lethal poxvirus challenge. Virology 266329-339. [DOI] [PubMed] [Google Scholar]
- 39.Janssen, E. M., N. M. Droin, E. E. Lemmens, M. J. Pinkoski, S. J. Bensinger, B. D. Ehst, T. S. Griffith, D. R. Green, and S. P. Schoenberger. 2005. CD4+ T-cell help controls CD8+ T-cell memory via TRAIL-mediated activation-induced cell death. Nature 43488-93. [DOI] [PubMed] [Google Scholar]
- 40.Janssen, E. M., E. E. Lemmens, T. Wolfe, U. Christen, M. G. von Herrath, and S. P. Schoenberger. 2003. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature 421852-856. [DOI] [PubMed] [Google Scholar]
- 41.Jing, L., T. M. Chong, B. Byrd, C. L. McClurkan, J. Huang, B. T. Story, K. M. Dunkley, L. Aldaz-Carroll, R. J. Eisenberg, G. H. Cohen, W. W. Kwok, A. Sette, and D. M. Koelle. 2007. Dominance and diversity in the primary human CD4 T cell response to replication-competent vaccinia virus. J. Immunol. 1786374-6386. [DOI] [PubMed] [Google Scholar]
- 42.Jing, L., T. M. Chong, C. L. McClurkan, J. Huang, B. T. Story, and D. M. Koelle. 2005. Diversity in the acute CD8 T cell response to vaccinia virus in humans. J. Immunol. 1757550-7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kennedy, R., and G. A. Poland. 2007. T-cell epitope discovery for variola and vaccinia viruses. Rev. Med. Virol. 1793-113. [DOI] [PubMed] [Google Scholar]
- 44.Kit, S., M. Kit, H. Qavi, D. Trkula, and H. Otsuka. 1983. Nucleotide sequence of the herpes simplex virus type 2 (HSV-2) thymidine kinase gene and predicted amino acid sequence of thymidine kinase polypeptide and its comparison with the HSV-1 thymidine kinase gene. Biochim. Biophys. Acta 741158-170. [DOI] [PubMed] [Google Scholar]
- 45.Koelle, D. M., H. B. Chen, M. A. Gavin, A. Wald, W. W. Kwok, and L. Corey. 2001. CD8 CTL from genital herpes simplex lesions: recognition of viral tegument and immediate early proteins and lysis of infected cutaneous cells. J. Immunol. 1664049-4058. [DOI] [PubMed] [Google Scholar]
- 46.Koelle, D. M., L. Corey, R. L. Burke, R. J. Eisenberg, G. H. Cohen, R. Pichyangkura, and S. J. Triezenberg. 1994. Antigenic specificities of human CD4+ T-cell clones recovered from recurrent genital herpes simplex virus type 2 lesions. J. Virol. 682803-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koelle, D. M., Z. Liu, C. L. McClurkan, R. C. Cevallos, J. Vieira, N. A. Hosken, C. A. Meseda, D. C. Snow, A. Wald, and L. Corey. 2003. Immunodominance among herpes simplex virus-specific CD8 T-cells expressing a tissue-specific homing receptor. Proc. Natl. Acad. Sci. USA 10012899-12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koelle, D. M., M. Schomogyi, C. McClurkan, S. N. Reymond, and H. B. Chen. 2000. CD4 T-cell responses to herpes simplex virus type 2 major capsid protein VP5: comparison with responses to tegument and envelope glycoproteins. J. Virol. 7411422-11425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larsson, P., P. C. Oyston, P. Chain, M. C. Chu, M. Duffield, H. H. Fuxelius, E. Garcia, G. Halltorp, D. Johansson, K. E. Isherwood, P. D. Karp, E. Larsson, Y. Liu, S. Michell, J. Prior, R. Prior, S. Malfatti, A. Sjostedt, K. Svensson, N. Thompson, L. Vergez, J. K. Wagg, B. W. Wren, L. E. Lindler, S. G. Andersson, M. Forsman, and R. W. Titball. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37153-159. [DOI] [PubMed] [Google Scholar]
- 50.Lefkowitz, E. J., C. Upton, S. S. Changayil, C. Buck, P. Traktman, and R. M. Buller. 2005. Poxvirus Bioinformatics Resource Center: a comprehensive Poxviridae informational and analytical resource. Nucleic Acids Res. 33D311-D316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu, F., and J. L. Whitton. 2005. Cutting edge: re-evaluating the in vivo cytokine responses of CD8+ T cells during primary and secondary viral infections. J. Immunol. 1745936-5940. [DOI] [PubMed] [Google Scholar]
- 52.Mannering, S. I., L. C. Harrison, N. A. Williamson, J. S. Morris, D. J. Thearle, K. P. Jensen, T. W. Kay, J. Rossjohn, B. A. Falk, G. T. Nepom, and A. W. Purcell. 2005. The insulin A-chain epitope recognized by human T cells is posttranslationally modified. J. Exp. Med. 2021191-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mathew, A., M. Terajima, K. West, S. Green, A. L. Rothman, F. A. Ennis, and J. S. Kennedy. 2005. Identification of murine poxvirus-specific CD8+ CTL epitopes with distinct functional profiles. J. Immunol. 1742212-2219. [DOI] [PubMed] [Google Scholar]
- 54.Mitra-Kaushik, S., J. Cruz, L. J. Stern, F. A. Ennis, and M. Terajima. 2007. Human cytotoxic CD4+ T cells recognize HLA-DR1-restricted epitopes on vaccinia virus proteins A24R and D1R conserved among poxviruses. J. Immunol. 1791303-1312. [DOI] [PubMed] [Google Scholar]
- 55.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 56.Moutaftsi, M., H. H. Bui, B. Peters, J. Sidney, S. Salek-Ardakani, C. Oseroff, V. Pasquetto, S. Crotty, M. Croft, E. J. Lefkowitz, H. Grey, and A. Sette. 2007. Vaccinia virus-specific CD4+ T cell responses target a set of antigens largely distinct from those targeted by CD8+ T cell responses. J. Immunol. 1786814-6820. [DOI] [PubMed] [Google Scholar]
- 57.Moutaftsi, M., B. Peters, V. Pasquetto, D. C. Tscharke, J. Sidney, H. H. Bui, H. Grey, and A. Sette. 2006. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat. Biotechnol. 24817-819. [DOI] [PubMed] [Google Scholar]
- 58.Oseroff, C., F. Kos, H. H. Bui, B. Peters, V. Pasquetto, J. Glenn, T. Palmore, J. Sidney, D. C. Tscharke, J. R. Bennink, S. Southwood, H. M. Grey, J. W. Yewdell, and A. Sette. 2005. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc. Natl. Acad. Sci. USA 10213980-13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pasquetto, V., H. H. Bui, R. Giannino, C. Banh, F. Mirza, J. Sidney, C. Oseroff, D. C. Tscharke, K. Irvine, J. R. Bennink, B. Peters, S. Southwood, V. Cerundolo, H. Grey, J. W. Yewdell, and A. Sette. 2005. HLA-A*0201, HLA-A*1101, and HLA-B*0702 transgenic mice recognize numerous poxvirus determinants from a wide variety of viral gene products. J. Immunol. 1755504-5515. [DOI] [PubMed] [Google Scholar]
- 60.Putz, M. M., I. Alberini, C. M. Midgley, I. Manini, E. Montomoli, and G. L. Smith. 2005. Prevalence of antibodies to Vaccinia virus after smallpox vaccination in Italy. J. Gen. Virol. 862955-2960. [DOI] [PubMed] [Google Scholar]
- 61.Reed, K. D., J. W. Melski, M. B. Graham, R. L. Regnery, M. J. Sotir, M. V. Wegner, J. J. Kazmierczak, E. J. Stratman, Y. Li, J. A. Fairley, G. R. Swain, V. A. Olson, E. K. Sargent, S. C. Kehl, M. A. Frace, R. Kline, S. L. Foldy, J. P. Davis, and I. K. Damon. 2004. The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 350342-350. [DOI] [PubMed] [Google Scholar]
- 62.Resch, W., K. K. Hixson, R. J. Moore, M. S. Lipton, and B. Moss. 2007. Protein composition of the vaccinia virus mature virion. Virology 358233-247. [DOI] [PubMed] [Google Scholar]
- 63.Rotz, L. D., D. A. Dotson, I. K. Damon, J. A. Becher, et al. 2001. Vaccinia (smallpox) vaccination: recommendations of the advisory committee on immunization practices (ACIP), 2001. MMWR Recommend. Rep. 50(RR-10)1-25. [PubMed] [Google Scholar]
- 64.Shedlock, D. J., and H. Shen. 2003. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science 300337-339. [DOI] [PubMed] [Google Scholar]
- 65.Stoeckert, C. J., Jr., S. Fischer, J. C. Kissinger, M. Heiges, C. Aurrecoechea, B. Gajria, and D. S. Roos. 2006. PlasmoDB v5: new looks, new genomes. Trends Parasitol. 22543-546. [DOI] [PubMed] [Google Scholar]
- 66.Stone, J. D., W. E. Demkowicz, Jr., and L. J. Stern. 2005. HLA-restricted epitope identification and detection of functional T cell responses by using MHC-peptide and costimulatory microarrays. Proc. Natl. Acad. Sci. USA 1023744-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sundaresh, S., D. L. Doolan, S. Hirst, Y. Mu, B. Unal, D. H. Davies, P. L. Felgner, and P. Baldi. 2006. Identification of humoral immune responses in protein microarrays using DNA microarray data analysis techniques. Bioinformatics 221760-1766. [DOI] [PubMed] [Google Scholar]
- 68.Sylwester, A. W., B. L. Mitchell, J. B. Edgar, C. Taormina, C. Pelte, F. Ruchti, P. R. Sleath, K. H. Grabstein, N. A. Hosken, F. Kern, J. A. Nelson, and L. J. Picker. 2005. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202673-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang, J., M. Murtadha, M. Schnell, L. C. Eisenlohr, J. Hooper, and P. Flomenberg. 2006. Human T-cell responses to vaccinia virus envelope proteins. J. Virol. 8010010-10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Terajima, M., J. Cruz, A. M. Leporati, W. E. Demkowicz, Jr., J. S. Kennedy, and F. A. Ennis. 2006. Identification of vaccinia CD8+ T-cell epitopes conserved among vaccinia and variola viruses restricted by common MHC class I molecules, HLA-A2 or HLA-B7. Hum. Immunol. 67512-520. [DOI] [PubMed] [Google Scholar]
- 71.Terajima, M., J. Cruz, G. Raines, E. D. Kilpatrick, J. S. Kennedy, A. L. Rothman, and F. A. Ennis. 2003. Quantitation of CD8+ T cell responses to newly identified HLA-A*0201-restricted T cell epitopes conserved among vaccinia and variola (smallpox) viruses. J. Exp. Med. 197927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Terajima, M., and F. A. Ennis. 2006. Using HLA-transgenic mice to identify immunodominant human CD8+ T cell epitopes: does (genome) size matter? Immunol. Lett. 10597-98. [DOI] [PubMed] [Google Scholar]
- 73.Tscharke, D. C., G. Karupiah, J. Zhou, T. Palmore, K. R. Irvine, S. M. Haeryfar, S. Williams, J. Sidney, A. Sette, J. R. Bennink, and J. W. Yewdell. 2005. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J. Exp. Med. 20195-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Upton, C. 2007. Vaccinia orthologs chart. http://www.poxvirus.org/vaccinia_orthologs.asp.
- 75.Viner, K. M., and S. N. Isaacs. 2005. Activity of vaccinia virus-neutralizing antibody in the sera of smallpox vaccinees. Microbes Infect. 7579-583. [DOI] [PubMed] [Google Scholar]
- 76.von Delwig, A., D. M. Altmann, J. D. Isaacs, C. V. Harding, R. Holmdahl, N. McKie, and J. H. Robinson. 2006. The impact of glycosylation on HLA-DR1-restricted T cell recognition of type II collagen in a mouse model. Arthritis Rheum. 54482-491. [DOI] [PubMed] [Google Scholar]
- 77.Wyatt, L. S., P. L. Earl, L. A. Eller, and B. Moss. 2004. Highly attenuated smallpox vaccine protects mice with and without immune deficiencies against pathogenic vaccinia virus challenge. Proc. Natl. Acad. Sci. USA 1014590-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu, R., A. J. Johnson, D. Liggitt, and M. J. Bevan. 2004. Cellular and humoral immunity against vaccinia virus infection of mice. J. Immunol. 1726265-6271. [DOI] [PubMed] [Google Scholar]
- 79.Yewdell, J. W. 2006. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity 25533-543. [DOI] [PubMed] [Google Scholar]
- 80.Yoder, J. D., T. S. Chen, C. R. Gagnier, S. Vemulapalli, C. S. Maier, and D. E. Hruby. 2006. Pox proteomics: mass spectrometry analysis and identification of vaccinia virion proteins. Virol. J. 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zaunders, J. J., W. B. Dyer, M. L. Munier, S. Ip, J. Liu, E. Amyes, W. Rawlinson, R. De Rose, S. J. Kent, J. S. Sullivan, D. A. Cooper, and A. D. Kelleher. 2006. CD127+ CCR5+ CD38+++ CD4+ Th1 effector cells are an early component of the primary immune response to vaccinia virus and precede development of interleukin-2+ memory CD4+ T cells. J. Virol. 8010151-10161. [DOI] [PMC free article] [PubMed] [Google Scholar]