Abstract
It is now well established that, for photosynthetic bacteria, the aerobic-to-microaerophilic transition activates the membrane-bound sensor kinase RegB, which subsequently phosphorylates the transcriptional activator RegA, thereby inducing elevated levels of intracellular photosynthetic membranes. The mechanism of RegB activation—in particular, the role of ubiquinone-10—is controversial at present. One problem here is that very limited quantitative in vivo data for the response of the ubiquinone redox state to different cultivation conditions exist. Here, we utilize Rhodospirillum rubrum to study the correlation of the quinone redox state to the expression level of photosynthetic membranes and determine an effective response function directly. Our results show that changes in the photosynthetic membrane levels between 50 and 95% of that maximally attainable are associated with only a twofold change in the ubiquinol/ubiquinone ratio and are not necessarily proportional to the total levels of either quinone or [NAD+ + NADH]. There is no correlation between the redox potentials of the quinone and pyridine nucleotide pools. Hill function analysis of the photosynthetic membrane induction in response to the quinone redox state suggests that the induction process is highly cooperative. Our results are probably generally applicable to quinone redox regulation in bacteria.
In addition to its well-known role as a redox mediator in respiratory and photosynthetic electron transport processes, ubiquinone (UQ) may be very important for gene regulation by several bacterial sensor kinase systems, as suggested by recent studies. At present, two UQ-dependent sensor kinase systems which play a primary role in O2 sensing, ArcB/ArcA in Escherichia coli (7) and RegB/RegA in photosynthetic bacteria (6), have been well characterized. Although Arc and Reg systems show no extensive sequence homology and molecular details of their mechanisms differ, several general features seem to be emerging. For the generalized sensor kinase, the autophosphorylated active form, present at low partial O2 pressure (pO2) levels, containing one or more reduced cysteines, transfers the phosphate to a water-soluble transcriptional activator (ArcA or RegA), thereby stimulating gene expression of genes containing the ArcA or RegA consensus sequences. At “high” pO2 levels, the sensor kinase is inactivated, thus causing ArcA- or RegB-mediated gene expression to cessate. The mechanisms of ArcB and RegB inactivation are highly controversial at the present time. On the one hand, early seminal studies by Kaplan and coworkers (16, 17) showed definitively that the reg-mediated (or, in Rhodobacter sphaeroides, prr-mediated) expression of the photosynthetic unit responds to the “electron flow” through the high-affinity oxidase cbb3. Recent studies (7, 23) have shown that both ArcB (in E. coli) and RegB (in Rhodobacter capsulatus) can be inactivated by oxidized UQ but not by ubiquinol (UQH2), thus implying that the sensor kinases respond to the initial input of reducing equivalents. This mechanism also allows a variation of the RegB response under completely anaerobic photosynthetic conditions. Unfortunately, the molecular details of the UQ-mediated inactivation process are so far unclear. For ArcB, Malpica and coworkers demonstrated in vitro that the cysteine redox pairs in the cytoplasmically located sensor domain are oxidized to cystine by UQ, thereby inducing an inactive conformational state of the sensor kinase (15). However, the Bauer group showed that, for RegB, the potentially redox-active cysteines are not essential for inhibition of the kinase activity by UQ in vitro, and a causal relation between UQ and the cysteine redox status is still unclear (22, 23).
The rather elegant idea that the UQ-mediated inhibition is the major mechanism for redox regulation in E. coli (ArcB) or photosynthetic bacteria (RegB) contains a number of thermodynamic as well as structural difficulties. First, for RegB, the standard redox potential of the conserved cysteines has been determined to be −294 mV (22), which probably corresponds closely to the cytoplasmic redox potential and is very distant from that of the UQH2/UQ redox pair (E°′ = +90 mV) (25) or that of the alternative quinone, menaquinone (MQ) (E°′ = −74 mV), of E. coli. The large discrepancy between the standard redox potentials of RegB (ArcB) and the putative oxidant UQ or MQ suggests that the regulatory processes must be operating continually at a state far from equilibrium. In addition, Kaplan and coworkers have very recently criticized a physiological role of UQ in regulating PrrB (14). In particular, UQ also inhibited a truncated form of PrrB which lacks the N-terminal transmembrane domain and also the putative UQ binding site. This result also implies that the cytoplasmic domain is the one which can be inhibited by UQ, which is not easily rationalized to the known exclusive localization of UQ to the membrane. These results have led Kaplan and coworkers to suggest that the cbb3-mediated pathway is the only one which is physiologically significant for PrrB regulation.
One of the fundamental conflicts between the UQ regulation hypotheses (23) and the cbb3 regulation hypothesis (14) can be assigned partly to the general lack of information as to the possible variation of the UQH2/UQ ratio under physiological conditions, its relation to the size of the total UQ (UQt) pool, and the response of the system to the UQH2/UQ ratio. The often-quoted “highly oxidized state of the quinone pool under aerobic conditions” can be traced back to a study of Takamiya and Takamiya (24), who demonstrated that in Chromatium vinosum (an obligate anaerobe) the UQ pool becomes highly oxidized in resting cell suspensions exposed to O2, and a study of Redfearn (18), who also exposed photosynthetic anaerobic cell suspensions of Rhodospirillum rubrum to oxygen by shaking them in a test tube for a period of 10 min. In the latter study, the UQH2/UQ ratio was determined exclusively by UV-visible spectroscopy of the isolated quinones (extracted with a procedure which also extracts rhodoquinone [RQ], the major alternative quinone present in R. rubrum). Finally, none of these early studies employed an endogenous monitor of the cellular redox potential(s) to estimate the gene expression response to various levels of UQ accurately. The latter is a major source of ambiguity; without an internal redox monitor, the terms “aerobic,” “semiaerobic” (or “microaerophilic”), and “anaerobic” are purely phenomenological and somewhat arbitrary. To confuse matters more, many studies of the regulatory function of UQ are derived from in vitro experiments with purified and often truncated sensor proteins lacking the membrane-spanning sensor domain and using water-soluble UQ analogues not naturally occurring in cells. Finally, a possible role of alternative quinone species present in the same membrane is completely unclear.
Facultative purple photosynthetic bacteria are particularly useful for studying the response function of UQ und UQH2/UQ to different redox regimes, as it is now well established that the activity of the RegB-type sensor kinase determines the levels of the light-harvesting 1 (LH1) complex (encoded by the puf operon, containing an upstream RegA consensus sequence), which is easily detected spectroscopically. In this study, we show that the LH1 levels observed in the facultative purple nonsulfur bacterium R. rubrum provide an excellent internal monitor of the membrane redox potential and allow an assessment of the UQ response function in vivo under aerobic, semiaerobic, and anaerobic conditions for the first time.
We show here that the “oxidizing” conditions induced by semiaerobic growth in conventional minimal medium and the “reducing” conditions obtained in fructose-containing M2SF medium (see below) are distinguished only by a very subtle change in the UQH2/UQ ratio, which is poised far above both the redox potential of the redox-active Cys265 of RegB from R. capsulatus in vitro (22) and that of the cytoplasmic NADH/NAD+ couple. In addition, we demonstrate that the UQH2/UQ ratio does not necessarily depend on the size of the total UQ pool. Finally, we employ a simple cooperativity analysis of the redox changes observed to predict the effective Hill function of the putative UQ-dependent inhibition of RegB in vivo.
MATERIALS AND METHODS
Cultivation and sampling conditions.
R. rubrum strain S1 (ATCC 11170) and the carotenoidless mutant R. rubrum G9 (DSM 468) were used in this study. R. rubrum G9 was cultivated chemoheterotrophically in a 7-liter bioreactor (Sartorius BBI Systems, Germany) with a 4-liter working volume, inoculated with a preculture grown under photosynthetic conditions. The cultivation conditions and semiaerobic process control used are described in reference 10. Cultures in shake flasks containing four slightly indented baffles were grown on a rotary shaker at 30°C with M (succinate, 20 mM), M2S (succinate, 40 mM), or M2SF (succinate, 40 mM, and fructose, 0.3%) medium. When alternative substrates where tested, succinate was replaced with 40 mM acetate, pyruvate, or fructose as the carbon source. For incubation of shake flask cultures under substrate-saturating conditions, exponentially growing cells were harvested by centrifugation and resuspended in fresh growth medium. Semiaerobic incubation was done in 100-ml shake flasks with 50 ml culture and parafilm-covered cellulose stoppers with slow aeration (100 rpm) on a rotary shaker for 1 h. For aerobic conditions, a smaller volume (10 ml) was incubated without parafilm and under rapid agitation (200 rpm) for 1 h.
For the determination of quinones, a rapid sampling procedure was employed, i.e., during sampling, 1 volume of the culture was transferred into 3 volumes of a methanol (60%):tricine (10 mM) buffered solution at −50°C. This quenching step stops all enzymatic reactions immediately. Tricine is a non-salt-based buffer more compatible with analysis by mass spectroscopy than HEPES and other buffers (3). No extraction of quinones was detected during this step, in accordance with the known insolubility of ubiquinone-10 (UQ10) in methanol. The whole sampling procedure was complete in less than 3 s. The cells were then centrifuged for 15 min at 15,000 × g at −10°C and extracted immediately.
Extraction of quinones.
Redox buffers such as pyrogallol were omitted because we found a significant bias by accelerated oxidation of ubiquinol-10 (UQ10H2) in the presence of this compound. Instead, the extraction procedure was done as follows. The harvested cells were extracted two to three times with methylene chloride:methanol (2:1, vol/vol). We found that when cells became visually completely depigmented after several extraction steps, i.e., all carotenoid and bacteriochlorophyll compounds were extracted, the quinones were also depleted completely from the extracted membranes, making the visual inspection valuable information about the extraction efficiency. Extracts were combined and evaporated to dryness under a stream of nitrogen. To prevent photodegradation, all steps were performed under dim light and in amber glass vials or aluminum foil-protected glass and polypropylene tubes. Prior to high-performance liquid chromatography-mass spectrometry (HPLC-MS) analysis, the dried samples were dissolved in methanol:2-propanol (4:1, vol/vol).
Preparation of standards.
Ubiquinol-10 (UQ10H2) standards were prepared freshly prior to analysis from a UQ10 (coenzyme Q10; Sigma) ethanolic stock solution by reduction with KBH4 and subsequent extraction (two times) of UQ10H2 with n-hexane. The hexane phase was separated and evaporated to dryness under a stream of nitrogen. The residue was dissolved in methanol:2-propanol (4:1, vol/vol) and analyzed by HPLC. For calibration with a standard, rhodoquinone-10 (RQ10) was purified from R. rubrum G9 by extraction with 2-propanol:heptan and subsequent chromatography on a silica gel column with methylene chloride:methanol (2:1, vol/vol) as the eluent. A rhodoquinol-10 (RQ10H2) standard was prepared from the purified RQ10 analogous to UQ10H2. The identity and purity of the standards were confirmed by diode array spectroscopy and atmospheric pressure chemical ionization (APCI)-MS.
HPLC-MS analysis.
The extracted quinones were separated using an HPLC system (Agilent 1100 series) equipped with a C8 column (Zorbax, Eclipse XDB-C8, 150 mm by 4.6 mm, 5 μm; Agilent). Separation of quinones was achieved by isocratic elution with methanol:2-propanol (4:1, vol/vol) and ammonium acetate (10 mM) in the mobile phase at a flow rate of 1 ml/min. For quantitative determination and identification of the resolved peaks, a photodiode array detector (DAD) (Agilent) and a single quadrupole MS detector (Agilent SL 1100) equipped with either an electrospray ionization (ESI) or an APCI source, connected in series, were used. MS detector settings were as follows: capillary voltage, 4,000 V; fragmentor voltage, 100 V; APCI source corona current, 40 μA (positive mode). Data analysis was performed using ChemStation software (Agilent).
Analytical procedures.
Cell density (A660) and formation of photosynthetic membranes (PM) (A880) were determined using a 4-mm path-length cuvette with a Jasco V-560 spectrophotometer. Oxygen consumption in the bioreactor was monitored by a fluorescence-based fiber optic oxygen meter (Trace oxygen optode, Presens, Germany). For monitoring oxygen consumption in shake flask cultures, a fiber optic oxygen microsensor (Presens, Germany) mounted in the stopper of the shake flask was used.
The redox potential in bioreactor cultures was measured with a redox electrode (Pt4805; Mettler-Toledo, Germany). The instrument reading was converted to the redox voltage of the standard hydrogen electrode (E°) by correction with the standard voltage of the reference electrode (Ag/AgCl2) according to the manufacturer's instructions. Fructose concentrations were determined enzymatically by using a test kit from Biopharm (Germany).
Determination of pyridine nucleotides.
The reduced coenzymes NADH and NADPH were extracted by rapid transfer of a culture aliquot to an equal volume of 1 M KOH. After incubation for 7 min at 60°C, the extracts were centrifuged (4,863 × g, 4°C, 10 min) and the supernatants were neutralized with 1 M HCl. Oxidized coenzymes (NAD+ and NADP+) were extracted by rapid transfer of cells to 1.5 M formic acid (4°C) and incubation for 1 h at 4°C. After centrifugation, the supernatant was lyophilized and the residue dissolved in 1 ml distilled water. All coenzymes were subsequently determined according to the redox cycling procedure described in reference 19, with phenazine methosulfate and dichlorphenolindophenol (DCPIP) as redox mediators. Briefly, the assay composition for determination of NAD+ and NADH was as follows: ethanol:phosphate buffer (6.5 ml ethanol plus 0.13 M K2HPO4, pH 7.4), 740 μl; DCPIP (51 μM), 200 μl; phenazine methosulfate (48 μM), 200 μl; sample extract, 200 μl; alcohol dehydrogenase (2.2 U/mg), 10 μl. For determination of NADP+ and NADPH, we used triethanolamine buffer (0.4 M, pH 7.6), 750 μl; glucose-6-phosphate (50 mM), 20 μl; and glucose-6-phosphate dehydrogenase (1.0 U/mg). Concentrations were determined by measuring the kinetics of DCPIP reduction at 578 nm with a Jasco V-560 spectrophotometer. For calibration, standard mixtures were used.
RESULTS
Preliminary considerations. (i) The LH1 complex of R. rubrum is a convenient endogenous monitor assessing the photosynthetic gene response to the cellular redox potential.
As mentioned in the introduction, it is now well accepted that the levels of intracytoplasmic PM in facultative photosynthetic bacteria are ultimately determined by the activity of the RegB (PrrB) sensor kinase. We have shown previously (8, 10) that the levels of LH1 complex of R. rubrum, measurable spectroscopically using the characteristic near-infrared absorption maximum (A882), correlate well with the PM levels. Thus, this parameter normalized to the cell density measured by the turbidity (using a 4-mm path-length cuvette to ensure a large linear dynamic range) at A660 allows the amount of PM/cell (determined as A882/A660) to be determined reliably throughout a growth profile. The reliability of this measurement is ensured by two additional factors. First, the protein composition of the PM is almost invariant with the growth media employed here. Second, only a single LH complex (LH1) is present under all growth conditions, and the ratio of this complex to the RC is essentially invariant. It is well documented (5, 13) that these factors are not true for the more well-studied bacteria R. sphaeroides and R. capsulatus, which renders the assessment of the correlation between LH levels and quinone ratios more difficult for these species.
(ii) The intracellular semiaerobic redox status of R. rubrum can be determined by a systematic and precise variation of the growth conditions.
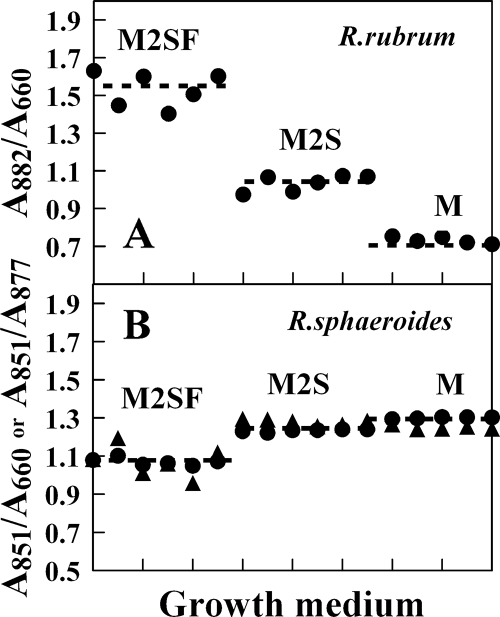
Phenomenologically, most facultative bacteria show levels of LH complexes corresponding to three redox regimes: (a) “aerobic” (high pO2), where no LH complexes are produced; (b) “semiaerobic” (low pO2 [<0.5% pO2 for R. rubrum]), where LH complexes are produced at levels lower than those observed under anaerobic photosynthetic conditions; and (c) “anaerobic” photosynthetic, where maximal LH levels are observed. A conceptual difficulty in correlating the quinone levels observed in these different regimes to the gene response function is that the general metabolic profiles in the various regimes are also significantly different. Clearly it would be useful to be able to vary the gene response function (here, the LH1 levels) in a single regime. This is exactly what can be achieved for R. rubrum, but so far not for R. capsulatus or R. sphaeroides, by using the minimal media M, M2S, and M2SF (see below), described by us previously (8, 10). Briefly, for R. rubrum, distinct modes of semiaerobic (microaerophilic) redox regimes (where the UQ-dependent sensor kinases are active) are accessible under dark conditions: (a) succinate-dependent growth in minimal Sistrom (M or M2S) medium (20) at pO2 levels of <0.5%, with only 50 to 60% of the PM levels of those maximally attainable under anaerobic photosynthetic conditions at low light (Fig. 1) (this redox regime, considered here to be “oxidizing” with respect to the UQ pool, corresponds approximately to that observed for other well-characterized organisms, such as R. sphaeroides and R. capsulatus, growing semiaerobically in minimal media), or (b) succinate plus fructose (M2SF medium)-dependent semiaerobic growth in the dark, which leads to (maximal) levels of LH1 equivalent to those obtained under low-light photosynthetic conditions (8, 10). This redox regime is considered here to be “reducing” with respect to the quinone pool and is so far not experimentally accessible for R. sphaeroides and R. capsulatus. As an example, Fig. 1B shows the levels of LH2/cell (A851/A660) of R. sphaeroides as well as the relative ratio of LH2/LH1 under the growth conditions used here. The levels of both parameters in M and M2S media correspond approximately to the LH1/cell level of R. rubrum in M2S medium, whereas in the presence of succinate plus fructose, both parameters are significantly reduced. The same reduction in the LH2/cell and LH2/LH1 levels was also observed when glucose (0.3%) was substituted for fructose (data not shown).
FIG. 1.
(A) Levels of LH1 complexes per R. rubrum cell (given as the ratio A882/A660), obtained with different media for 100-ml batch cultures grown under semiaerobic conditions at 30°C. Each point represents the A882/A660 ratio for a single culture at the growth curve maximum. (B) Levels of LH2/cell (given as the ratio A851/A660) (circles) and LH2/LH1 (given as the ratio A851/A877) (triangles) obtained for R. sphaeroides growing in the same media. A877 contains contributions from both LH1 and LH2 complexes and is thus only an empirical parameter.
We are also able to obtain completely aerobic conditions in the chemostat, where LH1 complexes are not expressed (completely white cells) without resorting to the use of rich medium (data not shown). This method is often used for R. capsulatus and R. sphaeroides to generate so-called “aerobic” conditions. However, as we show below, the results obtained with rich media are difficult to interpret, probably due to the poorly understood metabolic profile produced under these conditions.
Finally, during growth with acetate, which is significantly more oxidized than both succinate and fructose, the redox levels must be “highly oxidizing,” as only very low levels of PM are observed. Thus, levels of PM easily and sensitively detected by optical spectroscopy, together with the added redox dimension provided by M2SF medium, makes the R. rubrum system very useful for assessing the quantitative effects of redox parameters (such as quinone or pyridine nucleotide levels) upon functional gene expression. We thus employ different growth conditions to correlate changes in redox cofactors to the levels of key redox-regulated components (e.g., reaction centers and LH1).
HPLC-DAD-MS analysis of quinone extracts.
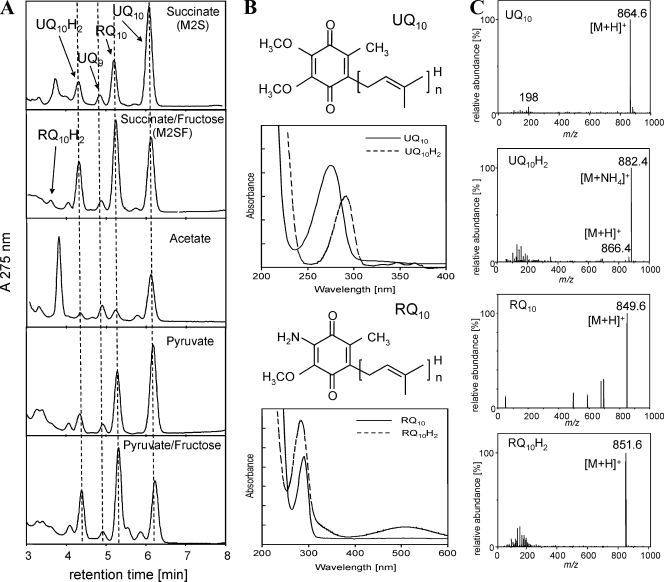
During this study, we utilized reversed-phase-HPLC in combination with photodiode array detection and APCI- or ESI-MS for the isocratic separation and identification of quinones in organic extracts of R. rubrum. The applied HPLC method allows the rapid elution of all quinones, including the separation of reduced and oxidized species within 8 min (Fig. 2A). No degradation or change of redox state of standard compounds was observed on the same time scale. Figure 2 illustrates the separation of extracts obtained from R. rubrum S1 cultures grown in modifications of the Sistrom medium (20) with different carbon sources. The UV absorption maxima of the quinone moieties of UQ (λmax = 275 nm) and RQ (λmax = 283 nm) (Fig. 2B) allowed the resolution of UQ and RQ species. The reduced quinol forms are accompanied by a shift of the UV maxima to 290 nm, which facilitates the detection of the reduced species by diode array spectroscopy. The combination of HPLC-MS detection with either ESI or APCI ionization allowed unambiguous identification including the isoprenoid side chain length according to the molecular ion (M+) and according to the formation of adduct ions ([M+H]+ and [M+NH4]+) (Fig. 2C). Both ionization techniques, ESI and APCI, can be applied for the ionization of quinones. However, due to the hydrophobic nature of the quinone structure, APCI tends to be more efficient and was used preferentially.
FIG. 2.
HPLC-DAD-MS analysis of quinone extracts. (A) HPLC profiles. Dashed lines are applied to facilitate the identification of quinone peaks. (B) DAD-UV spectra of UQ10 (λmax = 275 nm), UQ10H2 (λmax = 292 nm), RQ10 (λmax = 284 nm), and RQ10H2 (λmax = 290 nm). (C) MS signals (only the monoisotopic masses are indicated for clarity) obtained with APCI could be attributed to the following ions: for UQ10, m/z 864.6 to [M+H]+ and m/z 198.0 to a benzylium ion, a characteristic fragment of the UQ ring structure; for UQ10H2, m/z 882.4 to [M+NH4]+, m/z 866.4 to [M+H]+, and m/z 198.0 to a benzylium ion; for RQ10, m/z 849.6 to [M+H]+; and for RQ10H2, m/z 851.6 to [M+H]+. The last compound could be detected only when M2SF was used for cultivation under oxygen-limiting conditions. The signals obtained for UQ9 (not shown) were m/z 795.6 (M+) and m/z 812.8 ([M+NH4]+).
All of the known quinones (in order of elution, RQ10H2, UQ10H2, ubiquinone-9 [UQ9], RQ10, and UQ10) present in R. rubrum (12) could be identified in extracts from batch cultures. The additional major peak present in extracts of acetate-grown cultures at 3.7 min (Fig. 2A) revealed UV-visible and APCI-MS signals of the major carotenoid of R. rubrum, spirilloxanthin, and additional signals of an unidentified compound with an absorbance maximum at 272 nm and a single APCI-MS signal at m/z 647.2 (positive mode) (data not shown). In all cases, UQ10 was found to be the major ubiquinone, with UQ9 corresponding to only 10% of the total UQ10 concentration. Rhodoquinones were detected only as the decaterpenoid-derived species RQ10 and RQ10H2. The cellular concentrations of the total UQ10 pool (UQt = UQ10 + UQ10H2) in cultures grown under semiaerobic, dark conditions varied, depending on the carbon source employed, in the range of 1 nmol (acetate medium) to 3 nmol (M2SF medium)/mg (dry weight). The latter value corresponds closely to that (3.2 nmol/mg [dry weight]) obtained from isolated PM (chromatophores) of semiaerobically grown cells in M2SF medium (data not shown). We also found that the total amounts of UQ and RQ were significantly larger (approximately twofold) in the carotenoidless mutant R. rubrum G9 than in the wild-type S1 strain.
In control experiments, we also extracted and separated quinones from semiaerobic and photosynthetic batch cultures of R. capsulatus and R. sphaeroides. In the latter experiments, only UQ10 and UQ10H2 could be demonstrated, with no evidence for the presence of a second quinone. These observations are consistent with the literature (see reference 12).
Kinetics of quinone biosynthesis.
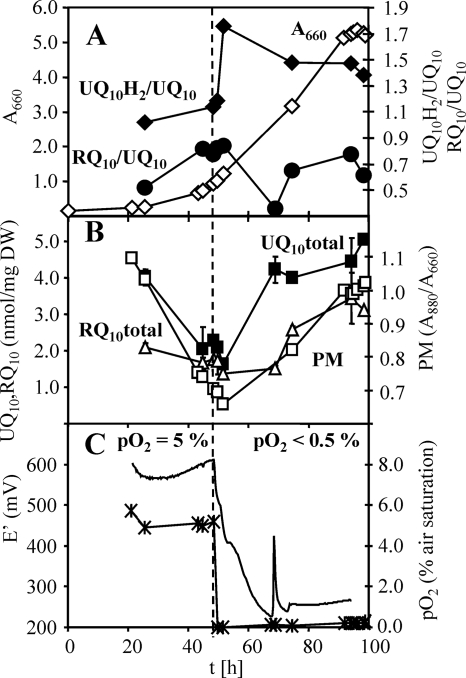
We performed a kinetic analysis of the quinone changes observed in a batch culture grown semiaerobically in M2SF (with succinate and fructose as substrates). Figure 3 shows a typical growth profile for R. rubrum G9 obtained using the process control strategy described in reference 10. R. rubrum G9 was chosen because the absence of carotenoids facilitates the extraction and separation of quinones. Control experiments with the wild type (S1) yielded essentially identical results. The characteristic hallmark of this type of cultivation is that the A880/A660 signal, which is a measure for the cellular level of PM, shows a typical time course with a sharp “kink” at the time point when photosynthetic gene expression is induced. This course (described in detail in references 8 and 10) is a result of cell growth with repressed photosynthetic gene expression during the initial aerobic growth phase. Initially (Fig. 3A), following inoculation with an anaerobic inoculum (PM maximal), oxygen is consumed rapidly due to aerobic growth, concurrent with dilution of PM until the semiaerobic regime (pO2 of <0.5% [Fig. 3C]) is reached, whereupon PM synthesis recommences, rising to achieve maximal (corresponding to the PM level of anaerobic cultures) levels at the end of the experiment. Figure 3B shows that the variation of the total UQ10 levels closely follows that of PM at all stages of the growth experiment and primarily reflects different levels of PM. This result, quantified by the ratio UQ10/(A880/A660) in Table 1, is also true for semiaerobic growth in M2S (succinate), M (succinate), and pyruvate media. The RQ10 level appears to show a similar behavior (Table 1). In these media, the total UQ10 and RQ10 levels, when based on the A880/A660 ratio, are also very similar to those found under photosynthetic growth conditions (Table 1). Strikingly, whereas membrane extracts from acetate-grown cells yielded the lowest UQ10 and RQ10 levels (related either to dry weight or to the A880/A660 ratio), PM levels of cultures grown with pyruvate or a combination of pyruvate and fructose as the carbon source were significantly enriched in RQ10. The pyruvate-fructose combination was unique in that it resulted in an RQ10/UQ10 ratio of >1, i.e., RQ10 was the predominant quinone under this condition. As both pyruvate and fructose can be catabolized by R. rubrum by pure fermentation, in contrast to succinate and acetate, we speculate that the stimulation of RQ10 biosynthesis is due to its probable role as a cofactor of fumarate reductase during anaerobic respiration.
FIG. 3.
Quinone profiles of a batch culture of R. rubrum G9. (A) A660, ⋄; UQ10H2/UQ10, ♦; RQ10/UQ10, •. (B) PM (A880/A660), □; UQ10 (total), ▪; RQ10 (total), ▵. DW, dry weight. (C) pO2, ×; E′, ---. The culture redox potential was calculated for pH 7 from the measured electrode potential. The peak at 68 min was caused by fluctuations of the process controller. The time point of switching from aerobic to semiaerobic conditions is indicated by the dashed line. Error bars represent standard deviations from three quinone extracts collected during one sampling.
TABLE 1.
Total (reduced plus oxidized species) levels of UQ10 and RQ10 after semiaerobic growth with different growth mediaa
| Growth substrate | Participation of pathway
|
PM content (A880/A660) | UQ10 total (nmol/mg [dry wt]) | UQ10/ (A880/A660) | RQ10 total (nmol/mg [dry wt]) | RQ10/ (A880/A660) | RQ10/UQ10 | |
|---|---|---|---|---|---|---|---|---|
| Aerobic | Fermentative | |||||||
| M (succinate) | + | − | 0.74 ± 0.07 | 2.81 ± 0.57 | 3.79 | 1.38 ± 0.29 | 1.86 | 0.49 (1:2.0) |
| M2S (succinate) | + | − | 0.78 ± 0.03 | 2.75 ± 0.89 | 3.53 | 1.14 ± 0.07 | 1.46 | 0.41 (1:2.4) |
| M2SF (succinate-fructose) | + | + | 0.92 ± 0.09 | 2.97 ± 0.40 | 3.24 | 1.81 ± 0.53 | 1.97 | 0.61 (1:1.6) |
| Pyruvate | + | + | 0.72 ± 0.06 | 2.86 ± 0.69 | 3.96 | 1.73 ± 0.54 | 2.39 | 0.60 (1:1.6) |
| Pyruvate-fructose | + | ++ | 0.84 | 1.77 | 2.10 | 2.30 | 2.74 | 1.30 (1.3:1) |
| Fructose | (+) | + | 0.60 ± 0.06 | 1.51 ± 0.35 | 2.51 | 0.59 ± 0.32 | 0.98 | 0.39 (1:2.6) |
| Acetate | + | − | 0.59 ± 0.03 | 1.09 ± 0.58 | 1.85 | 0.24 ± 0.15 | 0.41 | 0.22 (1:4.5) |
| M phototroph | − | + | 1.45 | 4.83 | 3.33 | 2.18 | 1.50 | 0.45 (1:2.2) |
| M2SF phototroph | − | ++ | 1.25 | 3.51 | 2.81 | 2.75 | 2.20 | 0.78 (1:1.3) |
| Fructose phototroph | − | ++ | 0.95 | 2.01 | 2.11 | 1.04 | 2.01 | 0.49 (1:2.0) |
Values represent mean values of 3 to 12 determinations. The known participation of “aerobic” (oxidative tricarboxylic acid cycle) and “fermentative” (reductive tricarboxylic acid pathway) pathways is also indicated. The symbols represent the possible metabolic bias under a given growth mode, as follows: ++, highly active; +, functional; (+), weakly functional; −, nonfunctional. For the RQ10/UQ10 ratios, the molar ratio is indicated in parentheses.
Redox states of the quinone pool.
The variation in UQ10H2/UQ ratio was also examined simultaneously in the experiment described above. Figure 3A shows that the aerobic-to-anaerobic transition (adjusted to coincide with the mid-exponential growth phase) is accompanied by an immediate (within less than one generation time) increase in UQ10H2/UQ10, with a concomitant decrease in RQ10/UQ10, with the final respective values decreasing asymptotically toward the end of the experiment. The biphasic behavior of the pools occurring at about 65 h, at a medium E′ of about 250 mV (Fig. 3C), corresponds to a significant depletion of fructose, followed by the transient appearance of acetate in the medium, as we have shown previously (10). However, it is particularly pertinent to note here that the absolute variation of UQ10H2/UQ10 at the redox transition is only within a factor of 2, i.e., 64% of the UQ10 pool (UQ10H2/UQ10 = 1.76) (Fig. 3A) was reduced after the transition to the semiaerobic growth condition. The decrease in the RQ10/UQ10 ratio is within the same order of magnitude. The changes in the UQ10H2/UQ10 and RQ10/UQ10 ratios here contrast strongly with the general monotonic increases in the pools of both quinones, the latter being well correlated to the levels of PM (Fig. 3B). Thus, the quinone pool sizes do not necessarily reflect their redox states. This is one of the major results of our studies, reported here for the first time. We note that we avoided the use of the widely used pyrogallol as a redox buffer, as its addition led to more oxidized UQ pools than in its absence (unpublished data).
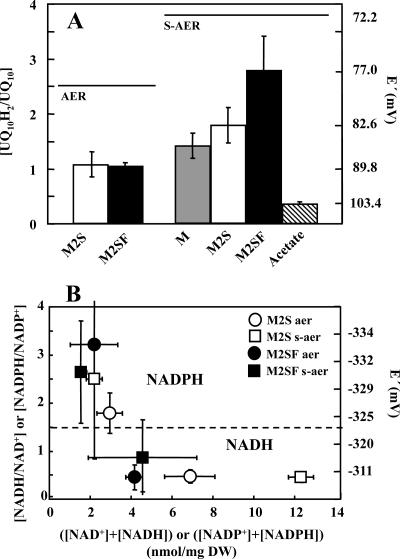
The second question we posed was whether growth substrates considered to be “reducing” lead to higher UQ10H2/UQ10 ratios than those considered to be “oxidizing.” In batch cultures, where the substrate concentration is not constant but decreases permanently, it is difficult to compare different medium compositions with respect to the reduction level of the quinones. We therefore chose a different strategy for comparison of M and M2S (low PM level) and M2SF (high PM level) as well as acetate (very low PM) media under substrate-saturating conditions. Cells were harvested during logarithmic growth, resuspended in fresh medium and incubated under aerobic (strong aeration, pO2 of >10%) and semiaerobic (low aeration, pO2 of <0.5%) conditions in the dark (measured with a miniaturized fiber optic oxygen sensor [Microx TX3; Presens, Germany]). After 1 h, quinones were extracted and analyzed immediately. When slow aeration was applied, oxygen-limiting conditions were reached 7 min after the cells were suspended in fresh medium. The results are shown in Fig. 4A and Table 2. For both M2S and M2SF media, about 51% of the UQ10 pool was reduced under conditions of high aeration (aerobic), which is identical to the level observed during aerobic growth at 5% pO2 in the bioreactor (Fig. 3A). However, under low-aeration (semiaerobic) conditions, cells incubated with M medium showed only a 1.4-fold increase in the reduction of the UQ10 pool compared to levels reached under high aeration. With M2S medium, the UQ10H2/UQ10 ratio was 1.7-fold higher than that for aerobic cells. The maximal increase (2.6-fold) was obtained with M2SF medium (Fig. 4A; Table 2). In these experiments, which were performed well within the generation time of about 3.5 to 5 h, the increases in the UQ10H2/UQ10 ratios correlate well with the relative amounts of PM that R. rubrum synthesizes under the respective medium composition (Fig. 1A). For comparison, incubation with the very low PM level substrate acetate resulted in the most oxidized pool (26% reduced) of all media tested. In contrast, the UQ10 pool obtained from cells grown aerobically with rich peptone-yeast extract complex medium was still 60% reduced, even though the cells were completely devoid of pigment. This observation indicates that the results obtained with rich medium, often assumed to be “maximally oxidizing,” cannot be interpreted in an unambiguous way, due to as yet undefined complex metabolic changes under these conditions.
FIG. 4.
(A) Redox states of UQ10 after 1 h of incubation under semiaerobic conditions in different growth media: M medium (20 mM succinate), M2S medium (40 mM succinate), M2SF medium (40 mM succinate, 0.3% [wt/vol] fructose), and acetate (40 mM acetate). AER, aerobic growth; S-AER, semiaerobic growth (see Materials and Methods for details). (B) Redox states and pool sizes of pyridine nucleotides. Data points represent mean values from three individual determinations, with the respective standard deviations shown as error bars.
TABLE 2.
UQ10 redox levels after aerobic or semiaerobic incubation with different growth mediaa
| Growth medium | UQ10H2/UQ10 ratio (% reduced) under aerobic conditions | E′ (mV) | UQ10H2/UQ10 ratio (% reduced) under semiaerobic conditions | E′ (mV) |
|---|---|---|---|---|
| M | NDb | 1.41 ± 0.23 (58.51) | 85.60 | |
| M2S | 1.07 ± 0.23 (51.69) | 89.13 | 1.78 ± 0.32 (64.03) | 82.61 |
| M2SF | 1.02 ± 0.08 (50.50) | 89.75 | 2.76 ± 0.64 (73.40) | 76.99 |
| Acetate | ND | 0.35 ± 0.02 (25.93) | 103.45 | |
| M2SF, dithionite reduced | 10.57 (91.35)c | 59.57d | ||
| Peptone-yeast extract mediume | 1.54 (60.58) | 84.47 | ND |
Data represent mean values from triplicate experiments±standard deviations.
ND, not determined.
The RQ pool was 49% reduced.
RQ = −62.50.
A880/A660 = 0.58.
Attempts to find culture conditions where the UQ pool was completely oxidized or completely reduced failed. Reduction levels higher than those obtained during semiaerobic growth with M2SF medium were found only when M2SF-grown cells were incubated under completely oxygen-free, anaerobic conditions. The maximal level of UQ10 pool reduction (91.4%) was observed only after incubation with sodium dithionite. In the latter extracts, it was also possible to determine the RQ10 redox state, as the reduced RQ10H2 species was found above the detection limit, resulting in a 49% reduced RQ10 pool (Table 2).
Redox state of the pyridine nucleotides.
The redox state of the pyridine nucleotides NADH/NAD+ and NADPH/NADP+ is connected to the quinone pool via the flow of reducing equivalents through the NADH dehydrogenase enzyme complex and therefore should exhibit responses similar to those exhibited under redox conditions of growth. Previously, it was demonstrated that in R. rubrum the redox states of both coenzymes are affected by the cultivation conditions and depend upon the supply of oxygen, substrates, and light (19). We have determined the pool sizes and redox states of the pyridine nucleotides in different media by using the same strategy as described above for the quinones (incubation of cells in fresh medium with subsequent rapid sampling and extraction), and the results are shown in Fig. 4B. A surprising result was that the relatively large variation (4.2 to 12.3 nmol/mg [dry weight]) of the total ([NAD]+ + [NADH]) pool in the different media under various growth conditions showed no correlation with the extent of reduction. Thus, the extent of NAD+ reduction appears to be maintained at approximately 30% for both M2S and M2SF under aerobic conditions, as well as for M2S under semiaerobic conditions, with only the semiaerobic extracts in M2SF medium showing a higher reduction level (46.5%). Thus, semiaerobic cultures in M2SF do show the largest level of reduction for both UQ10 (73.4%) (Table 2) and NAD+ (Fig. 4B) pools, concomitant with the highest level of PM.
In contrast, the total ([NADP+] + [NADPH]) levels in the different media and growth regimens showed little variation (approximately 2 to 3 nmol/mg [dry weight]), and their reduction levels varied significantly (64.5 to 76.3%), but these levels were consistently more reduced than those of the NADH/NAD+ pool. They showed no correlation to those of NADH/NAD+ or to the levels of PM. The higher reduction level suggests that the cells are able to maintain a highly charged “supply line” (NADPH/NADP+) of reducing equivalents for anabolic pathways even when the catabolic redox charge (NADH/NAD+) varies. Membrane-bound and soluble transhydrogenase activities may be critical for this function.
Figure 4B also indicates that under all conditions studied here, the [NAD+ + NADH] and [NADP+ + NADPH] pools are separated into two distinct redox regimes, with the “boundary” occurring at about −325 mV. We are not aware that this observation has been reported previously.
DISCUSSION
The variation of the quinone pools and their redox states is a cornerstone to the present controversy as to how the transmembrane sensor kinases ArcB and RegB and their homologs in other bacteria respond rapidly to the redox status of the cell. In this study, we have systematically examined the quinone levels and their relation to a reg-regulated endogenous marker, the LH1 (PM) levels in R. rubrum, under various growth conditions, employing substrates with different reduction levels. In addition, to our knowledge, very little data concerning the in vivo status of the redox couples NADH/NAD+, NADPH/NADP+, and UQ10H2/UQ10 have been reported under defined redox growth conditions, and the possible involvement of the alternative quinones (MQ8 in E. coli and RQ10 in R. rubrum), which have standard redox potentials significantly closer to the redox sensor kinases than that of UQ10, has been completely neglected.
R. rubrum is a particularly interesting model system, as it contains two quinones with rather different redox potentials (as in E. coli) and can express three clearly distinct PM levels (including the very high [maximal] level normally observed only under anaerobic conditions at low light) under semiaerobic conditions when grown in the appropriate media (Fig. 1). This contrasts strongly with the small variation in PM observed for R. sphaeroides (Fig. 1) under the same growth conditions. The reason for this difference is not yet clear, but it may be connected with the fact that R. sphaeroides possesses only a single quinone (UQ10). Another possibly relevant difference between the two organisms is that R. rubrum lacks a functional “low-affinity” cytochrome oxidase aa3 (26). We have also been unable to locate putative genes for cytochrome aa3 oxidase in the published R. rubrum genome sequence (data not shown), although the cbb3 and ubiquinol oxidases have been annotated.
The results of this study clarify a number of important points necessary for the mechanistic elucidation of redox regulation in bacteria. (i) The cytoplasmic redox potential of NADH/NAD+ varies only minimally between aerobic and semiaerobic conditions (Fig. 4B), hovering around the value of −310 mV. This value is very similar to those reported recently (4) for E. coli. (ii) The levels of PM observed with substrates at differing levels of reduction (Fig. 4) are well correlated with an increase in the UQ10H2/UQ10 ratio (Fig. 4A; Table 2). However, even at the extreme levels of PM, e.g., with M2SF (maximal PM) or acetate (minimal PM) as the growth medium, the UQ10H2/UQ10 ratios vary only twofold, corresponding to a redox range of +90 ± 15 mV (Fig. 5). This range corresponds perfectly to that reported by Takamiya and Dutton (25) for optimal turnover of the photocycle in isolated chromatophores and is also similar to that reported by Redfearn (18) for R. rubrum growing with malate aerobically (which we here interpret as semiaerobic growth) in the dark (26% UQ10 reduced) or under anaerobic, dark conditions (56% UQ10 reduced). Neither at very high aeration nor under phototrophic anaerobic growth conditions were we able to observe highly oxidized or reduced levels, respectively, of UQ10. In our study, the UQ10 pool still shows a reduction level of 30% even when cultures are completely depleted of substrate. This observation also applies for E. coli, as recent data show the identical values of the UQ8 pools of aerobic E. coli cultures at the stationary phase (1). Thus, the effective redox potentials of the membrane and cytoplasmic compartments are completely distinct and nonoverlapping. There is no apparent correlation between the redox potential of the [NAD+ + NADH] pool and that of the quinones, indicating no thermodynamic equilibrium between the two pools. This is presumably due to the proton motive force across the cytoplasmic membrane which balances the redox pressures exerted by the NADH dehydrogenases as well as transhydrogenase with those exerted by the Q cycle and membrane-bound oxidases. (iii) The size of the UQ10 pool varies proportionally with the PM level and shows no obligatory correlation to the UQ10H2/UQ10 ratio (Fig. 3A and B). Thus, variations of the quinone levels under different growth conditions reported previously (2, 18, 21) are probably unimportant for models of gene regulation by the UQ10 redox status. However, in experiments performed under carefully controlled conditions within the generation time of R. rubrum, there is a significant correlation between PM biosynthesis and redox status of the pool (Fig. 4A). The small decrease in the UQ10H2/UQ10 ratio observed in the growth profile (Fig. 3A) probably reflects the excretion of reducing equivalents due to fermentative metabolism toward the end of the batch run, as reported by us previously (10). (iv) RQ10 is present in significant amounts under all growth conditions examined here, and the RQ10/UQ10 ratio varies considerably with the growth substrate, suggesting an important role in metabolism and possibly redox regulation. Until now, a “photooxidase” function has been suggested for RQ10 in R. rubrum when grown photosynthetically (9). However, in our study we found that the RQ10/UQ10 ratio in R. rubrum is not a constant but is also affected by the nature of the growth substrate under semiaerobic, dark conditions. The highest values were observed with pyruvate-fructose medium, where both substrates can be fermented and oxidized aerobically, whereas very low values were obtained with acetate, which cannot be fermented (Table 1). This is consistent with the observation that RQ10H2 can serve as a substrate for the fumarate reductase, as a part of the reductively driven tricarboxylic acid pathway (11).
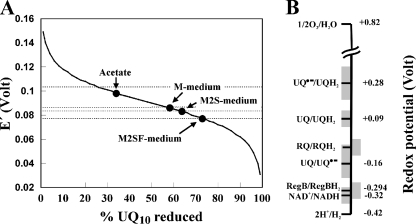
FIG. 5.
(A) Effective redox potential of R. rubrum cells grown in different media, determined by the UQ10H2/UQ10 ratios shown in Fig. 4A. (B) Redox potential of key components which may be involved in redox signaling. The gray boundaries represent the useful redox range (between 10% and 90% reduction) calculated using the Nernst equation: E′ = E°′ + (RT/nF)ln([oxidized redox compound)/(reduced redox compound]), where n is the number of electrons transferred R is the gas constant, T is the absolute temperature, and F is the Faraday constant.
Our in vivo data are completely consistent with the in vitro-derived hypothesis (23) that the relative level of UQ10 affects the level of activity of a RegB-type sensor kinase, thereby regulating the PM level via the phosphorylated transcriptional activator RegA. However, in light of our observation that the quinone pool never becomes either “strongly reduced” or “strongly oxidized” under comparable growth conditions, we favor a model that the cbb3 oxidase is more suited to being the primary switch for the shift between aerobic and anaerobic conditions, as elucidated in detail by Kaplan and coworkers (14), with the UQ10 levels playing a modulatory role.
How can relatively small changes in the UQ10 level affect the RegB activity significantly? So far, no data as to the response function of the RegB-type sensor kinase to changes in [UQ10] are available, i.e., is the response function hyperbolic or does it show a form of cooperativity? The rather small dynamic variation of the UQ redox state under different growth conditions suggests that if RegB is subject to significant physiological regulation by UQ, then the response function must be highly cooperative.
Here we consider a simple model for RegB regulation by UQ which allows us to estimate the degree of cooperativity which must be operating to explain the observed levels of PM in the different media. In our model, we assume that under oxidizing conditions RegB activity (v) is due to the (activating) interaction with cbb3 (via SenC) (17) combined with the inhibitory effect of UQ10. Thus, the level of RegB activity can be expressed as follows: v = VUQ(1 − Y) + VB, where Y is the fractional saturation of RegB with UQ10, VB is the basal RegB activity obtained at saturating (Y = 1) concentrations of UQ10, and VUQ is the maximal additional RegB activity attainable at very low concentrations (Y = 0) of UQ10. To estimate the possible cooperative response of RegB to UQ10, we must make the following reasonable assumptions. (i) The PM levels are directly related to the activity of RegB. For a complex process such as membrane biosynthesis, this assumption is not necessarily true. Nevertheless, in the simplest possible case, where RegB is essentially operating at Vmax(apparent), where the apparent level is modulated by UQ, then the amount of PM will be proportional to the amount of phosphorylated RegA, which is in turn proportional to RegB activity. However, the numerical values of the RegA-induced puf mRNA and the PM levels may differ significantly due to posttranscriptional events. (ii) The PM levels observed in M and M2SF media correspond to RegB bound by saturating UQ10 (Y = 1) and RegB nonsaturated (Y = 0) with UQ, respectively. (iii) The fractional saturation of RegB with UQ can be described by the Hill function Y = [UQ10]nα/(1 + [UQ10]nα), where α is an unknown association constant. To estimate the apparent cooperativity of UQ binding to RegB, we can employ the known limits of the Hill equation as follows. Standard binding theory states that if the product [UQ10]nα is about 100-fold less than 1, then Y = 0 within a 1% error margin. A similar argument states that Y = 1 when [UQ10]nα is about 100-fold greater than 1. Using these limits, we can express α as follows: α = (100/[UQ10]nM)M = (0.01/[UQ10]nM2SF)M2SF. This equation can be solved to obtain an estimate of the Hill coefficient n. For this calculation, we considered only the UQ data shown in Table 2 for growth in M and M2SF media, respectively. To normalize the amounts of [UQ10] obtained from different growth experiments, we employed the conservation equation [UQ10]t = [UQ10] + [UQ10H2], which can be rewritten as [UQ10]t = [UQ10]·{1 + ([UQ10H2]/[UQ10])}, where [UQ10]t is the total amount of quinone present under a given growth condition. The values for [UQ10]t in M2SF and M media were taken to be 2.97 and 2.81, respectively (Table 1). Calculation of n using the equation α = (100/[UQ10]nM)M = (0.01/[UQ10]nM2SF)M2SF then yielded a value of about 23.6, suggesting that if the M-to-M2SF transition is mediated by a fall in [UQ10], then this effect must be highly cooperative. It is interesting in this context that the active state of RegB appears to be a dimer whereas the inactive state is tetrameric (22). It is well known from cooperativity theory that, for equilibrium binding of a ligand to a multimeric catalytic complex, the Hill coefficient cannot exceed the number of binding sites, thus making it likely that the putative UQ regulation of RegB measured here as the change in PM is a complex interplay of several reactions, probably involving bistable states (corresponding to infinite cooperativity) of a type described recently (27). However, we emphasize that our calculation yields only a rough estimate of the Hill coefficient; a more reliable value must await precise binding experiments.
We note that our data are subject to a number of uncertainties and alternative interpretations. First, it is not known if there is a single, redox-equilibrated pool of UQ10 plus UQ10H2 in the cell or whether several rather localized UQ pools exist. We are presently examining this aspect. Second, our interpretation ignores heterogeneous responses caused by stochastic variations in the levels of signal transducers. However, in the absence of experimental data to the contrary, we are inclined to think that for an organism such as R. rubrum, which shows generation times from 3.5 to 5 h, the nondivision period is probably sufficient to allow most activators and repressors to achieve similar levels in active cells. So far, all studies of stochasticity in bacteria of which we are aware have employed rapidly dividing E. coli (generation time of 20 min).
Finally, on a speculative note, we note that the cytoplasmic redox potential of about −310 mV is very close to that reported (E°′ = −294 mV) for Cys265 of RegB (22), which makes it suitable for reversible oxidation within the reduced cytoplasmic compartment but far distant from the potential redox partner UQ10 (E°′ = +90 mV) (25) (Fig. 5B). Although the direct participation of NADH is a thermodynamic possibility, this has never been reported experimentally, nor is it consistent with the poor correlation to the PM level (Fig. 4B). We note also that although direct oxidation of Cys265 by UQ has never been proposed explicitly, it is nevertheless a possible scenario in analogy to the ArcB mechanism. It was demonstrated that although mutation of Cys265 of RegB did not abolish inhibition by UQ, a significant (20% [22]) diminution in activity of the mutant was observed in vitro. We wish therefore to raise the possibility that the UQ10/UQ10− (UQ10− is the semiquinone free radical) ratio is much more suited to reversible oxidoreduction close to the cytoplasmic redox potential and that the RQ10/RQ10− redox pair, where the redox potential is not known but which is probably even lower than that of UQ10/UQ10−, is also a potential player.
Acknowledgments
This work was supported by the FORSYS initiative of the German Federal Ministry of Education and Research and the Center Systems Biology at University Stuttgart.
We thank Ruxandra Rehner for technical assistance and Caroline Autenrieth for reading the manuscript.
Footnotes
Published ahead of print on 16 May 2008.
REFERENCES
- 1.Bekker, M., G. Kramer, A. F. Hartog, M. J. Wagner, C. G. de Koster, K. J. Hellingwerf, and M. J. Teixeira de Mattos. 2007. Changes in the redox state and composition of the quinone pool of Escherichia coli during aerobic batch-culture growth. Microbiology 1531974-1980. [DOI] [PubMed] [Google Scholar]
- 2.Carr, N. G., and G. Exell. 1965. Ubiquinone concentrations in Athiorodaceae grown under various environmental conditions. Biochem. J. 96688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castrillo, J. I., A. Hayes, S. Mohammed, S. J. Gaskell, and S. G. Oliver. 2003. An optimized protocol for metabolome analysis in yeast using direct infusion electrospray mass spectrometry. Phytochemistry 62929-937. [DOI] [PubMed] [Google Scholar]
- 4.de Graef, M. R., S. Alexeeva, J. L. Snoep, and M. J. Teixeira de Mattos. 1999. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J. Bacteriol. 1812351-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drews, G. 1978. Structure and development of the membrane system of photosynthetic bacteria. Curr. Top. Bioenerg. 8B161-207. [Google Scholar]
- 6.Elsen, S., L. R. Swem, D. L. Swem, and C. E. Bauer. 2004. RegB/RegA, a highly conserved redox-responding global two-component system. Microbiol. Mol. Biol. Rev. 68263-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georgellis, D., O. Kwon, and E. C. C. Lin. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 2922314-2316. [DOI] [PubMed] [Google Scholar]
- 8.Ghosh, R., A. Hardmeyer, I. Thoenen, and R. Bachofen. 1994. Optimization of the Sistrom culture medium for large-scale batch cultivation of Rhodospirillum rubrum under semiaerobic conditions with maximal yield of photosynthetic membranes. Appl. Environ. Microbiol. 601698-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gimenez-Gallego, G., M. P. Ramirez-Ponce, P. Lauzurica, and J. M. Ramirez. 1982. Photooxidase system of Rhodospirillum rubrum. III. The role of rhodoquinone and ubiquinone in the activity of preparations of chromatophores and photoreaction centers. Eur. J. Biochem. 121343-347. [DOI] [PubMed] [Google Scholar]
- 10.Grammel, H., E. D. Gilles, and R. Ghosh. 2003. Microaerophilic cooperation of reductive and oxidative pathways allows maximal photosynthetic membrane biosynthesis in Rhodospirillum rubrum. Appl. Env. Microbiol. 696577-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiraishi, A. 1988. Fumarate reduction systems in members of the family Rhodospirillaceae with different quinone types. Arch. Microbiol. 15056-60. [Google Scholar]
- 12.Imhoff, J. F., and U. Bias-Imhoff. 1995. Lipids, quinones and fatty acids of anoxygenic phototrophic bacteria, p. 179-205. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 13.Kiley, P., and S. Kaplan. 1988. Molecular genetics of photosynthetic membrane biosynthesis in Rhodobacter sphaeroides. Microbiol. Rev. 5250-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, Y. J., I. J. Ko, J. M. Lee, H. Y. Kang, Y. M. Kim, S. Kaplan, and J. I. Oh. 2007. Dominant role of the cbb3 oxidase in regulation of photosynthesis gene expression through the PrrBA system in Rhodobacter sphaeroides. J. Bacteriol. 1895617-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malpica, R., B. Franco, C. Rodriguez, O. Kwon, and D. Georgellis. 2004. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. USA 10113318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Gara, J. P., J. M. Eraso, and S. Kaplan. 1998. A redox-responsive pathway for aerobic regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1804044-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh, J. I., and S. Kaplan. 2000. Redox signalling: globalization of gene expression. EMBO J. 794237-42475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Redfearn, E. R. 1967. Redox reactions of ubiquinone in Rhodospirillum rubrum. Biochim. Biophys. Acta 131218-220. [Google Scholar]
- 19.Schön, G. 1971. Der Einfluβ der Kulturbedingungen auf den Nicotinamid-Adenin-Dinucleotid(phosphate)-Gehalt in Zellen von Rhodospirillum rubrum. Arch. Mikrobiol. 79147-163. [PubMed] [Google Scholar]
- 20.Sistrom, W. R. 1960. A requirement for sodium in the growth of Rhodopseudomonas sphaeroides. J. Gen. Microbiol. 22778-785. [DOI] [PubMed] [Google Scholar]
- 21.Sugimura, T., and H. Rudney. 1962. The effect of aerobiosis and diphenylamine on the content of ubiquinone in Rhodospirillum rubrum. Biochim. Biophys. Acta 62167-170. [DOI] [PubMed] [Google Scholar]
- 22.Swem, L. R., B. J. Kraft, D. L. Swem, A. T. Setterdahl, S. Masuda, D. B. Knaff, J. M. Zaleski, and C. E. Bauer. 2003. Signal transduction by the global regulator RegB is mediated by a redox-active cysteine. EMBO J. 224699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swem, L. R., X. Gong, C. A. Yu, and C. E. Bauer. 2006. Identification of a ubiquinone-binding site that affects autophosphorylation of the sensor kinase RegB. J. Biol. Chem. 2816768-6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takamiya, K., and A. Takamiya. 1969. Light-induced reactions of ubiquinone in a photosynthetic bacterium, Chromatium D. III. Oxidation reduction state of ubiquinone in intact cells of Chromatium D. Plant Cell Physiol. 10363-373. [Google Scholar]
- 25.Takamiya, K. I., and P. L. Dutton. 1979. Ubiquinone in Rhodopseudomonas sphaeroides some thermodynamic properties. Biochim. Biophys. Acta 5461-16. [DOI] [PubMed] [Google Scholar]
- 26.Taniguchi, S., and M. D. Kamen. 1965. The oxidase system of heterotrophically-grown Rhodospirillum rubrum. Biochim. Biophys. Acta 96395-428. [DOI] [PubMed] [Google Scholar]
- 27.Xiong, W., and J. E. Ferrell. 2003. A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature 426460-465. [DOI] [PubMed] [Google Scholar]