Abstract
The O antigens of outer membrane-bound lipopolysaccharides (LPS) in gram-negative bacteria are oligosaccharides consisting of repeating units with various structures and antigenicities. The O56 and O152 antigens of Escherichia coli both contain a Glc-β1-3-GlcNAc linkage within the repeating unit. We have cloned and identified the genes (wfaP in O56 and wfgD in O152) within the two O-antigen gene clusters that encode glucosyltransferases involved in the synthesis of this linkage. A synthetic substrate analog of the natural acceptor substrate undecaprenol-pyrophosphate-lipid [GlcNAc-α-PO3-PO3-(CH2)11-O-phenyl] was used as an acceptor and UDP-Glc as a donor substrate to demonstrate that both wfgD and wfaP encode glucosyltransferases. Enzyme products from both glucosyltransferases were isolated by high-pressure liquid chromatography and analyzed by nuclear magnetic resonance. The spectra showed the expected Glc-β1-3-GlcNAc linkage in the products, confirming that both WfaP and WfgD are forms of UDP-Glc: GlcNAc-pyrophosphate-lipid β-1,3-glucosyltransferases. Both WfaP and WfgD have a DxD sequence, which is proposed to interact with phosphate groups of the nucleotide donor through the coordination of a metal cation, and a short hydrophobic sequence at the C terminus that may help to associate the enzymes with the inner membrane. We showed that the enzymes have similar properties and substrate recognition. They both require a divalent cation (Mn2+ or Mg2+) for activity, are deactivated by detergents, have a broad pH optimum, and require the pyrophosphate-sugar linkage in the acceptor substrate for full activity. Substrates lacking phosphate or pyrophosphate linked to GlcNAc were inactive. The length of the aliphatic chain of acceptor substrates also contributes to the activity.
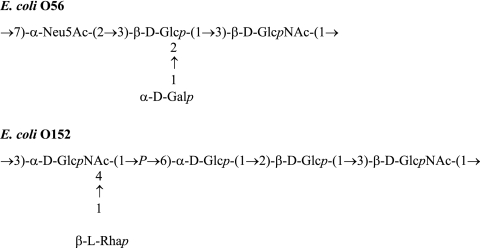
The O antigen (O polysaccharide) is the external part of the lipopolysaccharide (LPS) on the cell surface of gram-negative bacteria. It consists of oligosaccharide repeating units, normally containing two to eight sugar residues within the repeat (http://www.casper.organ.su.se/ECODAB/). Almost 190 serotypes with different O-antigenic oligosaccharide structures have been identified in Escherichia coli and Shigella. O antigens are major contributors to the antigenic variability of the bacterial cell surface; they are important virulence factors and confer resistance to complement-mediated killing (24, 26). Genes involved in O-antigen biosynthesis are normally clustered between galF and gnd in E. coli and Shigella and are classified into three different groups: (i) nucleotide sugar synthesis genes; (ii) glycosyltransferase genes; and (iii) O-antigen processing genes, including the flippase gene (wzx) and polymerase gene (wzy). With few exceptions (25, 27, 35), most of the glycosyltransferases that synthesize E. coli O antigens have not been characterized. The enzymes studied in this work are the glucosyltransferases that catalyze the second step in the synthesis of the O56 (30) and O152 (23) antigens (Fig. 1).
FIG. 1.
The O-antigen structures of E. coli O56 (30) and O152 (23). The O56 O-antigen structure has been reported previously (15) and was corrected by Torgov et al. (30).
GlcNAc-phospho-transferase WecA, encoded by a gene in an enterobacterial common antigen gene cluster, transfers GlcNAc-1-phosphate (or GalNAc-1-phosphate) from UDP-GlcNAc (or UDP-GalNAc) to an undecaprenolphosphate (UndP) carrier and initiates the repeating unit synthesis in many E. coli and Shigella strains (1). The glycosyltransferase genes within the O-antigen gene cluster are involved in the completion of the O repeating unit by addition of sugars to the nonreducing end of the oligosaccharide. These reactions take place at the cytoplasmic side of the inner membrane. There are three distinct processes for synthesis and translocation of O antigen, which are the Wzx/Wzy-dependent process, the ATP-binding cassette (ABC) transporter-dependent process, and the synthase-dependent process (8, 12, 14, 18, 26). Most E. coli and all the Shigella O antigens use the Wzx/Wzy process. The length of the polymer is determined by chain length regulator Wzz. It is known that Wzz associates into oligomers to accomplish the task of controlling oligosaccharide length. Recent research proposes a new model for Wzz function in which the external surfaces of Wzz oligomers at or near the base act as molecular scaffolds for multiple Wzy polymerase molecules (29). In this model, polymerization of an entire oligosaccharide would occur by transferring the growing chain from one Wzy molecule to an adjacent Wzy molecule. The completed polysaccharide would then be transferred by a ligase WaaL from UndP to the outer core structure of lipid A that is assembled simultaneously on the cytoplasmic face of the inner membrane to synthesize LPS. Finally, LPS would be transferred to the outer membrane where it could interact with the external environment. Recent work identified proteins LptA/LptB for transport of LPS from the inner membrane to the outer membrane of E. coli and the Imp/RlpB complex responsible for LPS reaching the outer surface of the outer membrane (28, 34).
Glycosyltransferases are classified into about 90 families (http://afmb.cnrs-mrs.fr/CAZy). The substrates that those glycosyltransferases utilize and the linkages they catalyze are various, and thus only a limited number of them have been functionally identified. Recently, we sequenced the O-antigen gene clusters of E. coli O56 (11) and O152 (our unpublished data). We found that the wfaP gene in the O56-antigen gene cluster and wfgD in the O152-antigen gene cluster are closely related; they share 59% and 46% identity at the DNA and amino acid levels, respectively. Since E. coli O56 and O152 O antigens share only one common linkage, Glc-β1-3-GlcNAc (Fig. 1), it is likely that both WfaP and WfgD are β-1,3-glucosyltransferases responsible for adding the second sugar (a Glc residue) to GlcNAc to form the Glc-β1-3-GlcNAc linkage.
We have previously used a synthetic acceptor substrate to characterize the β-1,3-galactosyltransferase WbbD that is involved in the second step of the synthesis of the E. coli O7-antigen repeating unit (22, 25). The synthetic acceptor substrate, GlcNAc-α-PO3-PO3-(CH2)11-O-phenyl (GlcNAc-PP-PhU), is an analog of the natural undecaprenol-pyrophosphate-sugar acceptor substrate for WbbD and has been shown to be very effective as an acceptor for Gal transfer. In this work, we demonstrate that wfaP of E. coli O56 and wfgD of E. coli O152 both encode β-1,3-glucosyltransferases. To assay and characterize the enzymes, we used GlcNAc-PP-PhU as an acceptor substrate. We found that WfaP and WfgD are similar to each other in their properties and acceptor specificities and similar to the previously characterized β-1,3-galactosyltransferase WbbD (25).
MATERIALS AND METHODS
Materials.
Reagents were obtained from Sigma, unless otherwise stated, or as reported elsewhere (25). Substrates and substrate analogs, as well as GlcNAc-terminating oligosaccharide derivatives, were synthesized as reported previously (6, 7, 22). The synthesis of GlcNAc-pyrophosphate-lipid derivatives will be described elsewhere. GlcNAc-α-PO3-PO3-(CH2)9-CH3 was kindly provided by Ole Hindsgaul, Carlsberg Laboratories, Copenhagen, Denmark.
Bacterial strains, plasmids, and cloning.
Strains and plasmids used in this study are listed in Table 1. Chromosomal DNA was prepared as previously described (3). E. coli O56 wfaP and E. coli O152 wfgD genes were amplified (using primer pairs wl-5344/wl-5345 and wl-5340/wl-5341, respectively) by PCR from strain G1068 and strain G1104, respectively. The primers used are listed in Table S1 in the supplemental material. A total of 30 cycles were performed using the following conditions: denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min, in a final volume of 25 μl. The amplified genes were cloned into pGEX-4T-1 to construct plasmids pLW1232 containing wfaP and pLW1235 containing wfgD (Table 1). The amplified genes (using primer pairs wl-13505/wl-13506 and wl-13507/wl-13508) were also cloned into pET-28a+ to construct plasmids pLW1448 containing wfaP and pLW1449 containing wfgD, which express the proteins without a tag (Table 1). The inserts were sequenced by Tianjin Biochip Corporation using an ABI 3730 sequencer. Plasmids were transformed into E. coli BL21. The strains H1514, H1517, H1827, and H1830 harbored the plasmids pLW1232, pLW1235, pLW1448, and pLW1449, respectively. The strains E. coli DH5α and E. coli BL21 (not containing plasmids) were used as controls.
TABLE 1.
Strains and plasmids used in this study
| Bacterial strain or plasmid | Description | Source |
|---|---|---|
| Strain | ||
| G1068 | E. coli O56 type strain | —a |
| G1104 | E. coli O152 type strain | —a |
| G1488 | E. coli BL21(DE3) F−ompT hsdSB(rB− mB−) gal dcm(DE3) | Novagen |
| H1517 | E. coli BL21 containing pLW1235 | This work |
| H1514 | E. coli BL21 containing pLW1232 | This work |
| H1827 | E. coli BL21 containing pLW1448 | This work |
| H1830 | E. coli BL21 containing pLW1449 | This work |
| G1370 | E. coli DH5α F− φ80lacZ M15 endA recA1 hsdR(rK− mK−) | —b |
| supE44 thi-1 gyrA96 relA1 (lacZYA-argF)U169 | ||
| Plasmid | ||
| pGEX-4T-1 | GST expression vector, Ampr | Amersham |
| pET28a+ | T7 expression vector, Kanr | Novagen |
| pLW1232 | pGEX-4T-1 containing wfaP at BamHI/EcoRI site | This work |
| pLW1235 | pGEX-4T-1 containing wfgD at BamHI/EcoRI site | This work |
| pLW1448 | pET28a+ containing wfaP at NcoI/EcoRI site | This work |
| pLW1449 | pET28a+ containing wfgD at NcoI/EcoRI site | This work |
The Institute of Medical and Veterinary Science, Adelaide, Australia.
Tianjin Biochip Corporation, Tianjin, China.
Bacterial growth.
For the induction of the plasmid-derived enzyme, bacteria were grown overnight at 37°C in 10 ml Luria broth containing 100 μg/ml ampicillin for H1514 and H1517 or 50 μg/ml kanamycin for H1827 and H1830. The bacterial suspension (6 ml) was transferred into 120 ml of Luria broth containing ampicillin or kanamycin and the mixture was incubated for 90 min at 37°C with shaking. Isopropyl-β-d-thiogalactopyranoside (IPTG; 1.2 ml, 100 mM) was added, and the mixture was incubated for another 4 h. Wild-type bacteria were grown without ampicillin, kanamycin, or IPTG. Cells were harvested by centrifugation. Pellets were resuspended in a total of 10 ml phosphate-buffered saline (PBS) and centrifuged again. Ten milliliters of 10% glycerol/PBS was added and stirred. Aliquots of bacterial suspensions were stored at −20°C.
Protein purification.
After IPTG induction, cells were harvested by centrifugation, washed with PBS, resuspended into the same buffer containing 0.1% Triton X-100, and sonicated. The cell debris was removed by centrifugation, and total soluble proteins in the supernatant were collected. The glutathione S-transferase (GST)-tagged fusion proteins in the supernatant were purified with a glutathione Sepharose high-performance (Amersham) column. Bound proteins were eluted with 10 mM reduced glutathione/50 mM Tris-HCl, pH 8.0, and stored in PBS. The fusion protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (8% gels).
Preparation of bacterial membranes for enzyme assays.
To prepare the enzyme source, an aliquot of bacterial suspension (1 ml) was thawed and centrifuged for 5 min at 12,000 rpm in a Centronics M-1200 centrifuge. The supernatant was removed, 500 μl PBS was added, and the suspension was recentrifuged. Five hundred microliters of sonication buffer 1 (10 mM Tris-acetate, pH 8.5, 50 mM sucrose, 1.2 mM EDTA), sonication buffer 2 (50 mM sucrose), or sonication buffer 3 (10 mM Tris-acetate, pH 8.5, 50 mM sucrose) was added and bacteria were sonicated twice with a sonic dismembrator M 100 (Fisher Scientific) for 15 s at setting 3, with 2 min of cooling on ice in between. The sonicate was diluted 6 to 10 times in the same sonication buffer. Protein concentration was assayed by the Bradford method (Bio-Rad), using bovine serum albumin as the standard.
Enzyme activity assays.
The standard incubation mixture contained the following in a total volume of 40 μl: 0.2 mM acceptor substrate GlcNAc-PP-PhU, 10% methanol, 5 mM MnCl2, 75 mM morpholineethanesulfonic acid (MES) buffer, pH 7, 10 μl sonication buffer, 0.2 mM UDP-[14C]Glc (2,000 to 6,000 cpm/nmol), and 10 μl enzyme sonicate diluted in sonication buffer (containing 1 to 12 μg protein). Control assays did not contain acceptor substrate. All assays were carried out in at least duplicate determinations. Results were calculated from assays with less than ±5% variation. Mixtures were incubated for 10 min at 37°C. Cold water (200 μl) was added and tubes were stored at −20°C. Water (500 μl) was added and mixtures applied to regenerated Sep-Pak C18 columns. Columns were washed with 4 ml water. Subsequently, fractions were eluted once with 1 ml water and three times with 1 ml methanol. The enzyme product in the methanol fractions was quantified by scintillation counting.
Production and analysis of enzyme products.
Radioactive products from fivefold scaled-up assays were isolated by the C18 Sep-Pak procedure. The first 2 ml of the methanol eluate were combined. An aliquot was counted by scintillation counting and characterized by high-pressure liquid chromatography (HPLC) and mass spectrometry. For nuclear magnetic resonance (NMR) analysis, large-scale nonradioactive products were produced by scaling up the small-scale assay 50 times, using nonradioactive UDP-Glc as the donor substrate. After the 10-min incubation, mixtures were centrifuged at 12,000 rpm for 6 min in a Centronics M-1200 centrifuge. The supernatant was further processed by C18 reversed-phase HPLC. The product was eluted at 1 ml/min with acetonitrile/water mixtures (10/90) as the mobile phase (25). Aliquots of pooled fractions were subjected to electrospray ionization mass spectrometry (negative ionization mode) using an Applied Biosystems/MDS QStar XL QqTOF mass spectrometer. For analysis by NMR, products were repeatedly dried in the presence of 99.96% D2O, dissolved in CD3OD, and analyzed by 600-MHz proton-NMR, using a Bruker-Avance instrument.
RESULTS
Overexpression and purification of enzymes.
The SDS-PAGE analysis indicated that most of the two recombinant proteins stayed in the membrane fraction after IPTG induction, and the amount of soluble recombinant protein was not enough for enzyme activity assays. To solubilize the protein, 0.1% Tritox-X100 was added in the process of protein purification. After purification, the apparent molecular masses of WfaP and WfgD (both with GST-tag) revealed by the SDS-PAGE analysis were 55 and 55.4 kDa, respectively, correlating well with the predicted molecular masses (54.7 and 54.9 kDa, respectively). However, the purified proteins had no enzyme activity. We also tried to purify the proteins by adding 0.1%, 0.5%, and 1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) or octyl-β-d-glucoside, but neither of the compounds improved the solubilization of the fusion proteins. These results suggested that the native enzymes were associated with membranes which may be essential for their function. It is also possible that the proteins were denatured by the purification procedure (see Discussion).
Enzyme reaction catalyzed by WfaP and WfgD.
Since both WfaP and WfgD are associated with the membrane, the enzyme assays were carried out using bacterial membranes as the source of the enzyme. The reaction progress curves of WfaP and WfgD (with GST-tag) are shown in Fig. 2. The specific activity of WfaP was 0.91 μmol/h/mg and of WfgD was 1.26 μmol/h/mg in the standard glucosyltransferase assay, using 50 mM sucrose for sonication and 1:6 dilution of the enzyme sonicate. Glc transfer was proportional to protein concentration (up to at least 1 μg protein in the assay) and linear with time (up to at least 10 min of incubation time). Both enzymes were fully active in different buffers used for sonication, i.e., buffer 1 (10 mM Tris-acetate, pH 8.5, 50 mM sucrose, 1.2 mM EDTA), buffer 2 (50 mM sucrose), or buffer 3 (50 mM sucrose, 10 mM Tris-acetate, pH 8.5). The control strains DH5α and BL21 were completely inactive in glucosyltransferase assays, indicating that the high glucosyltransferase activity in transformed cells was plasmid derived.
FIG. 2.
Dependence of activity on incubation time. Glucosyltransferase activity was assayed using UDP-[14C]Glc as the donor substrate and GlcNAc-PP-PhU as the acceptor substrate at conditions described in Materials and Methods. (A) Progress curve of WfaP. (B) Progress curve of WfgD.
We were unable to determine the accurate Km and Vmax values for the standard acceptor substrate GlcNAc-PP-PhU due to its hydrophobic character. It was clear, however, that a Vmax was not reached at 0.2 mM acceptor concentration in the assay. The kinetic parameters for UDP-Glc were established using ENZFITTER and were found to be similar for the two enzymes. For WfaP and WfgD, the apparent Km values were 0.5 and 0.12 mM, respectively, and the Vmax values were 2.2 and 0.18 μmol/h/mg, respectively. To test the effect of the GST domain on the enzyme activity, enzymes without GST-tag were also examined. The Km and Vmax values of WfaP and WfgD without GST-tag were 0.5 and 0.14 mM and 1.5 and 0.13 μmol/h/mg, respectively, very similar to the values of the corresponding GST-tagged enzymes. The GST-tagged enzymes showed relatively higher activity, suggesting that the presence of GST may offer some protection to the enzymes from being degraded.
Analysis of glucosyltransferase products.
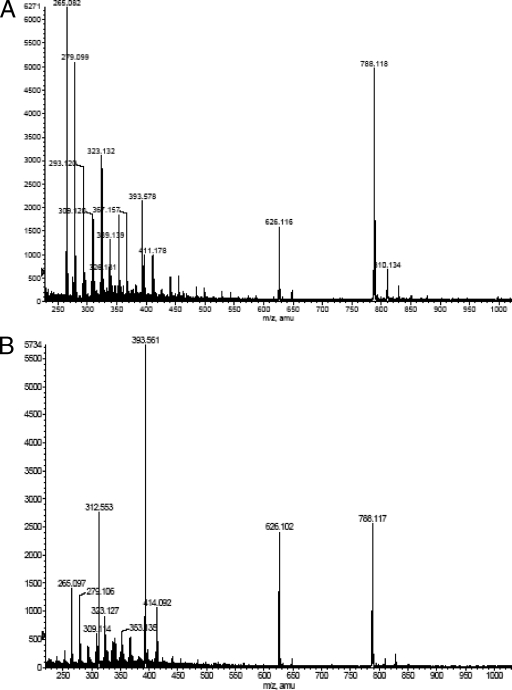
Enzyme products were isolated by reversed-phase HPLC. The matrix-assisted laser desorption ionization mass spectra of acceptor substrate GlcNAc-PP-PhU showed the expected mass/charge (m/z) of 626, corresponding to a molecular weight of 626. For the isolated enzyme product of WfaP, an m/z of 788, corresponding to Glc-GlcNAc-PP-PhU, and an m/z of 829, corresponding to Glc-GlcNAc-PP-PhU (plus K) (Fig. 3A) were found. Similarly, for the enzyme product of WfgD, m/z 788 and m/z 829 were found (Fig. 3B). This indicates that both glucosyltransferases added one Glc residue to GlcNAc-PP-PhU to form Glc-GlcNAc-PP-PhU.
FIG. 3.
Electospray mass spectrometry (negative ion mode) of WfaP and WfgD products. Products of WfaP and WfgD were produced as described in Materials and Methods, isolated by the Sep-Pak C18 procedure and HPLC, and analyzed by electrospray mass spectrometry (negative ion mode). (A) WfaP product. (B) WfgD product.
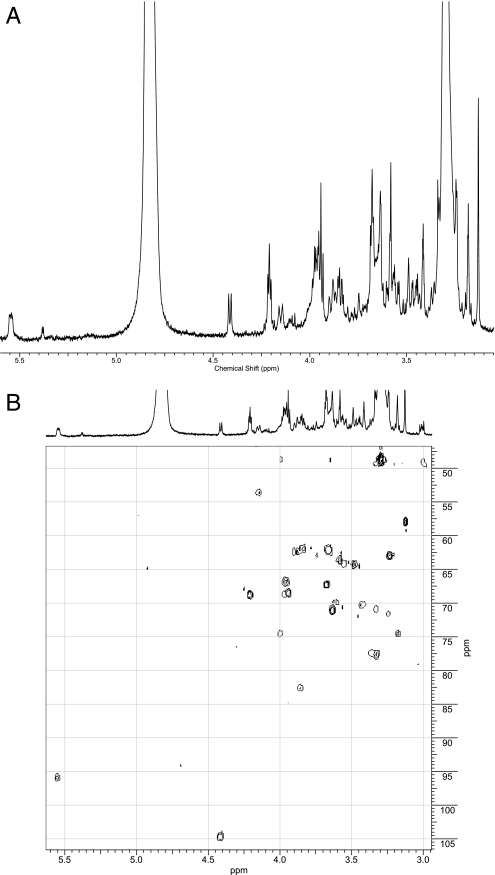
The 600-MHz NMR spectra of the products were compared to the spectrum of the substrate GlcNAc-PP-PhU. Both WfaP and WfgD products showed identical signals arising from Glc-GlcNAc-PP-PhU (Table 2), which differed from those in the acceptor substrate (Table 3). In the proton NMR spectrum of the WfaP product (Fig. 4), significant differences in chemical shifts between substrates and products were observed at H-2 (shift from 3.97 in the substrate to 4.14 ppm in the product), H-3 (from 3.75 to 3.86 ppm), and H-4 (from 3.33 to 3.42 ppm) of GlcNAc. In addition, the signals of a newly added β-linked Glc were seen (H-1, 4.41 ppm, J1,2 = 7.7 Hz; H-2, 3.18 ppm; H-3, 3.37 ppm; H-4, 3.33 ppm; 2H-6, 3.67 and 3.89 ppm). Similar shifts were observed in the WfgD product (Fig. 5; Table 2). The carbon shift in the HSQC (heteronuclear single-quantum coherence) spectrum showed that C-3 of GlcNAc in the WfaP was more deshielded than in the substrate, indicating the Glc-β1-3-GlcNAc linkage (shifting from 72.9 ppm in the substrate to 82.9 ppm in the product). In the WfgD product, both HSQC and ROESY (rotating-frame Overhauser effect spectroscopy) spectra confirmed the deshielding of C-3 of GlcNAc, due to the Glc-β1-3-GlcNAc linkage. These data indicate that both glucosyltransferases synthesized the Glc-β1-3-GlcNAc linkage.
TABLE 2.
NMR parameters of WfaP and WfgD products Glc-GlcNAc-PP-PhUa
| Position | δ H (ppm) (multiplet)
|
J (Hz)
|
δ C (ppm) from HSQC
|
|||
|---|---|---|---|---|---|---|
| WfaP | WfgD | WfaP | WfgD | WfaP | WfgD | |
| GlcNAc 1 | 5.54 | 5.54 | 6.6, 2.8* | 6.6, 2.8 | 95.8 | 94.4 |
| GlcNAc 2 | 4.14 | 4.14 | 11.0 | 11.0 | 53.8 | 53.7 |
| GlcNAc 3 | 3.86 | 3.87** | 82.9 | 82.7 | ||
| GlcNAc 4 | 3.42 | 3.44 | 70.4 | 70.3 | ||
| GlcNAc 5 | 4.00 | 4.00 | 74.8 | 74.6 | ||
| GlcNAc 6 | 3.66 | 3.66, 3.89* | 62.2 or 67.4* | 62.5 or 67.4* | ||
| Glc-1′ | 4.41 | 4.41 | 7.7 | 7.7 | 104.7 | 104.6 |
| Glc-2′ | 3.18 | 3.18 | 74.5 | 74.5 | ||
| Glc-3′ | 3.37 | 3.37** | 77.5 | 77.6 | ||
| Glc-4′ | 3.33 | 3.33 | 71.0 or 77.8* | 70.9 or 77.7* | ||
| Glc-5′ | 3.33* | 3.33* | 71.0 or 77.8* | 70.9 or 77.7* | ||
| Glc-6′ | 3.67 | 3.67 | 62.1 | 62.0 | ||
| 3.89 | 3.85 | |||||
Parameters were derived from 600-MHz proton NMR spectra, including COSY (correlation spectroscopy) and HSQC. The reference was CD3OH proton adjusted to 3.30 ppm and carbon adjusted to 49.0 ppm. *, shifts are uncertain; **, shifts were confirmed by ROESY.
TABLE 3.
Proton and C-13 NMR parameters of enzyme substrate GlcNAc-PP-PhUa
| Position | δ H (ppm) (multiplet) | δ C-13 (ppm) |
|---|---|---|
| GlcNAc 1 | 5.53* (J = 2.7 Hz) | 96.0 |
| GlcNAc 2 | 3.97 | 55.0 |
| GlcNAc 3 | 3.75 | 72.9 |
| GlcNAc 4 | 3.33 | 71.9* |
| GlcNAc 5 | 3.96 | 74.6 |
| GlcNAc 6 | 3.66 | 60.9 |
| PO-CH2 | 3.97 | 67.0 |
| CH2 | 1.63 | 31.5 |
| CH2 | 1.38 | 26.5 |
| CH2 … n | 1.31 | 30.3 |
| CH2 | 1.46 | 26.9 |
| CH2 | 1.77 | 30.0 |
| CH2-OPh | 3.94 | 68.6 |
| Ph-O | 6.88 | |
| m | 7.23 | |
| p | 6.87 | |
| N-Acetyl | 2.05 | 22.7 |
Parameters were derived from 600-MHz proton NMR spectra, COSY, TOCSY (total correlation spectroscopy), NOESY (nuclear Overhauser effect spectroscopy), ROESY, and proton-carbon correlation HSQC. The reference was CD3OH proton adjusted to 3.30 ppm and carbon adjusted to 49.0 ppm. Ph, phenyl; *, shifts are uncertain.
FIG. 4.
Six hundred-megahertz proton NMR spectrum of the WfaP product. The enzyme product of WfaP was isolated as described in Materials and Methods and analyzed by 600-MHz NMR. (A) Proton NMR spectrum. (B) HSQC spectrum.
FIG. 5.
Six hundred-megahertz proton NMR spectrum of the WfgD product. The enzyme product of WfgD was isolated as described in Materials and Methods and analyzed by 600-MHz NMR. (A) Proton NMR spectrum. (B) HSQC spectrum.
Properties of WfaP and WfgD.
The stability of the enzymes was tested after storage of the sonicates at 4°C and at −20°C. In both sonication buffer 1 (10 mM Tris-acetate, pH 8.5, 50 mM sucrose, 1.2 mM EDTA) and sonication buffer 2 (50 mM sucrose), the enzyme activities were stable for several days at 4°C as well as for several weeks after storage at −20°C.
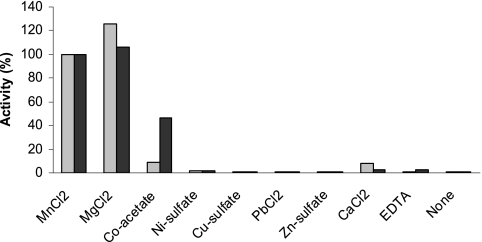
In order to determine the requirement of the enzymes for divalent metal ions, MnCl2 was substituted in the assay by salts of divalent metal ions or EDTA, or it was omitted (Fig. 6). Both MnCl2 and MgCl2 were efficient cofactors at 5 mM concentration in the assay. The enzyme was fully active at MnCl2 concentrations between 0.5 and 25 mM (data not shown). Cobalt(II)-acetate supported glucosyltransferase activity but much less than with MnCl2. The addition of 5 mM Co2+ also reduced the activation by 5 mM MnCl2 of WfaP (by 76%) and of WfgD (by 45%). None of the other metal ion salts (Ni2+, Cu2+, Pb2+, Zn2+, and Ca2+) significantly supported the glucosyltransferase activities. The enzymes were inactive in the absence of MnCl2 or with the inclusion of EDTA.
FIG. 6.
Metal ion requirements of WfaP and WfgD. Glucosyltransferase assays were performed as described in Materials and Methods, using 50 mM sucrose as the sonication buffer with 0.25 mM acceptor concentration and without the addition of MnCl2 in the assay. Different metal ions (from 0.1 M stock solutions adjusted to pH 6) were then added to the assays at 5 mM concentrations, as indicated. One hundred percent activity for WfaP corresponded to 1.26 μmol/h/mg and for WfgD to 0.91 μmol/h/mg. Light gray bars, WfaP; black bars, WfgD activity.
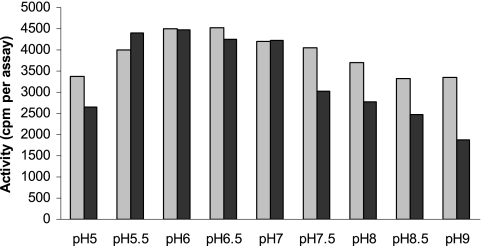
Both enzymes were active with assay buffers at pH values between 5 and 9 (Fig. 7). The optimum pH was found to be pH 6.5 for WfaP and pH 6 for WfgD.
FIG. 7.
Dependence of glucosyltransferase activities on buffer pH. Glucosyltransferase assays were performed as described in Materials and Methods, using 50 mM sucrose as the sonication buffer and 0.5 mM acceptor concentration in the assay. Buffers were added at different pH values (MES from pH 5 to 7 and Tris-HCl from pH 7 to 9.5). Activities are indicated as radioactivity of enzyme product per assay. Light gray bars, WfaP; black bars, WfgD activity.
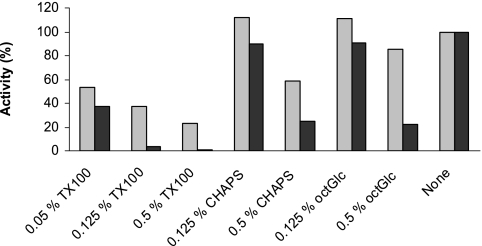
Detergents were added to the assay mixtures to determine their effects on glucosyltransferase activities (Fig. 8). At 0.125% in the assay, CHAPS had minor effects, but at 0.5% in the assay it inhibited WfaP by 41% and WfgD by 75%. Similarly, octyl-β-d-glucoside at 0.125% in the assay had little effect, but at 0.5% concentration it reduced WfaP activity by 15% and WfgD activity by 78%. Triton X-100 was more effective in reducing the activities, although WfaP was distinctly more resistant than WfgD to the deactivating effect of the detergent. The results suggest that detergents can inactivate the enzymes but that the concentrations and types of detergents are critical in this process.
FIG. 8.
Effect of Triton X-100 on glucosyltransferase activities. Glucosyltransferase assays were performed as described in Materials and Methods, using sonication buffer 1 (10 mM Tris-acetate, pH 8.5, 50 mM sucrose, 1.2 mM EDTA) and 0.25 mM acceptor concentration. Triton X-100 was added at the indicated assay concentrations. TX100, Triton X-100; octGlc, octyl-β-d-glucoside. Light gray bars, WfaP; black bars, WfgD activity.
Substrate specificities of WfaP and WfgD.
The acceptor substrate specificities of glucosyltransferase activities were tested with a series of synthetic acceptor substrate analogs (Table 4). The presence of the pyrophosphate linkage in the acceptor substrate was shown to be required for full activity of both WfaP and WfgD. An acceptor analog containing a single phosphate group (compound 604) showed 11% activity for WfgD and 1% activity for WfaP. Compounds lacking the pyrophosphate and having GlcNAc in either an α- or β-linkage were inactive (compounds 618 and 619, and GlcNAc-β-benzyl). Modification of the pyrophosphate group (compound 623) or replacement of pyrophosphate by malonate (compound 621) also produced inactive substrates.
TABLE 4.
Acceptor substrate specificities of WfaP and WfgDa
| Compound no. | Concentration (mM) | Acceptor substrate | Activity (%)
|
|
|---|---|---|---|---|
| WfaP | WfgD | |||
| 580 | 0.2 | GlcNAc-PP-PhU | 100 | 100 |
| 604 | 0.2 | GlcNAc-α-PO3-(CH2)11-OPh | 1 | 11 |
| 615 | 0.2 | GlcNAc-α-PO3-PO3-(CH2)6-OPh | 60 | 183 |
| 617 | 0.2 | GlcNAc-α-PO3-PO3-(CH2)16-OPh | 156 | 174 |
| 618 | 0.2 | GlcNAc-α-(CH2)11-OPh | <1 | <1 |
| 619 | 0.2 | GlcNAc-β-(CH2)11-OPh | <1 | <1 |
| 621 | 0.2 | GlcNAc-α-CO-CH2-CO-O-(CH2)11-OPh | <1 | <1 |
| 623 | 0.2 | GlcNAc-α-CO-CH2-PO3-(CH2)11-OPh | <1 | <1 |
| 628 | 0.2 | GlcNAc-α-PO3-PO3-(CH2)9CH3 | 32 | 150 |
| 2 | GlcNAc-β-Bn | <1 | 1 | |
| 612 | 2 | 1-Thio-N-butyrylGlcN-β-(2-naphthyl) | <1 | <1 |
| 580 + 612 | 0.2 + 0.5 | 94 | 96 | |
Glucosyltransferase assays were performed as described in Materials and Methods. The compounds indicated were used as acceptor substrates in the standard assays using sonication buffer 1. Ph, phenyl; Bn, benzyl.
The role of the hydrophobic moiety of the acceptor was studied using compounds with various lengths of the lipid chain. Chains of 6 carbons (compound 615) and 16 carbons (compound 617) in the acceptor supported activity. Compound 628 lacking the O-phenyl group and having a 10-carbon chain was 150% active with WfgD and 32% active with WfaP, compared to activity with the standard substrate GlcNAc-PP-PhU (Table 4). This shows that the phenyl group in the acceptor is not required for glucosyltransferase activity and that the length of the lipid chain influences the activity.
Compound 612 (N-butyryl-glucosamine-β-1-thio-2-naphthyl), which is an effective inhibitor of mammalian β4-galactosyltransferase (6), was not a substrate for glucosyltransferase. When tested at 0.5 mM concentration in the standard assay, no inhibition of either WfaP or WfgD activity was observed, showing that compound 612 did not compete with the substrate for binding to the enzyme.
The donor substrate specificity of glucosyltransferase was examined by replacing UDP-Glc with a number of other nucleotide sugars in the assay. No activity was observed using CMP-sialic acid, UDP-GlcNAc, UDP-GalNAc, or GDP-Man as the donor substrate. However, when UDP-Gal was used in the standard assay for glucosyltransferase, a low activity was noted. Compared to the glucosyltransferase activities, galactosyltransferase activities were only 0.6% (WfaP) and 0.2% (WfgD). Thus, galactosyltransferase activities in bacterial homogenates containing recombinant glucosyltransferase were relatively low, showing a distinct preference for UDP-Glc sugar donor substrate of both glucosyltransferases.
DISCUSSION
Glycosyltransferases are ubiquitous in eukaryotic and bacterial systems and are involved in the biosynthesis of protein-bound oligosaccharides, polysaccharides, teichoic acids, saccharolipids, and many other complex oligosaccharides. About 90 glycosyltransferase families have been identified (http://afmb.cnrs-mrs.fr/CAZy) by amino acid sequence similarity, and members of a glycosyltransferase family are predicted to have the same overall fold structure (4).
The majority of glycosyltransferase families belong to only two structural superfamilies, named GT-A and GT-B. The folding of the GT-A superfamily is characterized by an α/β/α sandwich, with several different strands in the central β-sheet, which has been confirmed by X-ray analysis of crystal structures of several mammalian and bacterial glycosyltransferases (4, 5, 9, 31). Glycosyltransferases in this superfamily often have a DxD motif to bind the divalent metal ion that has been shown to be involved in catalysis, for example, of mammalian GlcNAc-transferase I (31). Bacterial glycosyltransferases with the GT-A fold also have DxD motifs, such as WbbD which transfers the second sugar residue in the assembly of the O7 antigen (21). The GT-B superfamily contains two separate Rossmann domains with a connecting linker region and a catalytic site between the domains. The GT-B superfamily employs a different mechanism to catalyze glycosyltransfer and does not require a metal ion for activity; examples include the identified bacteriophage T4 β-glucosyltransferase that glucosylates DNA (33) and the α-1,3-N-acetylgalactosaminyl transferase WbnH involved in the second step of the assembly of the O86-antigen repeating unit (36).
WfaP and WfgD share a significant but low degree of similarity with many other prokaryotic enzymes but are clearly distinct from mammalian glucosyltransferases. Analysis of the sequences and comparison within the CAZy (carbohydrate-active enzymes) families of glycosyltransferases suggest that WfaP and WfgD could be associated with glycosyltransferase families GT2, -12, -21, -27, and -78 (http://afmb.cnrs-mrs.fr/CAZy). The folding of these two enzymes is expected to be consistent with a GT-A fold (Christelle Breton, personal communication). Both WfaP and WfgD have DxD motifs (see Fig. S1 in the supplemental material) that may serve as nucleophiles in catalysis and are dependent on a divalent metal ion for activity. Abundant short sequences in WfaP and WfgD containing Lys, Arg, and His (see Fig. S1 in the supplemental material) may also be used for binding to the negatively charged donor and acceptor substrates.
Many bacterial glycosyltransferases are found to be integral membrane proteins. For example, GtrII from Shigella flexneri, which adds a Glc residue to Rha of the O-antigen repeating unit, has nine transmembrane domains and three large periplasmic domains (16). Some other bacterial glycosyltransferases are also found to be membrane associated, but they do not have transmembrane regions (2, 19, 20, 32). Currently, the nature of the membrane-protein interaction for these glycosyltransferases is not clearly understood. WaaJ is a glycosyltransferase involved in the synthesis of the outer core region of LPS in E. coli, and it has a GT-A fold and belongs to the CAZy GT8 family. WaaJ also has a DxD motif and a hydrophobic segment at the C terminus, which was found to be critical for membrane association and catalytic activity (17). Both WfaP and WfgD lack transmembrane domains, i.e., there is no large hydrophobic domain; however, they have a short hydrophobic segment at the C terminus (see Fig. S1 in the supplemental material) which may enable the enzymes to associate with the inner leaflet of the membrane, with the catalytic domain exposed to the cytoplasm, the site of nucleotide sugar donor substrate synthesis. The cytoplasmic face of the inner membrane is thought to be the site of synthesis for the acceptor substrates, as well as the lipid A and oligosaccharide core structures of LPS and the peptidoglycans. This topology would give the enzymes access to the nucleotide sugar pool in the cytoplasm, and it would allow them to be close to the natural membrane-bound acceptor substrate. It is also possible that the enzymes form protein complexes for efficient glycosyltransfer.
The general properties, including the metal ion requirement, detergent sensitivity, as well as the acceptor specificities of WfaP and WfgD, are similar, and they are also similar to those of WbbD (25; I. Brockhausen, J. G. Riley, M. Joynt, X. Yang, and W. A. Szarek, submitted for publication). However, WfaP and WfgD have virtually no sequence similarity to WbbD, indicating that the similarity in properties is based on three-dimensional folding of the protein that could be revealed by analysis of the X-ray structures.
Both WfaP and WfgD have a distinct specificity for pyrophosphate in the acceptor. A slight difference is that WfgD had a much higher relative activity with GlcNAc-α-PO3-(CH2)11-OPh, GlcNAc-α-PO3-PO3-(CH2)6-OPh, and GlcNAc-α-PO3-PO3-(CH2)10 as substrates, indicating that the extended acceptor binding sites of these two enzymes may differ. The lipid moiety of the acceptor is less important but does have a significant influence on activity, possibly by regulating the exposure of the GlcNAc-PP acceptor in micelles or lipid vesicles.
Both WfaP and WfgD are specific for UDP-Glc as the donor substrate. However, low-level activity was observed when UDP-Gal was used as the donor substrate. It remains to be shown if this was due to a low galactosyltransferase activity of WfaP or WfgD or an active 4-epimerase that converted UDP-[3H]Gal to UDP-[3H]Glc, followed by transfer of [3H]Glc by WfaP and WfgD. The latter possibility is the most likely, since glycosyltransferases are usually very specific for the donor substrate, and E. coli are known to have active 4-epimerases (13).
It is interesting that the sugar at the reducing ends of the O56 and O152 repeating units has been designated as GlcNAcβ. However, the GlcNAc-phosphate transferred from UDP-GlcNAc as the first sugar of the repeating unit is always in α-linkage, and our standard substrate contains GlcNAc in α-linkage. It is therefore likely that during the polymerization of the O antigens the anomeric linkage is inverted.
Chen et al. studied the role of the lipid moiety in the acceptor substrates for several enzymes in the pathway of N glycosylation in Campylobacter jejuni (10). These enzymes add sugars to UndP in vivo. In vitro, the third enzyme in the assembly sequence, GalNAc-transferase PglJ, utilizes a number of sugar-pyrophosphate lipids in the presence of detergent and seems to be relatively insensitive to modifications of the lipid chain. The earlier-acting enzymes, however, seem to prefer a cis over a trans configuration of the double bond in the lipid, although it is not clear if the first or the second enzyme in the assembly has the cis specificity. GlcNAcα (or GalNAcα)-pyrophosphate-lipid acceptors have been applied successfully to characterize the functions of galactosyltransferase WbbD (25) and N-acetylgalactosaminyl transferase WbnH (36). In this study, we showed that this analog of the natural undecaprenol-pyrophosphate-sugar acceptor can be used to study the function of glucosyltransferases WfaP and WfgD. All of these four transferases catalyze the second step in the synthesis of E. coli O-antigen repeating units. It is highly likely that these synthetic substrates can also be used to produce O-antigenic oligosaccharides by chemo-enzymatic synthesis as substrates for other bacterial glycosyltransferases or for vaccine development.
Supplementary Material
Acknowledgments
This work was supported by the Canadian Cystic Fibrosis Foundation (to I.B.), the Natural Sciences and Engineering Research Council of Canada (to I.B. and W.A.S.), Tianjin Municipal Special Fund for Science and Technology Innovation Grant 05FZZDSH00800, the National Natural Science Foundation of China (NSFC) Key Programs Grants 30530010 and 20536040, NSFC General Program Grant 30670038, the Chinese National Science Fund for Distinguished Young Scholars (30788001), and the National 863 Program of China grants 2006AA020703 and 2006AA06Z409.
We thank John Riley and Pedro Montoya-Peleaz for the synthesis of substrate analogs and John Schutzbach for helpful discussions.
Footnotes
Published ahead of print on 16 May 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alexander, D. C., and M. A. Valvano. 1994. Role of the rfe gene in the biosynthesis of the Escherichia coli O7-specific lipopolysaccharide and other O-specific polysaccharides containing N-acetylglucosamine. J. Bacteriol. 1767079-7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barreras, M., P. L. Abdian, and L. Ielpi. 2004. Functional characterization of GumK, a membrane-associated beta-glucuronosyltransferase from Xanthomonas campestris required for xanthan polysaccharide synthesis. Glycobiology 14233-241. [DOI] [PubMed] [Google Scholar]
- 3.Bastin, D. A., and P. R. Reeves. 1995. Sequence and analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene 16417-23. [DOI] [PubMed] [Google Scholar]
- 4.Breton, C., L. Snajdrova, C. Jeanneau, J. Koca, and A. Imberty. 2006. Structures and mechanisms of glycosyltransferases. Glycobiology 1629R-37R. [DOI] [PubMed] [Google Scholar]
- 5.Brockhausen, I. 2007. Structure and function of glycosyltransferases specific for O-glycan processing, p. 217-234. In C. Sanson and O. Markman (ed.), Glycobiology. Scion Publishing Ltd., Woodbury NY.
- 6.Brockhausen, I., M. Benn, S. Bhat, S. Marone, J. G. Riley, P. Montoya-Peleaz, J. Z. Vlahakis, H. Paulsen, J. S. Schutzbach, and W. A. Szarek. 2006. UDP-Gal: GlcNAc-R beta1,4-galactosyltransferase—a target enzyme for drug design. Acceptor specificity and inhibition of the enzyme. Glycoconj. J. 23525-541. [DOI] [PubMed] [Google Scholar]
- 7.Brockhausen, I., E. A. Larsson, and O. Hindsgaul. 2007. A very simple synthesis of GlcNAc-alpha-pyrophosphoryl-decanol: a substrate for the assay of a bacterial galactosyltransferase. Bioorg. Med. Chem. Lett. 18804-807. [DOI] [PubMed] [Google Scholar]
- 7a.Brockhausen, I., J. G. Riley, M. Joynt, X. Yang, and W. A. Szarek. Acceptor specificity of UDP-Gal:GlcNAc-R1,3-galactosyltransferase (WbbD) from Escherichia coli O7:K1. Glycoconj. J., in press. [DOI] [PubMed]
- 8.Bronner, D., B. R. Clarke, and C. Whitfield. 1994. Identification of an ATP-binding cassette transport system required for translocation of lipopolysaccharide O-antigen side-chains across the cytoplasmic membrane of Klebsiella pneumoniae serotype O1. Mol. Microbiol. 14505-519. [DOI] [PubMed] [Google Scholar]
- 9.Charnock, S. J., and G. J. Davies. 1999. Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry 386380-6385. [DOI] [PubMed] [Google Scholar]
- 10.Chen, M. M., E. Weerapana, E. Ciepichal, J. Stupak, C. W. Reid, E. Swiezewska, and B. Imperiali. 2007. Polyisoprenol specificity in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry 4614342-14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, J., Q. Wang, W. Wang, Y. Wang, L. Wang, and L. Feng. 2006. Characterization of E. coli O24 and O56 O antigen gene clusters reveals a complex evolutionary history of the O24 gene cluster. Curr. Microbiol. 53470-476. [DOI] [PubMed] [Google Scholar]
- 12.Daniels, C., C. Vindurampulle, and R. Morona. 1998. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy). Mol. Microbiol. 281211-1222. [DOI] [PubMed] [Google Scholar]
- 13.Guo, H., W. Yi, L. Li, and P. G. Wang. 2007. Three UDP-hexose 4-epimerases with overlapping substrate specificity coexist in E. coli O86:B7. Biochem. Biophys. Res. Commun. 356604-609. [DOI] [PubMed] [Google Scholar]
- 14.Keenleyside, W. J., and C. Whitefield. 1996. A novel pathway for O-polysaccharide biosynthesis in Salmonella enterica serovar Borreze. J. Biol. Chem. 27128581-28592. [DOI] [PubMed] [Google Scholar]
- 15.Kogan, G., A. S. Shashkov, B. Jann, and K. Jann. 1993. Structure of the O56 antigen of Escherichia coli, a polysaccharide containing 7-substituted alpha-N-acetylneuraminic acid. Carbohydr. Res. 238261-270. [DOI] [PubMed] [Google Scholar]
- 16.Lehane, A. M., H. Korres, and N. K. Verma. 2005. Bacteriophage-encoded glucosyltransferase GtrII of Shigella flexneri: membrane topology and identification of critical residues. Biochem. J. 389137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leipold, M. D., N. A. Kaniuk, and C. Whitfield. 2007. The C-terminal domain of the Escherichia coli WaaJ glycosyltransferase is important for catalytic activity and membrane association. J. Biol. Chem. 2821257-1264. [DOI] [PubMed] [Google Scholar]
- 18.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 285-13. [DOI] [PubMed] [Google Scholar]
- 19.Liu, D., A. M. Haase, L. Lindqvist, A. A. Lindberg, and P. R. Reeves. 1993. Glycosyl transferases of O-antigen biosynthesis in Salmonella enterica: identification and characterization of transferase genes of groups B, C2, and E1. J. Bacteriol. 1753408-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, D., L. Lindquist, and P. R. Reeves. 1995. Transferases of O-antigen biosynthesis in Salmonella enterica: dideoxyhexosyltransferases of groups B and C2 and acetyltransferase of group C2. J. Bacteriol. 1774084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marolda, C. L., J. Welsh, L. Dafoe, and M. A. Valvano. 1990. Genetic analysis of the O7-polysaccharide biosynthesis region from the Escherichia coli O7:K1 strain VW187. J. Bacteriol. 1723590-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montoya-Peleaz, P. J., J. G. Riley, W. A. Szarek, M. A. Valvano, J. S. Schutzbach, and I. Brockhausen. 2005. Identification of a UDP-Gal: GlcNAc-R galactosyltransferase activity in Escherichia coli VW187. Bioorg. Med. Chem. Lett. 151205-1211. [DOI] [PubMed] [Google Scholar]
- 23.Olsson, U., K. Lycknert, R. Stenutz, A. Weintraub, and G. Widmalm. 2005. Structural analysis of the O-antigen polysaccharide from Escherichia coli O152. Carbohydr. Res. 340167-171. [DOI] [PubMed] [Google Scholar]
- 24.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riley, J. G., M. Menggad, P. J. Montoya-Peleaz, W. A. Szarek, C. L. Marolda, M. A. Valvano, J. S. Schutzbach, and I. Brockhausen. 2005. The wbbD gene of E. coli strain VW187 (O7:K1) encodes a UDP-Gal: GlcNAc{alpha}-pyrophosphate-R {beta}1,3-galactosyltransferase involved in the biosynthesis of O7-specific lipopolysaccharide. Glycobiology 15605-613. [DOI] [PubMed] [Google Scholar]
- 26.Samuel, G., and P. Reeves. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 3382503-2519. [DOI] [PubMed] [Google Scholar]
- 27.Shao, J., M. Li, Q. Jia, Y. Lu, and P. G. Wang. 2003. Sequence of Escherichia coli O128 antigen biosynthesis cluster and functional identification of an alpha-1,2-fucosyltransferase. FEBS Lett. 55399-103. [DOI] [PubMed] [Google Scholar]
- 28.Sperandeo, P., R. Cescutti, R. Villa, C. Di Benedetto, D. Candia, G. Deho, and A. Polissi. 2007. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J. Bacteriol. 189244-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tocilj, A., C. Munger, A. Proteau, R. Morona, L. Purins, E. Ajamian, J. Wagner, M. Papadopoulos, L. Van Den Bosch, J. L. Rubinstein, J. Fethiere, A. Matte, and M. Cygler. 2008. Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat. Struct. Mol. Biol. 15130-138. [DOI] [PubMed] [Google Scholar]
- 30.Torgov, V. I., A. S. Shashkov, B. Jann, and K. Jann. 1995. NMR reinvestigation of two N-acetylneuraminic acid-containing O-specific polysaccharides (O56 and O24) of Escherichia coli. Carbohydr. Res. 27273-90. [DOI] [PubMed] [Google Scholar]
- 31.Unligil, U. M., S. Zhou, S. Yuwaraj, M. Sarkar, H. Schachter, and J. M. Rini. 2000. X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily. EMBO J. 195269-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van den Brink-van der Laan, E., J. W. Boots, R. E. Spelbrink, G. M. Kool, E. Breukink, J. A. Killian, and B. de Kruijff. 2003. Membrane interaction of the glycosyltransferase MurG: a special role for cardiolipin. J. Bacteriol. 1853773-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vrielink, A., W. Ruger, H. P. Driessen, and P. S. Freemont. 1994. Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. EMBO J. 133413-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, T., A. C. McCandlish, L. S. Gronenberg, S. S. Chng, T. J. Silhavy, and D. Kahne. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 10311754-11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi, W., J. Shao, L. Zhu, M. Li, M. Singh, Y. Lu, S. Lin, H. Li, K. Ryu, J. Shen, H. Guo, Q. Yao, C. A. Bush, and P. G. Wang. 2005. Escherichia coli O86 O-antigen biosynthetic gene cluster and stepwise enzymatic synthesis of human blood group B antigen tetrasaccharide. J. Am. Chem. Soc. 1272040-2041. [DOI] [PubMed] [Google Scholar]
- 36.Yi, W., Q. Yao, Y. Zhang, E. Motari, S. Lin, and P. G. Wang. 2006. The wbnH gene of Escherichia coli O86:H2 encodes an alpha-1,3-N-acetylgalactosaminyl transferase involved in the O-repeating unit biosynthesis. Biochem. Biophys. Res. Commun. 344631-639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.