Abstract
The Streptococcus pyogenes genome harbors two clusters of class Ib ribonucleotide reductase genes, nrdHEF and nrdF*I*E*, and a second stand-alone nrdI gene, designated nrdI2. We show that both clusters are expressed simultaneously as two independent operons. The NrdEF enzyme is functionally active in vitro, while the NrdE*F* enzyme is not. The NrdF* protein lacks three of the six highly conserved iron-liganding side chains and cannot form a dinuclear iron site or a tyrosyl radical. In vivo, on the other hand, both operons are functional in heterologous complementation in Escherichia coli. The nrdF*I*E* operon requires the presence of the nrdI* gene, and the nrdHEF operon gained activity upon cotranscription of the heterologous nrdI gene from Streptococcus pneumoniae, while neither nrdI* nor nrdI2 from S. pyogenes rendered it active. Our results highlight the essential role of the flavodoxin NrdI protein in vivo, and we suggest that it is needed to reduce met-NrdF, thereby enabling the spontaneous reformation of the tyrosyl radical. The NrdI* flavodoxin may play a more direct role in ribonucleotide reduction by the NrdF*I*E* system. We discuss the possibility that the nrdF*I*E* operon has been horizontally transferred to S. pyogenes from Mycoplasma spp.
Ribonucleotide reductases (RNRs) are a group of enzymes that carry out the only metabolic pathway for providing living cells with optimal amounts of the necessary buildings blocks for DNA replication and DNA repair (29). Three classes of RNRs (I, II, and III) have been characterized to date according to their different mechanisms for radical generation, allosteric regulation, and oxygen dependence and for their quaternary structural differences, but all of them have in common a reaction mechanism based on radical chemistry and the catalytic core structure (29).
Class I RNRs (which are further subdivided into class Ia and class Ib) are composed of two homodimeric proteins, R1 (α2) and R2 (β2). The larger α subunit contains the catalytic and allosteric sites, while the smaller β subunit harbors a stable free radical at a tyrosine residue and a diiron center bridged by an oxo (O2−) ion. Class Ib shares with class Ia this subunit composition as well as all conserved residues, including the tyrosyl radical and the iron ligands in the β subunit and the active-site cysteines in the α subunit, but all class Ib α subunits lack approximately 50 N-terminal amino acids that constitute the allosteric-activity site. The α and β subunits are encoded by the nrdA and nrdB genes for class Ia and the nrdE and nrdF genes for class Ib. Class Ib operons contain two additional nrd genes: nrdH, which encodes an electron donor glutaredoxin-like protein (19), and nrdI, which encodes a small flavoprotein whose function remains unknown (18). Typically, class Ib nrd genes are arranged as an nrdHIEF operon, although examples of bacteria bearing class Ib genes in slightly different patterns have been described as well (43).
The genome of the gram-positive bacterium Streptococcus pyogenes (group A Streptococcus) harbors three different clusters of genes involved in ribonucleotide reduction (10). One cluster contains the genes for the anaerobic class III RNR and its cognate activase (nrdDG), but surprisingly, each of the two other clusters contains genes to form unique class Ib operons (nrdHEF and nrdF*I*E*). The S. pyogenes genome also contains a second stand-alone copy of an nrdI gene (designated nrdI2 in this work) elsewhere in the chromosome. Two features make the class Ib operons in S. pyogenes unique. First, the operons are arranged in a way that deviates from the common nrdHIEF operon structure, and second, three residues involved in iron binding are not conserved within the β subunit of the nrdF*I*E* operon.
Although it is common to find multiple RNR classes within a single microorganism (41), just a few cases of potential redundancy, i.e., with a duplication of nrd genes belonging to the same class, have been characterized. Saccharomyces cerevisiae has a class Ia redundancy (Rnrp1 to Rnrp4), and Mycobacterium tuberculosis has a second class Ib-like β subunit gene (46, 48; also see http://rnrdb.molbio.su.se for additional candidates). S. pyogenes is one of a few bacteria (all belonging to the Firmicutes) that have two distinct class Ib RNR gene clusters. A redundancy of RNR genes might be of great importance under different environmental conditions. In this study, we have analyzed both the in vivo and the in vitro functionality of the S. pyogenes class Ib nrd genes in an attempt to provide an explanation for this atypical redundancy.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli strains were routinely grown in Luria-Bertani (LB) broth at 15, 30, or 37°C. Streptococcus pyogenes Rosenbach M1 (ATCC 700294, SF370) was grown in Todd-Hewitt broth (Oxoid) supplemented with 0.2% yeast extract (THY) at 37°C and in Columbia sheep blood agar plates (BioMérieux) whenever needed. Streptococcus pneumoniae (Klein) Chester R6 (ATCC BAA-255) was grown on Columbia sheep blood agar plates (BioMérieux) at 37°C. Antibiotics and chromogenic substrates, when required, were included in the culture media or on agar plates at the following concentrations for E. coli: 100 μg/ml carbenicillin, 100 μg/ml kanamycin, 34 μg/ml chloramphenicol, and 30 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Sigma).
The E. coli strain IG101 [nrdA(Ts) nrdB1 thyA thr leu thi deoC and/or deoD tonA lacY supE44 gyrA nrdH::Spc] was constructed by introducing the nrdH::Spc allele (IG100 [this laboratory]) into KK450 (31) by P1 transduction.
Sequence analysis and DNA manipulations.
Genomic DNA from S. pyogenes was extracted as described previously (11). Electroporation of S. pyogenes was performed as described previously (25), and other general DNA manipulations were done by standard procedures (34). DNA sequence determination was carried out using the BigDye Terminator kit and an ABI Prism 310 sequencer (Applied Biosystems) according to the manufacturer's specifications. Images of gels were captured with a Gel Doc 2000 station and the Quantity One software package (Bio-Rad Laboratories).
All nrd sequences were from RNRdb, the RNR database (http://rnrdb.molbio.su.se). The Clustal W 1.8 (40) and GeneDoc 2.6.001 (28) programs were used to perform global alignments.
RNA isolation.
S. pyogenes was grown in 10 ml of THY until the A600 was approximately 0.4. Ten-milliliter cultures were centrifuged and resuspended in 200 μl of diethyl pyrocarbonate (Sigma)-treated distilled water and quick-frozen in a dry ice bath. Pellets were thawed on ice and added to 2 ml FastPrep Blue tubes containing zirconite/silica beads, 500 μl of CPRS-Blue, and 500 μl of acid phenol, as described by the manufacturer (Bio101, La Jolla, CA). Bacteria were lysed with a FastPrep FP120 instrument (Bio101, La Jolla, CA) at setting 6 for 11 s and immediately placed on ice after disruption. Samples were incubated at 65°C for 10 min and centrifuged at 10,000 × g for 5 min at 4°C. The upper aqueous phase was recovered and extracted with an equal volume of acid phenol heated to 65°C. The phases were separated by centrifugation as described above, and the extraction was repeated with acid phenol-chloroform (1:1, vol/vol) and chloroform-isoamyl alcohol (24:1, vol/vol). The recovered aqueous phase was treated with 50 U of DNase I (Ambion, Inc.) for 30 min at 37°C, and then 0.2 volume of inactivating reagent (Ambion, Inc.) was added to eliminate the enzyme. RNA concentrations were determined by measuring the A260. Samples were immediately stored at −80°C. Reverse transcription-PCR (RT-PCR) was performed as described in reference 43, and identical aliquots were processed in parallel without the addition of RT in order to ensure that residual chromosomal DNA was not being used as the template for PCR amplification.
Cloning and expression.
The nrdF, nrdE, nrdF*, and nrdE* genes were amplified by PCR using primer pairs containing NdeI (upper primers) and BamHI (lower primers) restriction sites and S. pyogenes genomic DNA as a template (see Table S1 in the supplemental material). The resulting fragments were purified and digested with NdeI and BamHI. The digested nrdF, nrdE, and nrdE* fragments were inserted into pET22b(+) (Novagen) cut with the same enzymes to give p22-FSpyo, p22-ESpyo, and p22-E*Spyo, respectively. The digested nrdF* fragment was inserted into pET24a(+) (Novagen) cut with the same enzymes to give p24-F*Spyo. The sequences of all of the above-named constructs were verified by sequencing.
E. coli BL21(DE3) (Novagen) cells transformed with one of the expression plasmids (p22-FSpyo, p22-ESpyo, or p22-E*Spyo) were grown overnight in LB medium containing carbenicillin at 30 or 37°C. This culture was then used to inoculate 5-liter flasks containing 2 liters of LB medium with the same concentration of antibiotic as noted above (1:100). Cells were then grown at 30°C under vigorous shaking to an A600 of ∼0.5 to 0.6 and subsequently induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside; final concentration) for 4 hours.
BL21(DE3) transformed with plasmid p24-F*Spyo was grown overnight in LB medium containing kanamycin at 30°C. This culture was then used to inoculate 5-liter flasks containing 2 liters of LB medium with the same concentration of antibiotic as noted above (1:100). Cells were then grown at 30°C with vigorous shaking. When they reached an A600 of ∼0.5 to 0.6, the cultures were chilled to 15°C before being induced with 0.3 mM IPTG (final concentration) and then grown for 15 to 17 h at 15°C. After induction, cells were harvested by centrifugation and the pellets quickly frozen and stored at −80°C.
Protein purifications.
Frozen pellets were disintegrated by X-press (Biox AB) and extracted in 50 mM Tris-HCl (pH 8), 50 mM KCl, 5 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride (PMSF) (buffer L), after which nucleic acids were precipitated by dropwise addition of streptomycin sulfate to a final concentration of 1%. The precipitate was removed by centrifugation, and solid ammonium sulfate was added to the supernatant to 45% saturation for NrdE and NrdE* and to 60% saturation for NrdF and NrdF*. The precipitate was recovered by centrifugation.
The NrdF and NrdF* pellets were dissolved in buffer L and desalted by dialysis against 2 liters of buffer L for 4 to 5 h. The dialyzed solution was diluted with buffer L to a 5-mg/ml protein concentration and loaded onto a DEAE-Sepharose column (GE Healthcare) coupled to a fast-performance liquid chromatography instrument (Biologic DuoFlow Systems, Bio-Rad). After a washing step with 140 ml of 0.15 M KCl in buffer L, elution was continued with a 0.15 to 0.6 M KCl linear gradient in buffer L in a total volume of 300 ml. Fractions checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis containing the NrdF or NrdF* protein were pooled and concentrated by ultrafiltration on a Centriplus YM-30 instrument (Millipore) against buffer L and stored at −80°C for further purification. The NrdF proteins eluted between 0.3 to 0.36 M KCl. The concentrated enzyme solution was loaded onto a Superdex 75 10/300 GL column (GE Healthcare) previously equilibrated against buffer L. Proteins were eluted with the same buffer at a constant flow rate of 0.4 ml/min. Fractions containing NrdF or NrdF* were pooled, concentrated, and stored as described above.
For the purification of the NrdE and NrdE* proteins, the ammonium sulfate-precipitated pellets were dissolved in 50 mM Tris-HCl (pH 8), 0.75 M ammonium sulfate, 2.5 mM DTT, and 1 mM PMSF and loaded onto a HiLoad 16/10 phenyl-Sepharose high-performance column (GE Healthcare) equilibrated with the same buffer. Elution was performed with a 0.75 to 0 M ammonium sulfate linear gradient in 50 mM Tris-HCl (pH 8), 2.5 mM DTT, and 1 mM PMSF. NrdE and NrdE* eluted between 0.44 to 0.35 M and 0.03 to 0 M ammonium sulfate, respectively. The enzyme-containing fractions were pooled and concentrated in Centriplus YM-50 (Millipore) and then adsorbed to a 2 ml MonoQ anion-exchange column (GE Healthcare) equilibrated with buffer L. Proteins were eluted with 20 ml of a 0.05 to 0.5 M KCl linear gradient in buffer L at a flow rate of 0.1 ml/min. NrdE eluted between 0.21 and 0.28 M KCl and NrdE* between 0.19 and 0.27 M KCl. Fractions of 0.4 ml were pooled and concentrated as described above and then stored at −80°C.
Analytical methods and enzyme activity assays.
Iron reconstitution assays were performed anaerobically as described in reference 17, and iron content was determined as described in reference 33. Protein concentration was determined by the Bradford method (6).
RNR activity was assayed in 50-μl mixtures containing 50 mM Tris-HCl (pH 8), 0.2 mM dATP as an effector, 20 mM magnesium acetate, and 30 mM DTT. The reaction was started by the addition of [3H]CDP (specific activity, 10,000 to 20,000 cpm/nmol) to a final concentration of 1 mM. Assay mixtures were incubated for 10 min at 29°C, and the reaction was stopped by the addition of 0.5 ml of 1 M ice-cold perchloric acid. The amount of dCDP formed was determined by the standard method as described in reference 39. One unit of enzyme activity corresponds to 1 nmol of dCDP formed per minute of incubation. Km, Vmax, and KL (ligand binding constant) values were obtained by direct curve fitting as described previously (3, 7) and with the Kaleida-Graph software (Synergy Software).
UV-visual-light (UV-vis) absorption spectroscopy and decay of the tyrosyl radical site.
Scanning spectra were registered on a Perkin Elmer Lambda2 spectrophotometer at 25°C, and a 20 μM concentration of the monomer in 50 mM Tris-HCl, pH 7.6, was used. The radical decay of the nrdF protein was studied after the addition of hydroxyurea (HU) at final concentrations ranging from 3.5 to 60 mM. Spectra were typically recorded between 300 and 750 nm in cycles of 150 s, with a scanning rate of 240 nm/min. The endpoint spectrum, when radical absorption at 407 nm had decayed, was subtracted from the start spectrum to give the radical concentration using the extinction coefficient 3.25 mM−1 cm−1 (30). Alternatively, the radical concentration was determined by calculating the dropline between 402 and 417 nm and using an ɛ407 of 2.11 mM−1 cm−1 in the equation {A407 − [A402 − (5/15)·(A402 − A417)]}/ɛ. Kinetic traces at 407 nm were also run to determine the decay rate; incubations of E. coli with HU were run at 410 nm as controls. Spectra were run before and after the kinetic trace to measure the degree of completeness of the reaction after subtractions as described above.
EPR spectroscopy.
Electron paramagnetic resonance (EPR) measurements at X-band were performed either at ≥90 K on a Bruker ESP300 spectrometer using a nitrogen flow system or at <60 K on a Bruker ESP500 using a Oxford helium flow cryostat. Radical concentrations were determined, under unsaturated conditions, using an E. coli NrdB sample with known tyrosyl radical concentrations as the standard. The error limits for radical quantitations are at most ±10%. Reconstituted samples for EPR measurements were either frozen with liquid nitrogen 6 min after the addition of 2 Fe ions/polypeptide or frozen after the addition of 4 Fe ions/monomer followed by desalting on a NAP-5 column to remove unbound iron.
Heterologous complementation assay.
Class Ib nrd genes were amplified by PCR using primers pairs containing a ribosomal binding site sequence from E. coli and the SacI (upper primers) and PstI (lower primers) restriction sites (see Table S1 in the supplemental material). The resulting fragments were purified and digested with SacI and PstI and inserted into pBAD18Km or pBAD33Cm (14) to generate the final constructs. p18-FI*Spyo, p18-IE*Spyo, and p18-FE*Spyo were derived from p18-FIE*Spyo by enzymatic digestion.
In the heterologous complementation assay, constructs were transformed into the E. coli strain IG101 and grown overnight at 30°C in LB medium containing 50 μg/ml thymine plus the corresponding antibiotics. The following day, serial dilutions of the liquid cultures were plated onto LB agar plates containing the corresponding antibiotics plus 50 μg/ml thymine and either 0.3% l-arabinose or 0.3% d-glucose and were incubated at both 30 and 42°C for 24 to 48 h. Attempts to obtain S. pyogenes chromosomal mutants either by insertional or recombinant inactivation in all class Ib nrd genes were unsuccessful, which was initially attributed to the essentiality of these genes. However, when we used the same mutagenic strategies to obtain a chromosomal mutant of the previously described nonessential covR gene (9), mutant colonies were recovered at extremely low frequencies (1 × 10−8); therefore, the essentiality of the S. pyogenes class Ib nrd genes could not be assessed by direct complementation.
RESULTS
S. pyogenes encodes two redundant class Ib gene clusters.
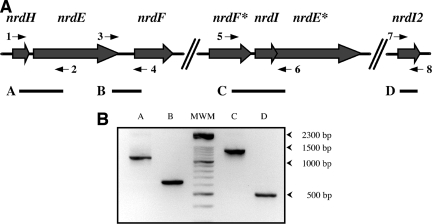
The complete S. pyogenes Rosenbach M1 genome (10) contains seven genes putatively encoding class Ib RNR-related proteins in three different regions of its genome (Fig. 1A). All 10 hitherto-completed S. pyogenes genomes encode the same seven class Ib genes (see the RNRdb at http://rnrdb.molbio.su.se).
FIG. 1.
(A) Schematic organization of the S. pyogenes class Ib nrd genes. Arrows indicate the positions and numbers of the primers used for RT-PCR amplification, and black lines labeled A, B, C, and D show the positions and lengths of the expected transcripts. (B) Linkage RT-PCR of the nrdHE (lane A), nrdEF (lane B), and nrdFIE (lane C) junctions and the nrdI2 gene (lane D). The linkage strategy is outlined in panel A. MWM, molecular weight markers.
The first class Ib region spans over 4.5 kbp and comprises the nrdHEF cluster, which codes for a putative glutaredoxin-like protein (NrdH) as well as the large (NrdE) and small (NrdF) subunits of the holoenzyme. Both the NrdE and NrdF proteins contained all the conserved residues necessary for ribonucleotide reduction (see Table S2 and S3 in the supplemental material).
The second Ib region comprises the nrdF*I*E* cluster and spans over 4 kbp. The nrdI* gene encodes an unknown protein of 18 kDa and 162 amino acids which is 75% similar to E. coli NrdI (19). S. pyogenes nrdE* encodes an RNR large subunit with all the essential cysteines in the active site, the C-terminal cysteine pair, and the two conserved C-terminal tyrosines (required for the transfer of the radical from the small subunit) as well as the residues that form the allosteric-specificity site (see Table S2 in the supplemental material). Finally, the nrdF* gene encodes an RNR small subunit that lacks some important residues for the proper function of the enzyme. Compared to the E. coli NrdF protein, S. pyogenes NrdF* turned out to have amino acid substitutions in three of the six important residues involved in iron binding as well as in one of the residues involved in radical stability. Instead of Glu115 in E. coli NrdB, S. pyogenes NrdF* has Val116, instead of Glu204 it has Pro176, instead of Glu238 it has Lys210, and instead of Phe208 it has Leu180. All other residues important for radical stability and transfer are conserved (see Table S3 in the supplemental material).
A second stand-alone nrdI gene (nrdI2) was annotated elsewhere in the genome (10). S. pyogenes nrdI2 encodes a protein of 160 amino acids and 17.7 kDa which is 36% similar to nrdI*.
The two redundant class Ib operons are expressed simultaneously.
To investigate whether all class Ib RNR regions were simultaneously transcribed and to assess the operonic organization of each cluster, total RNA from strain SF370 was extracted at early exponential phase and used to perform nonquantitative RT-PCR experiments. Primer pairs were selected to bracket the intergenic junctions between nrdHE, nrdEF, and nrdF*I*E* (Fig. 1A). Any PCR fragment generated by these oligonucleotides is therefore derived from a polycistronic transcript.
An amplicon spanning the nrdHE junction was detected by using primer 2 to generate the cDNA and primers 1 and 2 (see Table S1 in the supplemental material) for the PCR (Fig. 1B, lane A). Similarly, a PCR product bracketing the junction between nrdE and nrdF could also be detected with primers 3 and 4 (Fig. 1B, lane B). Using primer 6 to generate a reversed transcribed cDNA template and the primer pair 5-6 to amplify this template, we were also able to detect an amplicon spanning the nrdF*I*E* intergenic junction (Fig. 1B, lane C). An RT-PCR for the second nrdI2 gene was also carried out with primers 7 and 8, and the corresponding transcript was readily detected (Fig. 1B, lane D). The nrdI2 product was sequenced to ensure its correct amplification.
Control experiments conducted concurrently with each set of RT-PCR assays confirmed the quality of the samples and guaranteed the specificity of each component of the RT-PCR. (i) Our inability to amplify the desired product when RT was omitted validated the compulsory requirement of this enzyme for the initiation of cDNA synthesis and ensured that no residual DNA contaminated the starting RNR preparations; (ii) the expected amplicons were detected when genomic DNA acted as the template, demonstrating the fidelity of the PCR primers (data not shown). These results indicated the presence of a polycistronic transcript for each cluster of genes as well as their simultaneous transcription under standard laboratory growth conditions.
Spectroscopic characterization of NrdF and NrdF*.
Once it was demonstrated that all class Ib nrd genes in S. pyogenes were actively transcribed, we decided to check whether they were capable of forming an active holoenzyme. The nrdE and nrdF genes from both the nrdHEF and nrdF*I*E* operons were cloned, overproduced, and purified as described in Materials and Methods. After different purification steps, all purified proteins showed the expected theoretical molecular mass. The purity of the samples in all cases was over 90%. Attempts to overproduce and purify the NrdI* and NrdI2 proteins were unsuccessful since the overproduced proteins were continuously recovered in the insoluble fraction of the bacterial extract, regardless of the incubation temperature or the IPTG concentration used (not shown).
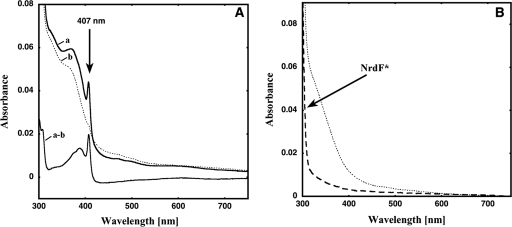
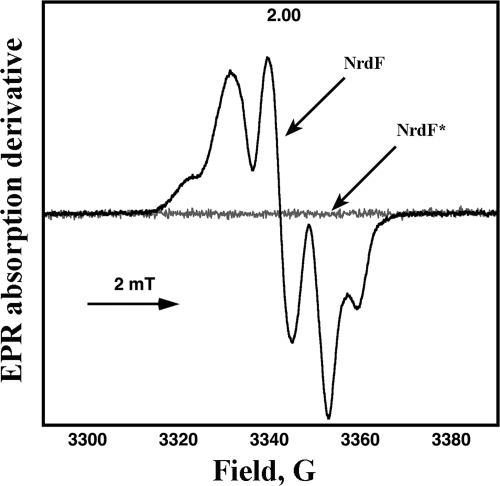
Purified NrdF protein showed a light absorption spectrum typical of a diiron-tyrosyl radical RNR. Charge transfer bands of the μ-oxobridged diferric center were observed at 325 and 370 nm, as were weaker bands at 500 and 600 nm (Fig. 2A). The tyrosyl radical showed a sharp peak at 407 nm. We also registered the EPR spectrum of the NrdF protein (Fig. 3), and the tyrosyl radical proved to be a typical class Ib radical similar to those found in NrdF proteins from S. enterica serovar Typhimurium, Corynebacterium ammoniagenes, and M. tuberculosis (17, 21, 24). At 95 K, the radical saturated at 1 order of magnitude less power than does the E. coli NrdB protein (4.7 mW compared to 54.0 mW [data not shown]) but similarly to the S. enterica serovar Typhimurium, C. ammoniagenes, and M. tuberculosis NrdF proteins (3.7 mW, 1.3 mW, and 1.3 mW, respectively) (17, 44). The S. pyogenes NrdF radical signal had the same hyperfine structure at 10 to 20 K and at 95 K as that previously observed for the class Ib radicals mentioned above, in contrast to the E. coli Ia signal, which is considerably broadened at 95 K compared to its structure at 10 to 20 K (32). The radical saturation behavior reflects its environment, in this case plausibly coupling to the iron, and its geometry. The difference from the E. coli Ia radical may indicate that the S. pyogenes NrdF radical has a weaker coupling to the iron site, perhaps with a water molecule between the radical and the closest iron, as for other class Ib tyrosyl radicals. Addition of the radical scavenger HU to reduce the tyrosyl radical gave the spectrum shown in Fig. 2A (dotted line), where it is shown that the diiron site remained essentially unchanged. Subtraction of the endpoint spectrum gives a typical tyrosyl radical spectrum (Fig. 2A, lowest spectrum) with a sharp peak at 407 nm. Using an extinction coefficient (ɛ) of 3.25 mM−1 cm−1 (30) in the subtracted spectra or using the dropline as specified in Materials and Methods, we calculated, for the best preparation, a radical content of 0.51 per monomer, in good agreement with the 0.57 radical per monomer determined by EPR. Analysis of the NrdF iron content showed approximately 1.2 Fe ions/monomer (Table 1). It is not uncommon that RNRs purify with suboptimal iron content and, consequently, a small amount of radical when they are overexpressed in E. coli. Hence, we added ferrous ions to the protein to investigate if we could increase the yield of the diiron-radical site. Addition of ferrous ions, however, did not increase the radical yield of NrdF in the radical-containing samples. To assess the stability of the tyrosyl radical, we monitored its reactivity toward the radical scavenger HU. S. pyogenes NrdF reacted at a considerably lower rate (k1 [first order rate constant] of 0.12 min−1 with 60 mM HU) than E. coli NrdB (k1 of 0.50 min−1 with 15 mM HU) at 25°C; i.e., it is at least an order of magnitude less sensitive than the E. coli enzyme to reduction by HU.
FIG. 2.
(A) Line a, UV-vis spectra of the S. pyogenes NrdF protein. The arrow indicates the tyrosyl radical. Dotted line b, HU-treated sample. Line a-b, subtraction of the HU-treated spectrum from the purified NrdF spectrum. (B) UV-vis spectra of the S. pyogenes NrdF* protein upon purification (dashed line) and after the addition of 2 Fe ions/monomer.
FIG. 3.
First-derivative EPR spectra of NrdF and NrdF*, as indicated in the figure.
TABLE 1.
Iron and radical contents of S. pyogenes NrdF and NrdF* before and after iron reconstitutiona
| Protein | No. of Fe ions/monomer | No. of tyrosyl radicals/monomer |
|---|---|---|
| NrdF | 1.2 | 0.5 |
| NrdFR | 2.1 | 0.5 |
| NrdF* | 0.05 | ND |
| NrdFR* | 0.47 | ND |
R, iron was reconstituted; ND, not detected.
On the other hand, purified NrdF*, which was obtained in the apoform, showed no prominent features in light absorption spectra (Fig. 2B, dashed line) and no EPR signal (Fig. 3); i.e., it had no diiron-tyrosyl radical site. As expected, analysis of the purified NrdF* iron content showed only 0.05 Fe ion/monomer (Table 1). The addition of 2 Fe ions/monomer gave a spectrum without radical absorption and without the characteristics of the diiron site observed in the NrdF protein but with absorption in the 325- to 450-nm region, similar to what has been observed in iron ligand mutants in E. coli NrdB (Fig. 2B, dotted line) (ɛ350 ≈ 1,800 M−1 cm−1) (1, 45). Iron analysis of the reconstituted and desalted samples gave values of ∼0.47 Fe ion/monomer. Presumably, the altered residues within the iron binding center of this protein impair the formation of a dinuclear iron center and a tyrosyl radical.
The NrdEF enzyme is enzymatically active, whereas the NrdE*F* enzyme is not.
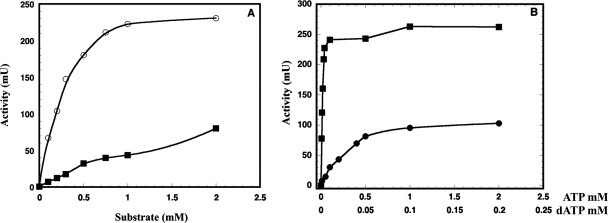
When assayed for enzymatic activity, none of the four proteins had any activity on its own. When mixed and assayed together, the NrdE and NrdF proteins purified from the nrdHEF system were active and showed proportional dCDP formation, with one protein in excess and increasing amounts of its partner. Specific activities, in the presence of dATP as a positive allosteric effector, were 169 nmol/min/mg for the NrdF protein and 45 nmol/min/mg for the NrdE protein. Activity was dependent on the addition of magnesium and DTT at an optimal pH of 8.0 and required CDP as a substrate, with a Km of 0.34 mM (Fig. 4A). CDP reduction was strongly stimulated by dATP as an effector and to a lesser extent by ATP. Optimal activity with dATP was obtained at nucleotide concentrations as low as 0.01 mM, and no significant inhibition was seen even with 2 mM dATP. When ATP was used, concentrations up to 2 mM were not enough to achieve optimal activity. From Fig. 4B, apparent KL values of 0.0011 mM for dATP and approximately 0.3 mM for ATP can be calculated. No significant gain in specific activity was detected for the iron-reconstituted NrdF protein, in agreement with the data provided by EPR and UV-vis spectroscopy.
FIG. 4.
(A) Comparison of CDP and CTP as substrates for the NrdF/NrdE reductase. Assay mixtures were made under standard conditions with 1.2 μM NrdE and 5 μM NrdF (○, CDP; ▪, CTP). (B) Effect of ATP and dATP on CDP reduction. Incubation was with 1.2 μM NrdE and 5 μM NrdF, replacing the standard concentration of 0.2 mM dATP by the indicated concentrations of either ATP (•) or dATP (▪).
The NrdE* and NrdF* proteins had no activity when assayed together, nor when NrdF* was combined with the catalytically competent NrdE protein, which agrees with the lack of a tyrosyl radical in NrdF*. One might, on the other hand, expect some activity when combining the NrdE* and NrdF proteins, since NrdE* seems to have all the catalytically important residues and NrdF harbors a tyrosyl radical and is catalytically competent. However, the mixing of NrdE* and NrdF did not result in any enzyme activity. One explanation for the lack of activity in this case is that NrdE* and NrdF cannot form a holoenzyme complex. To test this, we performed competition assays to detect protein interactions between subunits from the different systems. Fixed concentrations of NrdF and NrdE were assayed against increasing amounts of either NrdE* or NrdF*. As shown in Fig. 5, concentrations of up to 2 μM NrdF* or NrdE* had no inhibitory effect on NrdF/NrdE holoenzyme activity, suggesting that subunits from the two systems do not cross-react.
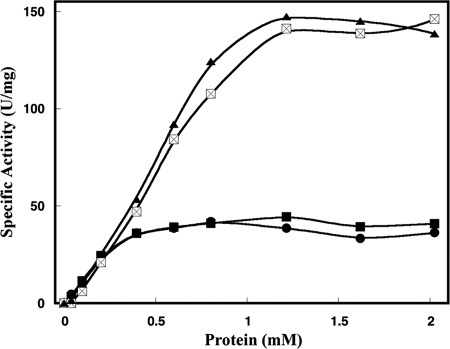
FIG. 5.
Assay for the competitive inhibition of both NrdE and NrdF. Incubations were carried out under standard conditions (i) with increasing concentrations of NrdF in the presence of 0.6 μM NrdE (•) and 0.6 μM NrdE together with 2 μM NrdF* (▪) or (ii) with increasing concentrations of NrdE in the presence of 0.6 μM NrdF (⊠) and 0.6 μM NrdF together with 2 μM of NrdE* (▴).
Complementation assays show that nrdF*I*E* is functional in vivo and that nrdI is essential for complementation.
We also decided to compare the in vitro results of the nrdHEF and nrdF*I*E* systems with the results of complementation assays of the in vivo functionality of each operon. To do so, we used the E. coli IG101 strain, which is an nrdA(Ts)-nrdB1/nrdH::Spc double mutant unable to reduce ribonucleotides aerobically at 42°C unless a functional RNR is provided to complement the temperature-sensitive phenotype. The entire S. pyogenes nrdHEF and nrdF*I*E* sequences were cloned separately into the pBAD18 vector under the control of the E. coli arabinose promoter (PBAD), an on/off switch promoter repressed by glucose and induced by arabinose, generating plasmids p18-HEFSpyo and p18-FIE*Spyo, which were then transformed into the E. coli IG101 strain. Complementation was determined by plating serial dilutions of liquid cultures on LB agar plates supplemented with either 0.3% l-arabinose or 0.3% d-glucose at both 30 and 42°C. As an independent positive control, we used the Salmonella enterica serovar Typhimurium nrdHIEF sequence cloned into the pBAD18 vector (plasmid p18-HIEFSal). Unexpectedly, the IG101 strain transformed with p18-HEFSpyo was not able to grow at 42°C in the presence of either arabinose or glucose, whereas IG101 transformed with p18-FIE*Spyo gave 1.56 × 108 CFU/ml after 24 to 48 h at 42°C in plates supplemented with arabinose, but not with glucose, indicating that the nrdF*I*E* cluster was capable of complementing the temperature-sensitive phenotype (Table 2). The same results were obtained with constructs in the low-copy-number plasmid pBAD33 instead of pBAD18 (data not shown). Neither pBAD18 nor pBAD33 alone was able to support the growth of the IG101 strain at the restrictive temperature.
TABLE 2.
CFU/ml and plating efficiencies for pBAD constructs in the heterologous complementation assay
| pBAD construct | Mean no. of CFU/ml ± SD at 42°C with 0.3 % arabinose | Mean plating efficiency ± SDa |
|---|---|---|
| p18-HIEFSal | 1.56 × 108 ± 9.60 × 105 | 0.214 ± 1.31 × 10−3 |
| p18-HEFSpyo | <10 | <1 × 10−8 |
| p18-FIE*Spyo | 1.56 × 108 ± 8.10 × 105 | 0.214 ± 1.11 × 10−3 |
| p18-FI*Spyo | <10 | <1 × 10−8 |
| p18-FE*Spyo | <10 | <1 × 10−8 |
| p18-IE*Spyo | <10 | <1 × 10−8 |
| p18-FI*Spyo/p33-E*Spyo | 1.56 × 108 ± 5.69 × 105 | 0.214 ± 7.78 × 10−4 |
| p18-FE*Spyo/p33-I*Spyo | 1.56 × 108 ± 4.24 × 105 | 0.213 ± 5.80 × 10−4 |
| p18-IE*Spyo/p33-F*Spyo | 1.54 × 108 ± 7.78 × 105 | 0.211 ± 1.06 × 10−3 |
| p18-HEFSpyo/p33-I2Spyo | <10 | <1 × 10−8 |
| p18-HEFSpyo/p33-I*Spyo | <10 | <1 × 10−8 |
| p18-HEFSpn/p33-I2Spn | 1.55 × 108 ± 9.74 × 105 | 0.212 ± 1.33 × 10 −3 |
| p18-HEFSpn/p33-I2Spyo | <10 | <1 × 10−8 |
| p18-HEFSpn/p33-I*Spyo | <10 | <1 × 10−8 |
| p18-HEFSpyo/p33-I2Spn | 9.66 × 104 ± 3.21 × 103 | 0.00013 ± 4.39 × 10−6 |
Plating efficiencies were determined as the fraction of CFU/ml under restrictive conditions compared to the number of CFU/ml under nonrestrictive conditions. Values are based on results of at least six independent experiments.
Our next step was to determine if all three genes within the nrdF*I*E* operon were required for the observed in vivo complementation. A set of pBAD18/pBAD33 vectors were constructed so that each gene within the nrdF*I*E* operon could be evaluated in pairwise combinations with the other genes. No complementation was detected when any of the three nrdF*I*E* genes was missing (Table 2), suggesting that the entire operon is needed for complementation. On the other hand, complementation was restored when the missing gene was provided in trans.
These results stress the essential role of the nrdI* gene for in vivo activity. In view of these results, both the S. pyogenes nrdI* and nrdI2 genes cloned into pBAD33 were cotransformed with the nrdHEF operon into the IG101 strain. However, and as shown in Table 2, neither S. pyogenes nrdI* nor nrdI2 could complement S. pyogenes nrdHEF to overcome the temperature-sensitive phenotype. We then checked whether an nrdI gene from the closely related bacterium Streptococcus pneumoniae could restore complementation when combined with S. pyogenes nrdHEF. S. pneumoniae harbors an nrdHEF operon as well as an isolated nrdI2 gene but lacks an nrdFIE operon (16). Plasmids p18-HEFSpn and p33-I2Spn were generated by cloning the nrdHEF and nrdI2 genes from S. pneumoniae into the pBAD vectors. When assayed alone, p18-HEFSpn did not complement IG101 at 42°C. Transformants with both p18-HEFSpn and p33-I2Spn, however, restored the growth of the IG101 strain at 42°C in arabinose-LB plates (but not in glucose), showing that the S. pneumoniae nrdHEF operon can complement if it is supplemented with S. pneumoniae nrdI2. When we combined the S. pneumoniae nrdHEF operon with either the S. pyogenes nrdI* or nrdI2 gene, however, we were unable to detect complementation at the restrictive temperature, suggesting that neither S. pyogenes NrdI nor NrdI2 can successfully interact with the S. pneumoniae system. In contrast, growth at 42°C was indeed detected when we combined the S. pyogenes nrdHEF operon with the S. pneumoniae nrdI2 gene, although the plating efficiency was low (Table 2). These results show that the S. pyogenes nrdHEF operon has functionality also in vivo, given that it is supplemented with a functional nrdI gene.
DISCUSSION
Genomes that display multiple RNRs are found in almost every group of bacteria (41). The concurrence of an aerobic enzyme (class I) and an anaerobic one (class III) in facultative microorganisms reflects a metabolic need. In species that have combinations of RNRs that can be used under the same environmental conditions (such as classes I and II, Ia and Ib, or II and III), it has been proposed that one of the enzymes might support normal cell growth while the other would support DNA repair and growth recovery (4, 5, 8, 26, 37, 42). The genome of S. pyogenes is one of a few genomes that contain redundant RNR operons—here, two different class Ib operons. Moreover, only three genes are found within each of these operons (instead of the four nrdHIEF genes in the classical class Ib systems). The S. pyogenes nrdHEF cluster lacks an nrdI gene, and the nrdF*I*E* cluster lacks an nrdH gene. A second nrdI gene (nrdI2) is found elsewhere in the S. pyogenes chromosome. Both of the Ib gene clusters are transcribed simultaneously with two independent polycistronic operons. In addition, Western blotting against rabbit-raised nrdF antibodies demonstrated the presence of both nrdF and nrdF* products within this microorganism (data not shown), suggesting that both enzyme systems are expressed simultaneously.
Only one of the two redundant class Ib RNRs in S. pyogenes is active in vitro.
The NrdF subunit from the S. pyogenes nrdHEF cluster is a classical class Ib protein with a dinuclear iron center and a stable tyrosyl radical (30). It was catalytically competent when assayed in vitro in the presence of its corresponding NrdE subunit. Specific activities for this system were within the range of those described in class Ib systems from M. tuberculosis (48) and C. ammoniagenes (11). The nrdHEF-encoded enzyme also displayed dATP insensitivity, which is a common feature in class Ib enzymes since their α subunit lacks 50 to 60 N-terminal residues which in class Ia enzymes constitute the ATP/dATP allosteric-activity site (29). Collectively these results show that the nrdHEF system behaves as a functional class Ib enzyme in vitro.
The NrdF* subunit from the nrdF*I*E* cluster lacks three of the six conserved side chains that normally ligate the iron center. It has a low iron content and lacks the tyrosyl radical as well as enzyme activity in vitro in mixtures with the NrdE* subunit. Yet, NrdE* seems to retain all catalytically important residues. Components from one class Ib cluster do not cross-react with components from the other cluster.
In vivo heterologous complementation highlights the essentiality of nrdI.
Our in vivo results showed a completely different picture. Both S. pyogenes gene clusters proved to be functional in heterologous complementation assays even though the nrdHEF operon required coexpression of an nrdI gene, in this case from S. pneumoniae. To our surprise, the addition of either nrdI* or nrdI2 from S. pyogenes did not render nrdHEF functional in vivo. Moreover, in vivo complementation by nrdF*I*E* was achieved only when all three genes were simultaneously present, thus indicating the compulsory requirement for the nrdI* gene product. In a similar way, the S. pneumoniae nrdHEF operon was able to complement only if it was coexpressed with its own nrdI gene. These results show that S. pyogenes nrdI* is functional in vivo and suggest that S. pyogenes nrdI2 is not.
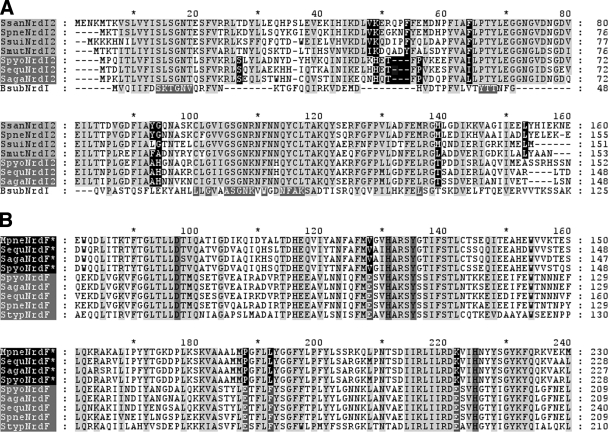
An nrdI gene can be found in every class Ib-containing microorganism (http://rnrdb.molbio.su.se/), and preliminary assays in our laboratory with the Salmonella enterica serovar Typhimurium class Ib RNR indicate that this gene is required for in vivo class Ib ribonucleotide reduction in this microorganism (I. Sala, unpublished results). The Bacillus subtilis nrdI gene has also been shown to be essential (22, 36). Protein comparison between S. pyogenes NrdI* and streptococcal NrdI2 showed that they are distant relatives (36% similarity). Sequence comparison between the streptococcal NrdI2 proteins showed that two different groupings could be distinguished according to small differences in their primary sequences (Fig. 6A). The first group comprised the NrdI2 proteins from S. pneumoniae, Streptococcus mutans, and Streptococcus suis, this being the only NrdI-like protein in their genomes, while the second group included NrdI2 proteins from S. pyogenes, Streptococcus agalactiae, and Streptococcus equi, all of which posses an nrdF*I*E* cluster. As shown in Fig. 6A, clustering of these sequences into two separate groups is due mainly to a 9-bp (3 amino acids) indel and approximately 10 substituted residues differing between the two groups. Comparison of B. subtilis NrdI and streptococcal NrdI2 proteins indicates that none of these differences affect the residues inferred in flavin mononucleotide (FMN) binding (see below), as these regions of NrdI2 seem to be fully conserved among all the streptococcal polypeptide chains. It is remarkable that the three streptococcal species that bear the nrdI2 gene with the indel also have in common a redundant class Ib operon.
FIG. 6.
(A) Clustal W alignment of deduced amino acid fragments of the nrdI gene. Ssan, Streptococcus sanguis; Spne, Streptococcus pneumoniae; Ssui, Streptococcus suis; Smut, Streptococcus mutans; Spyo, Streptococcus pyogenes; Sequ, Streptococcus equi; Saga, Streptococcus agalactiae; Bsub, Bacillus subtilis. Shading of the sequence names indicates common clustering. Those residues unique to each group are highlighted in black. Conserved residues among all streptococci are highlighted in light gray, and the FMN binding residues in B. subtilis are shown in dark gray. (B) Clustal W alignment of deduced amino acid fragments of the nrdF gene. Mpne, Mycoplasma pneumoniae; Styp, Salmonella enterica serovar Typhimurium. Conserved residues are highlighted in light gray, and the conserved radical tyrosyl residue and conserved iron binding residues are highlighted in dark gray. Substituted residues in S. pyogenes, S. equi, S. agalactiae, Mycoplasma pulmonis, Mycoplasma genitalium, and M. pneumoniae are highlighted in black. All sequences were retrieved from the RNRdb at http://rnrdb.molbio.su.se.
Interestingly, an nrdFIE gene cluster with high levels of similarity to the streptococcal nrdF*I*E* operon (87%, 57%, and 76% similarity for NrdF*, NrdI*, and NrdE*, respectively) can also be found in all Mycoplasma species. In addition, the substitution of three of the iron binding residues described for S. pyogenes NrdF* is also conserved in all Mycoplasma NrdFs (Fig. 6B). Mycoplasma organisms are intracellular parasites and among the simplest prokaryotes capable of self-replication (23). Although Mycoplasma organisms may import deoxyribonucleotides from their host, the nrdFIE genes encode its only putative RNR. One possible explanation for the striking similarities between the nrdFIE operons of Mycoplasma and S. pyogenes could be that this operon has been horizontally transferred to S. pyogenes to compensate for the loss of the in vivo functionality of the streptococcal nrdHEF system, due to the lack of a functional NrdI2 protein. Further analyses are needed to clarify this suggestion, since the codon usage of all current class Ib nrd genes in S. pyogenes is in agreement with that calculated for the rest of the S. pyogenes genome (2, 10, 38), and we found no phagic signatures or mobile elements flanking the streptococcal nrdF*I*E* genes. Interestingly, our results also suggest that nrdH is dispensable for nrdF*I*E* function. This is not surprising since an nrdH gene is also lacking in Mycoplasma and in B. subtilis, both of which contain a class Ib enzyme. In E. coli, it has been shown that NrdEF can be reduced by glutaredoxin-1 (Grx1) in the absence of NrdH (13), and in B. subtilis, the essential thioredoxin-1 (TrxA) has been proposed as the electron donor for class Ib RNR (15). We have not found grx homologues in S. pyogenes or Mycoplasma, but they do contain a trx homologue, which might supply the role of nrdH.
NrdI is a member of the flavodoxin family.
The three-dimensional structure of the NrdI protein from B. subtilis is available (Protein Data Bank accession no. 1RLJ) and provides evidence that NrdI contains a flavodoxin-like fold, and thus, NrdIs are classified as the flavodoxin NrdI family (Pfam accession no. PF07972). Flavodoxins are small electron transfer proteins that utilize a noncovalently bound FMN as the redox-active component (35). In E. coli, flavodoxin and NADPH:flavodoxin oxidoreductase are essential for class III RNR activation (27). Likewise, the flavin reductase Fre (12) and the YfaE ferredoxin (47) can reactivate a non-radical-containing met-β2 of E. coli class Ia RNR. We suggest that the role of the NrdI flavodoxin is to reduce the iron cluster in class Ib NrdF, thereby allowing the reactivation of the essential tyrosyl radical. The iron center of Bacillus anthracis NrdF (44) has in preliminary experiments been efficiently reduced by incubation with B. anthracis NrdI (M. Sahlin, E. Torrents, and B.-M. Sjöberg, unpublished data). Class Ib operons may thus carry two specific electron donors, NrdH, which provides reducing power to the α subunit, and NrdI, which supplies electrons to the β subunit.
Jordan et al. (19) showed that the activity of the S. enterica serovar Typhimurium NrdEF enzyme could be slightly stimulated by the addition of a partially purified recombinant NrdI protein, but all classical NrdEF enzymes analyzed to date are active in vitro without the addition of an NrdI protein (17, 20, 21, 44, 48). Likewise, NrdI is not essential for the in vitro activity of the S. pyogenes NrdEF enzyme. In sharp contrast to this, the S. pyogenes NrdE*F* mixture had no in vitro enzyme activity, likely relating to the lack of three of the six iron binding side chains in NrdF*. The obvious in vivo activity of the S. pyogenes nrdF*I*E* operon suggests that NrdI* may play a more direct role in ribonucleotide reduction than in activating a reduced met-β2 protein.
Supplementary Material
Acknowledgments
This work was supported by grants BFU2004-03383 (Ministerio de Educación y Ciencia, Spain), 2005SGR-00956 (Generalitat de Catalunya, Spain), and 621-2004-2864 (Swedish Science Research Council). I.G. and I.R. were also recipients of a grant from Fundación Maria Francisca de Roviralta. E.T. is supported by the Ramón y Cajal program and the Jeansson Foundations.
Footnotes
Published ahead of print on 23 May 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andersson, M. E., M. Högbom, A. Rinaldo-Matthis, W. Blodig, Y. Liang, B. O. Persson, B.-M. Sjöberg, X. D. Su, and P. Nordlund. 2004. Structural and mutational studies of the carboxylate cluster in iron-free ribonucleotide reductase R2. Biochemistry 437966-7972. [DOI] [PubMed] [Google Scholar]
- 2.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 9910078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birgander, P. L., S. Bug, A. Kasrayan, S.-L. Dahlroth, M. Westman, E. Gordon, and B.-M. Sjöberg. 2005. Nucleotide-dependent formation of catalytically competent dimers from engineered monomeric ribonucleotide reductase protein R1. J. Biol. Chem. 28014997-15003. [DOI] [PubMed] [Google Scholar]
- 4.Borovok, I., B. Gorovitz, M. Yanku, R. Schreiber, B. Gust, K. Chater, Y. Aharonowitz, and G. Cohen. 2004. Alternative oxygen-dependent and oxygen-independent ribonucleotide reductases in Streptomyces: cross-regulation and physiological role in response to oxygen limitation. Mol. Microbiol. 541022-1035. [DOI] [PubMed] [Google Scholar]
- 5.Borovok, I., R. Kreisberg-Zakarin, M. Yanko, R. Schreiber, M. Myslovati, F. Åslund, A. Holmgren, G. Cohen, and Y. Aharonowitz. 2002. Streptomyces spp. contain class Ia and class II ribonucleotide reductases: expression analysis of the genes in vegetative growth. Microbiology 148391-404. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 7.Climent, I., B.-M. Sjöberg, and C. Y. Huang. 1992. Site-directed mutagenesis and deletion of the carboxyl terminus of Escherichia coli ribonucleotide reductase protein R2. Effects on catalytic activity and subunit interaction. Biochemistry 314801-4807. [DOI] [PubMed] [Google Scholar]
- 8.Dawes, S. S., D. F. Warner, L. Tsenova, J. Timm, J. D. McKinney, G. Kaplan, H. Rubin, and V. Mizrahi. 2003. Ribonucleotide reduction in Mycobacterium tuberculosis: function and expression of genes encoding class Ib and class II ribonucleotide reductases. Infect. Immun. 716124-6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 1813649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 984658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fieschi, F., E. Torrents, L. Toulokhonova, A. Jordan, U. Hellman, J. Barbé, I. Gibert, M. Karlsson, and B.-M. Sjöberg. 1998. The manganese-containing ribonucleotide reductase of Corynebacterium ammoniagenes is a class Ib enzyme. J. Biol. Chem. 2734329-4337. [DOI] [PubMed] [Google Scholar]
- 12.Fontecave, M., R. Eliasson, and P. Reichard. 1989. Enzymatic regulation of the radical content of the small subunit of Escherichia coli ribonucleotide reductase involving reduction of its redox centers. J. Biol. Chem. 2649164-9170. [PubMed] [Google Scholar]
- 13.Gon, S., M. J. Faulkner, and J. Beckwith. 2006. In vivo requirement for glutaredoxins and thioredoxins in the reduction of the ribonucleotide reductases of Escherichia coli. Antioxid. Redox Signal. 8735-742. [DOI] [PubMed] [Google Scholar]
- 14.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Härtig, E., A. Hartmann, M. Schätzle, A. M. Albertini, and D. Jahn. 2006. The Bacillus subtilis nrdEF genes, encoding a class Ib ribonucleotide reductase, are essential for aerobic and anaerobic growth. Appl. Environ. Microbiol. 725260-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 1835709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huque, Y., F. Fieschi, E. Torrents, I. Gibert, R. Eliasson, P. Reichard, M. Sahlin, and B.-M. Sjöberg. 2000. The active form of the R2F protein of class Ib ribonucleotide reductase from Corynebacterium ammoniagenes is a diferric protein. J. Biol. Chem. 27525365-25371. [DOI] [PubMed] [Google Scholar]
- 18.Jordan, A., E. Aragall, I. Gibert, and J. Barbé. 1996. Promoter identification and expression analysis of Salmonella typhimurium and Escherichia coli nrdEF operons encoding one of two class I ribonucleotide reductases present in both bacteria. Mol. Microbiol. 19777-790. [DOI] [PubMed] [Google Scholar]
- 19.Jordan, A., F. Åslund, E. Pontis, P. Reichard, and A. Holmgren. 1997. Characterization of Escherichia coli NrdH. A glutaredoxin-like protein with a thioredoxin-like activity profile. J. Biol. Chem. 27218044-18050. [DOI] [PubMed] [Google Scholar]
- 20.Jordan, A., E. Pontis, F. Åslund, U. Hellman, I. Gibert, and P. Reichard. 1996. The ribonucleotide reductase system of Lactococcus lactis. Characterization of an NrdEF enzyme and a new electron transport protein. J. Biol. Chem. 2718779-8785. [DOI] [PubMed] [Google Scholar]
- 21.Jordan, A., E. Pontis, M. Atta, M. Krook, I. Gibert, J. Barbé, and P. Reichard. 1994. A second class I ribonucleotide reductase in Enterobacteriaceae: characterization of the Salmonella typhimurium enzyme. Proc. Natl. Acad. Sci. USA 9112892-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 1004678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koonin, E. V. 2000. How many genes can make a cell: the minimal-gene-set concept. Annu. Rev. Genomics Hum. Genet. 199-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, A., S. Pötsch, A. Davydov, A. L. Barra, H. Rubin, and A. Gräslund. 1998. The tyrosyl free radical of recombinant ribonucleotide reductase from Mycobacterium tuberculosis is located in a rigid hydrophobic pocket. Biochemistry 3716369-16377. [DOI] [PubMed] [Google Scholar]
- 25.McLaughlin, R. E., and J. J. Ferretti. 1995. Electrotransformation of streptococci. Methods Mol. Biol. 47185-193. [DOI] [PubMed] [Google Scholar]
- 26.Monje-Casas, F., J. Jurado, M. J. Prieto-Álamo, A. Holmgren, and C. Pueyo. 2001. Expression analysis of the nrdHIEF operon from Escherichia coli. Conditions that trigger the transcript level in vivo. J. Biol. Chem. 27618031-18037. [DOI] [PubMed] [Google Scholar]
- 27.Mulliez, E., D. Padovani, M. Atta, C. Alcouffe, and M. Fontecave. 2001. Activation of class III ribonucleotide reductase by flavodoxin: a protein radical-driven electron transfer to the iron-sulfur center. Biochemistry 403730-3736. [DOI] [PubMed] [Google Scholar]
- 28.Nicholas, K. B., and H. B. J. Nicholas. 1997. GeneDoc: a tool for editing and annotating multiple sequence alignments v. 2.6.001. K. B. Nicholas and H. B. J. Nicholas (http://www.nrbsc.org/gfx/genedoc/index.html).
- 29.Nordlund, P., and P. Reichard. 2006. Ribonucleotide reductases. Annu. Rev. Biochem. 75681-706. [DOI] [PubMed] [Google Scholar]
- 30.Petersson, L., A. Gräslund, A. Ehrenberg, B.-M. Sjöberg, and P. Reichard. 1980. The iron center in ribonucleotide reductase from Escherichia coli. J. Biol. Chem. 2556706-6712. [PubMed] [Google Scholar]
- 31.Platz, A., and B.-M. Sjöberg. 1980. Construction and characterization of hybrid plasmids containing the Escherichia coli nrd region. J. Bacteriol. 143561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahlin, M., L. Petersson, A. Gräslund, A. Ehrenberg, B.-M. Sjöberg, and L. Thelander. 1987. Magnetic interaction between the tyrosyl free radical and the antiferromagnetically coupled iron center in ribonucleotide reductase. Biochemistry 265541-5548. [DOI] [PubMed] [Google Scholar]
- 33.Sahlin, M., B.-M. Sjöberg, G. Backes, T. Loehr, and J. Sanders-Loehr. 1990. Activation of the iron-containing B2 protein of ribonucleotide reductase by hydrogen peroxide. Biochem. Biophys. Res. Commun. 167813-818. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 35.Sancho, J. 2006. Flavodoxins: sequence, folding, binding, function and beyond. Cell. Mol. Life Sci. 63855-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scotti, C., A. Valbuzzi, M. Perego, A. Galizzi, and A. M. Albertini. 1996. The Bacillus subtilis genes for ribonucleotide reductase are similar to the genes for the second class I NrdE/NrdF enzymes of Enterobacteriaceae. Microbiology 1422995-3004. [DOI] [PubMed] [Google Scholar]
- 37.Smalley, D., E. R. Rocha, and C. J. Smith. 2002. Aerobic-type ribonucleotide reductase in the anaerobe Bacteroides fragilis. J. Bacteriol. 184895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 994668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thelander, L., B.-M. Sjöberg, and S. Eriksson. 1978. Ribonucleoside diphosphate reductase (Escherichia coli). Methods Enzymol. 51227-237. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torrents, E., P. Aloy, I. Gibert, and F. Rodríguez-Trelles. 2002. Ribonucleotide reductases: divergent evolution of an ancient enzyme. J. Mol. Evol. 55138-152. [DOI] [PubMed] [Google Scholar]
- 42.Torrents, E., A. Poplawski, and B.-M. Sjöberg. 2005. Two proteins mediate class II ribonucleotide reductase activity in Pseudomonas aeruginosa: expression and transcriptional analysis of the aerobic enzymes. J. Biol. Chem. 28016571-16578. [DOI] [PubMed] [Google Scholar]
- 43.Torrents, E., I. Roca, and I. Gibert. 2003. Corynebacterium ammoniagenes class Ib ribonucleotide reductase: transcriptional regulation of an atypical genomic organization in the nrd cluster. Microbiology 1491011-1020. [DOI] [PubMed] [Google Scholar]
- 44.Torrents, E., M. Sahlin, D. Biglino, A. Gräslund, and B.-M. Sjöberg. 2005. Efficient growth inhibition of Bacillus anthracis by knocking out the ribonucleotide reductase tyrosyl radical. Proc. Natl. Acad. Sci. USA 10217946-17951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voegtli, W. C., M. Sommerhalter, L. Saleh, J. Baldwin, J. M. Bollinger, Jr., and A. C. Rosenzweig. 2003. Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2. J. Am. Chem. Soc. 12515822-15830. [DOI] [PubMed] [Google Scholar]
- 46.Wang, P. J., A. Chabes, R. Casagrande, X. C. Tian, L. Thelander, and T. C. Huffaker. 1997. Rnr4p, a novel ribonucleotide reductase small-subunit protein. Mol. Cell. Biol. 176114-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, C. H., W. Jiang, C. Krebs, and J. Stubbe. 2007. YfaE, a ferredoxin involved in diferric-tyrosyl radical maintenance in Escherichia coli ribonucleotide reductase. Biochemistry 4611577-11588. [DOI] [PubMed] [Google Scholar]
- 48.Yang, F., S. C. Curran, L. S. Li, D. Avarbock, J. D. Graf, M. M. Chua, G. Lu, J. Salem, and H. Rubin. 1997. Characterization of two genes encoding the Mycobacterium tuberculosis ribonucleotide reductase small subunit. J. Bacteriol. 1796408-6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.