Abstract
The Sec-dependent translocation pathway that involves the essential SecA protein and the membrane-bound SecYEG translocon is used to export many proteins across the cytoplasmic membrane. Recently, several pathogenic bacteria, including Mycobacterium tuberculosis, were shown to possess two SecA homologs, SecA1 and SecA2. SecA1 is essential for general protein export. SecA2 is specific for a subset of exported proteins and is important for M. tuberculosis virulence. The enzymatic activities of two SecA proteins from the same microorganism have not been defined for any bacteria. Here, M. tuberculosis SecA1 and SecA2 are shown to bind ATP with high affinity, though the affinity of SecA1 for ATP is weaker than that of SecA2 or Escherichia coli SecA. Amino acid substitution of arginine or alanine for the conserved lysine in the Walker A motif of SecA2 eliminated ATP binding. We used the SecA2(K115R) variant to show that ATP binding was necessary for the SecA2 function of promoting intracellular growth of M. tuberculosis in macrophages. These results are the first to show the importance of ATPase activity in the function of accessory SecA2 proteins.
Mycobacterium tuberculosis is the causative agent of the disease tuberculosis (TB). TB kills about 2 million people annually, and approximately one-third of the world's population is currently infected with M. tuberculosis (40). A serious problem in the worldwide fight against TB is the emergence of multidrug-resistant strains of M. tuberculosis. To develop logical targets for new, effective drugs, the physiology of mycobacteria must be better understood.
As with all bacterial pathogens, the majority of M. tuberculosis virulence factors are extracytoplasmic proteins exported to the bacterial cell surface or secreted further to the extracellular milieu (11, 21, 24). Bacteria possess several different pathways for exporting proteins from the cytoplasm, including the highly conserved Sec pathway (8, 24). The Sec pathway uses the SecA protein and the membrane-integrated SecYEG translocon to transport precursor proteins that contain a characteristic amino-terminal signal sequence across the cytoplasmic membrane (18). SecA, an essential ATPase found in all bacteria, undergoes conformational changes upon ATP binding and hydrolysis that drive the transport of unfolded precursor proteins through the SecYEG translocon (36, 37). The well-characterized ATPase activity of Escherichia coli SecA is absolutely required for protein export and is stimulated by the addition of phospholipids and by the presence of precursor protein (27, 29, 39).
Most bacteria, including the model organisms E. coli and Bacillus subtilis, possess a single, essential SecA protein. Recently, several microorganisms, including M. tuberculosis, Listeria monocytogenes, corynebacteria, and some streptococci, were found to carry two SecA proteins, SecA1 and SecA2 (2-5, 7, 26). At the amino acid level, SecA1 and SecA2 of M. tuberculosis are about 50% similar to each other and 61% and 50% similar to E. coli SecA, respectively (3). The mycobacterial SecA1 protein is essential and is thought to function similarly to the single SecA proteins of E. coli and B. subtilis (3, 16). The mycobacterial SecA2 protein is not essential for growth in culture but is required for exporting a subset of proteins (3, 4, 14). Furthermore, the secA2 deletion mutant of M. tuberculosis is attenuated in virulence, suggesting that some of the SecA2-dependent exported proteins are virulence factors (4, 22). Interestingly, the proteins exported by SecA2 systems in different bacteria include examples with and without amino-terminal signal sequences (1, 2, 4, 7, 22, 25).
The features that distinguish the function of SecA1 from that of SecA2 in a single bacterial species are not known, nor have the biochemical properties of each SecA been studied previously. Here, we report that the SecA1 and SecA2 proteins of M. tuberculosis are present at comparable levels, indicating that expression levels do not explain the different functions of these proteins.
Both SecA1 and SecA2 exhibit high sequence homology with other SecA proteins in the Walker A and B motifs commonly found in ATPases (38). The Walker motifs are part of the motor domain of SecA. In structural studies, the motor domain of M. tuberculosis SecA1 also shows the highest similarity to that of B. subtilis SecA (32). Using purified M. tuberculosis SecA1 and SecA2 proteins, we show that both proteins exhibit fully functional ATPase activities. Moreover, replacement of the conserved lysine residue in the Walker A motif of SecA2 to produce the SecA2(K115R) or the SecA2(K115A) variant eliminates ATP binding. This amino acid replacement in the Walker A motif also affects the biological activity of SecA2, as the secA2(K115R) variant fails to complement the intracellular growth defect of the M. tuberculosis secA2 mutant in macrophages. Our data present the first report of the characterization of the ATPase activity for any SecA2 protein and show that ATP binding is necessary for SecA2 function. This work represents an important first step toward understanding how the two SecA proteins in mycobacteria function in protein translocation.
MATERIALS AND METHODS
Plasmids and strains.
The E. coli SecA expression vector was a gift from Linda Randall (31). The M. tuberculosis SecA1 expression vector was a gift from James Sacchettini. The SecA1 plasmid was generated by PCR amplification of the secA1 gene from M. tuberculosis H37Rv genomic DNA into pET29a (Novagen). A stop codon was added to the 3′ end of the gene to avoid addition of the C-terminal hexahistidine tag from the plasmid. The SecA2 expression plasmid was constructed by PCR amplification of the secA2 gene from M. tuberculosis H37Rv genomic DNA, using the primers 5′ CATTAATGGTGCCCAAGACCACCCGCGCTCA 3′ and 5′ TAAGCTTCAGCGGAACACCCCGGGCAGACT 3′. The resulting PCR product was cloned into pET41b (Novagen) to create pNR14. This SecA2 expression vector uses an internal GTG start codon located at nucleotide position 91 relative to the annotated start site (NCBI accession number NP_216337). We believe this represents the true start codon. Translation from this codon results in a protein whose sequence more closely resembles that of SecA2 from other mycobacteria, and the corresponding shorter SecA2 sequence can complement the secA2 mutant phenotypes in Mycobacterium smegmatis and M. tuberculosis (data not shown). Plasmids that overexpress SecA1 or SecA2 were transformed into E. coli BL21(DE3).
The Walker A motif of M. tuberculosis SecA2 was mutated in plasmid pNR14, using a QuikChange site-directed mutagenesis kit (Stratagene). The primers used to change lysine 115 (AAG) to arginine (AGG) (underlined) were 5′ CGGTGAGGGCAGAACCCTTGCC 3′ and 5′ CGGCAAGGGTTCTGCCCTCACC 3′. The resulting SecA2(K115R) expression vector for E. coli is plasmid pNR24. To create the same substitution in a mycobacterial expression vector, pMB162 was used as a template to create pNR7. Plasmid pMB162 contains the M. tuberculosis secA2 open reading frame, as annotated in the genome database, under control of the constitutive mycobacterial hsp60 promoter and an attachment site that enables stable integration into the mycobacterial genome in a single copy (4). To generate the SecA2(K115A) pNR60 expression vector, site-directed mutagenesis was performed with pNR24 with the primers 5′ CGGTGGCGGCAGAACCCTTGCC 3′ and 5′ CGGCAAGGGTTCTGCCGCCACC 3′. Sequencing was used to confirm the appropriate secA2 sequence in all vectors used in this study (Eton Biosciences).
Purification of proteins.
E. coli SecA was expressed and purified as described previously (10). Purification of M. tuberculosis SecA1 and SecA2 followed essentially the same protocol. Freshly transformed cells were selected for the ability to overproduce each protein. The cultures that overproduced the SecA proteins were pooled and used for the protein preparation. The induced cells were suspended in 25 mM Tris (pH 7.6), 10 mM magnesium acetate, and 10 mM NaCl and lysed, using a French pressure cell at 15,000 lb/in2, into a 2 μg/ml final concentration of pepstatin A, leupeptin, and aprotinin, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma). The lysate was clarified by low-speed centrifugation, and the supernatants were centrifuged in a Sorvall T-865 rotor at 115,000 × g for 3 h to remove the cell membranes.
For the purification of SecA1 and SecA2, the supernatant was applied to a Blue-Sepharose CL-6B column (Pharmacia Biotech) equilibrated with 25 mM Tris at pH 7.6, 0.5 mM EDTA, 100 μM dithiothreitol, and 0.5 mM PMSF. The protein was eluted with a step gradient consisting of a low-salt (the same buffer supplemented with 10 mM NaCl), a medium-salt (0.4 M NaCl), and a high-salt (1.3 M NaCl) buffer. Fractions containing SecA1 were collected, precipitated with ammonium sulfate at 60%, and dialyzed against 10 mM HEPES (pH 7.6), 25 mM potassium acetate, 0.5 mM EDTA, and 100 μM tris(2-carboxyethyl)phosphine (TCEP)-HCl (Pierce). SecA2 was concentrated by using an Amicon ultrafiltration stirred cell and a membrane with a molecular weight (MW) cutoff of 30,000. The purified protein was stored in aliquots at −80°C. The SecA2(K115R) and SecA2(K115A) variant proteins were purified by the same method as that used for the wild-type (WT) SecA2.
Quantification of the ratio of SecA2 to SecA1 in vivo.
Whole-cell lysates of exponential phase M. tuberculosis cultures grown in Middlebrook 7H9 medium were prepared following fixation in an equal volume of 10% formalin for 1 h. Fixed cells were pelleted by centrifugation, resuspended in extraction buffer, and lysed by bead beating. For quantitative SecA1 and SecA2 Western blot analysis, 100 μg of formalin-fixed whole-cell lysates were run on a 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel alongside a standard curve of known amounts of purified SecA1 or SecA2 protein. Proteins were transferred to nitrocellulose membranes and probed with rabbit polyclonal anti-SecA1 (1:50,000 dilution) or anti-SecA2 (1:20,000 dilution) antibody (16). The secondary antibody was goat anti-rabbit conjugated to alkaline phosphatase (1:20,000 dilution), and detection of the fluorescence from ECF (Amersham/GE Healthcare) was done with a PhosphorImager (Molecular Dynamics). The signal intensity values from six independently prepared whole-cell lysates were quantified by comparison with standard curve values to determine the number of moles of SecA1 and SecA2 per mg of cellular protein.
Circular dichroism spectra.
Circular dichroism (CD) was measured with an Applied Photophysics Pi-Star 180 circular dichroism spectrapolarimeter, with the cuvette maintained at 20°C with a circulating water bath. A 1-cm-path-length quartz cuvette was used. The protein concentration was 0.1 mg/ml in 20 mM phosphate buffer. The slits were set at 2 nm. The CD spectrum was taken from 250 to 200 nm with a 1-nm step size. The signal was averaged for 500,000 samples for a total of 12.5 s at each wavelength.
Determination of melting temperature.
CD at 222 nm was used to monitor the secondary structure of the SecA proteins. The protein concentration was 0.1 mg/ml. To determine the melting temperature, the temperature was ramped from 8°C to 65°C with a step size of 0.5°C. At each temperature, the signal at 222 nm was averaged for 100,000 points, which required around 10 s. The raw data were normalized (12) to show the fraction unfolded at each temperature.
ATP binding assay.
Nitrocellulose membrane filtration assays were used to measure the ATP binding affinities of the SecA proteins as described in reference 42. The 0.2-μM-pore-size nitrocellulose membranes (Whatman) were incubated briefly with 0.5 N NaOH, rinsed extensively with deionized H2O, and then equilibrated with binding buffer [50 mM HEPES-NaOH (pH 7.5), 30 mM KCl, 10 mM Mg(OAc)2] for at least 1 h. ATP stocks were prepared using Sigma Ultrapure ATP and GE Healthcare [α-32P]ATP. The protein concentration in each assay was held between 3.7 and 4.5 μM of monomer, and increasing concentrations of ATP were added. The protein-ATP mixtures were incubated on ice, which slows the ATPase activity to a rate undetectable by the use of the malachite green assay, as described below. The solutions were filtered through the nitrocellulose membrane to separate the bound ATP from the free ATP. Because the apparent Kd (dissociation constant) values of E. coli SecA and M. tuberculosis SecA2 for ATP were lower than the protein concentrations used in the assays, the binding data were fitted with an equation used when there is ligand depletion (15, 33), using the following equation: [LR] = [(Kd + [Rt] + [Lt]) − √{(Kd + [Rt] + [Lt])2 − 4[Rt][Lt]}]/2, where Rt is the total protein concentration and Lt is the total ATP added. The Kd value for SecA1 was obtained by fitting the data to the standard equation for binding data, since here the apparent Kd value was above the protein concentration used in the assay equation [LR] = (Rt[L])/(Kd + [L]), where L is the free ligand. The data were fitted with KaleidaGraph software (Synergy Software). The Kd value was determined for each assay performed in triplicate, and the average Kd value and the standard error were determined.
Analysis of ATPase activity.
The protocol used to measure SecA ATPase activity was a modification of the malachite green assay described previously (23, 27). A reaction mixture contained 40 μg/ml of the SecA proteins, 1 mg/ml bovine serum albumin, and 4 mM ATP in reaction buffer [50 mM Tris-HCl (pH 7.0), 30 mM KCl, 30 mM NH4Cl, 1 mM dithiothreitol, and 5 mM Mg(OAc)2]. The assay was conducted at 25°C. Formation of inorganic phosphate was monitored spectrophotometrically by the increase in absorbance at 660 nm at each time point. The inorganic phosphate concentration generated in the reaction mixture with time was calculated using a standard curve. Each assay was done at least four times with each protein. The rate of hydrolysis was determined for each assay done in duplicate, and the averages and standard errors were determined.
Macrophage infections.
Bone marrow macrophages were elicited from femurs of C57BL/6 mice, as described previously (22, 28), and 2.5 × 105 macrophages were seeded into wells of 8-well-chamber slides 24 h prior to infection. The M. tuberculosis strains were grown to mid-exponential phase, washed with phosphate-buffered saline containing 0.05% Tween 80, diluted in tissue culture medium (Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum, 2 mM glutamine, and 1× nonessential amino acids [Gibco]) and added to the macrophage monolayer to achieve a multiplicity of infection of 1.0. Macrophage monolayers were infected with M. tuberculosis strains for 4 h at 37°C in 5% CO2. On days 0, 1, and 5 postinfection, the contents of triplicate wells for each infection were washed and then lysed with 0.05% SDS. The resulting lysates were diluted and plated on Middlebrook 7H10 agar to enumerate intracellular bacteria during the course of infection.
RESULTS
SecA1 and SecA2 proteins are present in equivalent amounts in M. tuberculosis.
The identification of bacteria that possess two SecA proteins, such as M. tuberculosis, is a relatively new discovery. As a starting point to understanding the contribution of each SecA protein to the process of protein export in M. tuberculosis, we determined the relative amounts of SecA1 and SecA2 inside the bacterial cell. Quantitative immunoblot analysis was performed with whole-cell lysates prepared from the virulent M. tuberculosis strain H37Rv, using antibodies specific to each SecA protein. These antibodies were raised against peptides specific for SecA1 or SecA2, and they recognize only the respective protein (16). These antibodies were used to determine the number of moles of SecA1 or SecA2 protein per mg of cellular protein by comparison to a standard curve for the purified M. tuberculosis SecA1 protein or to that of the SecA2 protein. The results, from an evaluation of six independent whole-cell lysates, revealed nearly equivalent amounts of each protein. The ratio of the average number of moles per mg of protein of SecA2 (2.07 × 10−11) to that of SecA1 (2.25 × 10−11) across these experiments was 0.93 ± 0.12 (standard error). This indicates that the differences between the functions of SecA1 and those of SecA2 in M. tuberculosis are not due to different levels of expression, at least under standard laboratory conditions.
Purification of M. tuberculosis SecA1 and SecA2.
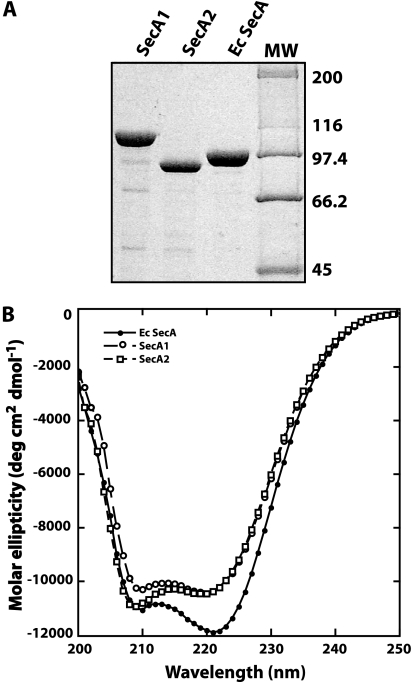
The two SecA proteins from any bacteria have not been biochemically characterized. The SecA1 and SecA2 proteins of M. tuberculosis were expressed from a plasmid and purified from E. coli as soluble proteins. SecA1 has a predicted MW of 106,000/monomer, while SecA2 is smaller, with a predicted MW of 85,400/monomer (3). E. coli SecA has a MW of 101,000/monomer. All the proteins were purified to >90% using Blue-Sepharose column chromatography (Fig. 1A). SecA2 bound less tightly to this column than SecA1 or E. coli SecA did, eluting with 0.5 M salt instead of 1.2 M. This change in binding to a pseudo-affinity column for nucleotide-binding proteins suggested that SecA2 might have an altered affinity for nucleotides.
FIG. 1.
(A) SDS gel of the purified SecA proteins. SecA1, SecA2, and E. coli SecA were purified as described in Materials and Methods. The proteins were separated on a 10% SDS polyacrylamide gel. Molecular weight markers (MW, in thousands) are indicated on the right side of the gel. (B) CD spectra of each SecA protein. The spectra were obtained as described in Materials and Methods. The protein concentration was 0.1 mg/ml in 20 mM phosphate buffer (pH 7.6).
We next determined if the purified M. tuberculosis SecA1 and SecA2 proteins had CD spectra consistent with folded secondary and tertiary structures (Fig. 1B). Each of the proteins exhibited the characteristic minima at 209 and 222 nm associated with proteins with significant levels of helical structure. Tryptophan fluorescence spectra for all the proteins were also consistent with a native tertiary structure with emission maxima around 340 nm, which is typical for folded proteins (data not shown). These results were expected for properly folded SecA1 and SecA2 proteins, as the three-dimensional structure of M. tuberculosis SecA1 is similar to that of B. subtilis SecA and E. coli SecA (17, 30, 32) and the sequences of the mycobacterial SecA proteins are highly homologous.
SecA1 and SecA2 are less stable to heat denaturation than E. coli SecA.
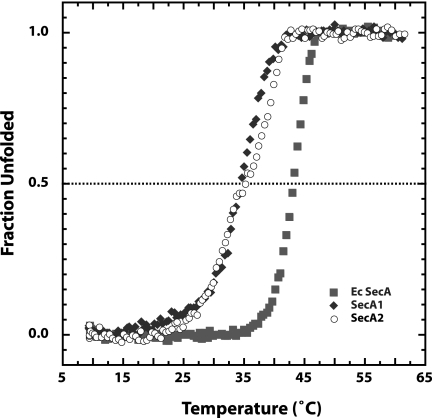
At the standard temperature (37°C) used for ATP hydrolysis assays, we saw little ATPase activity for the M. tuberculosis SecA proteins. Since the E. coli SecA ATPase activity is temperature dependent because the protein is thermolabile (27), we reasoned that the low activity levels of SecA1 and SecA2 proteins could be due to lower thermal stability. To characterize the thermal stability of the M. tuberculosis SecA1 and SecA2 proteins, we denatured the proteins with heat and followed the melting transition by CD at 222 nm (Fig. 2). The data were normalized to show the fraction unfolded at each temperature (12). The melting temperature (Tm) for E. coli SecA was ∼43°C, as shown previously (34). Both M. tuberculosis SecA1 and SecA2 were less stable to heat denaturation, with Tms of ∼35°C, indicating a moderate decrease in the stability of the M. tuberculosis SecA proteins compared to that of E. coli SecA. Our data show that measurements of activity of the recombinant M. tuberculosis SecA proteins must be done at temperatures lower than 30°C to maintain their fully native structures.
FIG. 2.
M. tuberculosis SecA proteins have decreased thermostability compared to that of E. coli SecA. To determine the melting temperatures, the SecA proteins (0.1 mg/ml) were ramped from 8 to 65°C. At each 0.5 °C increment in temperature, the CD at 222 nm was measured. The CD signal was normalized to show the fraction unfolded at each temperature.
M. tuberculosis SecA1 and SecA2 bind ATP with different affinities.
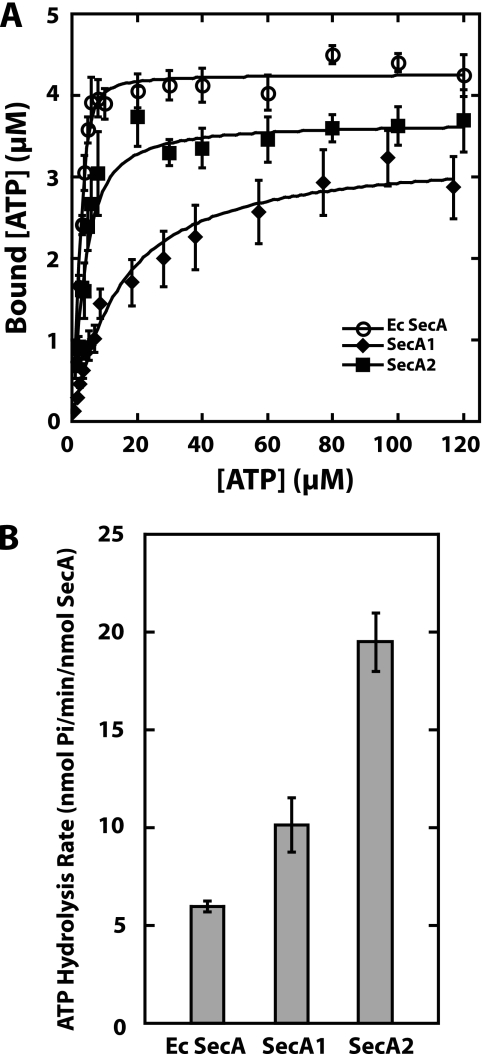
To determine if SecA1 and SecA2 bind ATP as anticipated for proteins carrying canonical Walker A motifs, we used a standard nitrocellulose membrane binding assay with [α-32P]ATP to determine the affinity of each protein for ATP (42). The protein concentration in each assay was held constant, and increasing concentrations of ATP were added. The solutions were filtered through a nitrocellulose membrane to separate the bound ATP from the free ATP. Because the apparent Kd values of E. coli SecA and M. tuberculosis SecA2 for ATP were lower than the protein concentrations used in the assays, the binding data were fitted with an equation used when there is ligand depletion, as described in Materials and Methods and previously (15, 33). The Kd value of SecA1 for ATP was obtained by fitting the data to the standard equation for binding data, since here the apparent Kd value was above the protein concentration used in the assay. Analysis of the binding data (Fig. 3A) showed that E. coli SecA bound ATP with a Kd value of 0.46 ± 0.11 μM, which is within the standard error for a previously published value (42). M. tuberculosis SecA2 bound ATP with a somewhat weaker affinity of 1.9 ± 0.3 μM. M. tuberculosis SecA1 bound ATP ∼10-fold more weakly than SecA2, with a Kd value of 21.8 ± 4.0 μM.
FIG. 3.
(A) SecA2 binds ATP with high affinity. The affinity of ATP binding by SecA proteins was determined by a nitrocellulose membrane filtration assay. The concentrations of bound and free ATP were determined. The data were fitted using the binding equations described in Materials and Methods to determine the Kd values. Since the data were fitted using two different equations, the ATP concentration on the x axis is the total ATP for the E. coli SecA and SecA2 proteins and the free ATP for SecA1. The binding assays were repeated at least three times for each protein. Shown here are the averages and standard errors of the averages determined for the combined data. (B) Endogenous ATPase activities of the M. tuberculosis SecA proteins. The endogenous activity levels of the SecA proteins were determined using a malachite green assay for the formation of free inorganic phosphate. Each assay was done in duplicate and repeated at least four times to determine the average rates and standard errors of the averages.
An interesting observation is that SecA2 binds more weakly to the Blue-Sepharose column than either E. coli SecA or SecA1, even though its Kd value for ATP binding is similar to that of E. coli SecA, which binds tightly to this column. This observation suggests that the SecA2 binding site for the Cibacron Blue dye, presumably the ATP binding site, may have a different conformation than SecA1 or E. coli SecA.
SecA1 and SecA2 exhibit ATPase activity.
To determine if M. tuberculosis SecA1 and SecA2 hydrolyze ATP, we used a malachite green assay, which measures the amount of free inorganic phosphate produced over time (6, 9, 23, 29) (Fig. 3B). Because each of the Tms of the M. tuberculosis SecA proteins was lower than that of E. coli SecA, all ATPase assays were done at 25°C. The ATPase activity level of each protein was determined in duplicate from at least four separate experiments. While we did see some variability in the results of the ATPase assays, the trends were the same for all assays. With assays done at 25°C, each of the SecA proteins was able to hydrolyze ATP; the endogenous ATPase activity level of M. tuberculosis SecA1 was similar to that of E. coli SecA, while that of SecA2 was about threefold higher than E. coli SecA.
Amino acid substitutions in the Walker A motif of SecA2 affect ATP binding in vitro.
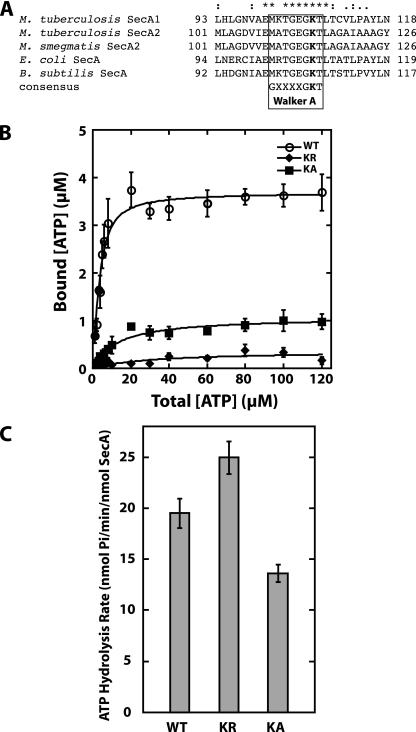
Alignment of M. tuberculosis SecA1 and SecA2 with the SecA of E. coli and B. subtilis reveals high sequence identity across the characteristic Walker A motif of ATPases (Fig. 4A) (3). Structural studies demonstrate that the amino acids of this motif participate in ATP binding (20, 37). In previous studies, site-directed mutagenesis of the conserved lysine in the Walker A motif (K108 in E. coli SecA and K106 in B. subtilis SecA) has shown it to be important for SecA ATP binding (29, 35). The E. coli SecA ATPase K108R mutant is defective in ATP binding and protein translocation in vitro, as well as biologically inactive in vivo as demonstrated by the inability to complement an E. coli temperature-sensitive secA allele (29). The B. subtilis SecA(K106N) ATPase mutant is also unable to translocate precursor proteins in vivo (20).
FIG. 4.
Effect of the K115R and K115A substitutions in the Walker A motif of SecA2 in vitro. (A) Alignment of Walker A motifs in SecA proteins. Sequence alignment of the highly conserved Walker A motif from mycobacterial SecA1 and SecA2 to the well-characterized SecA proteins of E. coli and B. subtilis. The conserved lysine residue that was mutated to generate SecA2(K115R) or SecA2(K115A) is shown in bold. (B) ATP binding by SecA2(K115R) (KR) and SecA2(K115A) (KA). The ATP binding was measured as described in the legend to Fig. 3. The binding data for WT SecA2 is the same as that shown in Fig. 3. (C) Endogenous ATPase activity of SecA2(K115R) and SecA2(K115A). The average rates and standard deviations of the ATP hydrolysis by each protein were determined by using the malachite green assay for determination of free inorganic phosphate, as described in the legend to Fig. 3. The WT SecA2 hydrolysis rate is the same as that shown in Fig. 3. The average rates and standard deviations from all of the experiments are shown.
To determine if ATP binding and ATPase activity in M. tuberculosis SecA2 depend similarly on the Walker A motif, we substituted arginine or alanine for the conserved lysine K115 in the Walker A motif of M. tuberculosis SecA2 and purified the corresponding proteins as described above. We tested the ability of the SecA(K115R) and SecA2(K115A) variants to bind ATP (Fig. 4B). Both the K115R and the K115A substitutions significantly decrease the ability of SecA2 to bind ATP. We then tested the ATP hydrolysis activity of the SecA(K115R) variant, using the malachite green assay. The endogenous ATPase activity for the SecA2(K115R) variant was somewhat higher than that for the WT SecA2. The SecA2(K115A) variant had an ATP hydrolysis rate lower than that of WT SecA2 (Fig. 4C). These in vitro results show that the Walker box of SecA2 is required for efficient ATP binding, as predicted and shown to be the case for other SecA proteins (20, 29, 35). These data also indicate that a SecA2 variant in which lysine, K115, has been replaced can be used to assess the importance of ATP binding for the biological functions of SecA2 in M. tuberculosis.
A substitution in the Walker A motif of SecA2 affects biological activity in M. tuberculosis.
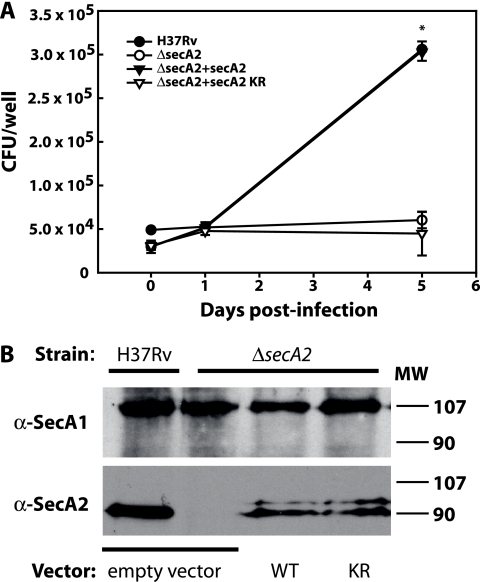
A ΔsecA2 mutant of M. tuberculosis is attenuated for growth in macrophages and in mice (3, 4). The biochemical data presented above and the results of previous studies suggest that M. tuberculosis SecA2 functions as an ATPase to promote the export of a specific subset of proteins that are important to pathogenesis. To test this idea, we asked whether the secA2(K115R) allele encodes a functional protein that is able to fulfill the function of SecA2 in promoting the growth of M. tuberculosis in macrophages. The secA2(K115R) allele was introduced into the M. tuberculosis ΔsecA2 mutant in a single copy at the chromosomal attB site. Murine bone marrow-derived macrophages were infected in parallel with the parental M. tuberculosis strain H37Rv, the ΔsecA2 mutant of M. tuberculosis, the ΔsecA2 mutant complemented with the wild-type secA2 gene inserted at the attB locus, or the ΔsecA2 mutant with secA2(K115R) (Fig. 5A). As shown previously, H37Rv and the secA2-complemented strain grew at similar rates in macrophages over a 5-day period of infection, while the ΔsecA2 mutant failed to grow in macrophages (22). Unlike introduction of the WT secA2 gene at the attB site, which promoted M. tuberculosis growth in macrophages, the introduction of secA2(K115R) in the ΔsecA2 mutant failed to promote this growth. Importantly, Western blot analysis confirmed that the ΔsecA2 mutant strains carrying WT secA2(K115R) or secA2(K115R) expressed the SecA2 protein at similar levels (Fig. 5B). In addition, the growth defect of the secA2(K115R) strain in macrophages was specific, as this strain did not exhibit a general growth defect when tested in 7H9 liquid medium (data not shown). These results indicate that replacing the conserved lysine in the Walker A motif of SecA2 renders SecA2 inactive in its biological role of promoting M. tuberculosis growth in macrophages.
FIG. 5.
(A) The secA2(K115R) allele does not complement the M. tuberculosis secA2 mutant phenotype in macrophages. Murine bone marrow-derived macrophages were infected at a multiplicity of infection of 1.0 with the strain H37Rv, the ΔsecA2 mutant, the ΔsecA2 mutant complemented by the addition of WT secA2, (ΔsecA2+secA2), and the ΔsecA2 mutant with secA2(K115R) (ΔsecA2+secA2 KR). CFU were determined by plating macrophage lysates at various times postinfection. The infection was performed with triplicate wells for each strain per time point, and the error bars represent means ± standard deviations for the triplicate wells. The symbols for H37Rv and the complemented strain are overlapping at most time points in the graph presented. Data are representative of three independent experiments. *, data are statistically significantly different (P < 0.05). (B) Expression of SecA1 and SecA2 in strains with different secA2 alleles. Equal amounts of formalin-fixed whole-cell lysates of the M. tuberculosis H37Rv strain carrying an empty vector, the M. tuberculosis ΔsecA2 mutant with an empty vector, the ΔsecA2 mutant with the WT secA2 integrated at the chromosomal attB site (WT), and the ΔsecA2 mutant with the secA2(K115R) allele integrated at the attB site (KR) were run on SDS-polyacrylamide gels and subjected to Western blot analysis with anti-SecA1 or anti-SecA2 antibodies. The top panel shows SecA1 protein and the lower panel shows SecA2 protein. MW, molecular weight (in thousands).
DISCUSSION
Here we report the first description of ATPase activities for two SecA proteins from the same bacterium and the first description of the ATPase activity for any SecA2 protein. Based on amino acid sequence analysis, M. tuberculosis SecA1 and SecA2 possess the consensus Walker A and B motifs (3) that are found in ATP hydrolyzing enzymes and are essential to E. coli and B. subtilis SecA functions (20, 29). However, for SecA2, which currently has an undefined role in protein export and differs from the well-characterized E. coli SecA protein in being nonessential in mycobacteria, the question of whether it functions as an ATPase remains unanswered (4).
Here we demonstrated that both SecA proteins of M. tuberculosis are ATP binding and hydrolyzing proteins. SecA2 bound ATP with an affinity that was somewhat weaker than that of E. coli SecA, and SecA1 bound ATP about 10-fold more weakly than SecA2. Since the ATP concentration within an M. tuberculosis cell is approximately 1 mM (13, 19), even with these different affinities, both SecA proteins should be fully saturated with ATP in vivo. Therefore, the physiological significance of these differences in affinity is unclear.
The M. tuberculosis SecA proteins showed a lower melting temperature than E. coli SecA under our buffer conditions, suggesting that the M. tuberculosis SecA proteins are less thermostable than E. coli SecA. Therefore, all of the ATP hydrolysis assays were done at a temperature at which the proteins maintained their native state. At 25°C, each of the SecA proteins was able to hydrolyze ATP. Although it is difficult to directly compare results of ATP hydrolysis activities of SecA proteins from different species because of the use of different experimental conditions, our results are in general agreement with the ATP hydrolysis rates of SecA proteins from Pseudomonas aeruginosa (41), E. coli (29), and B. subtilis (35). Nevertheless, our data suggest that there may be some functional differences between SecA1 and SecA2 with regard to ATP affinities and hydrolysis rates. The endogenous ATPase activity of M. tuberculosis SecA2 was found to be higher than that of M. tuberculosis SecA1 and E. coli SecA.
The K115R substitution in the SecA2 Walker A motif clearly establishes the fact that ATP binding of SecA2 is vital for its function in vivo, as this substitution in SecA2 abolished the ability of M. tuberculosis to grow within macrophages. In an ATP binding assay, the SecA2(K115R) and SecA2(K115A) variants exhibited significantly reduced ATP binding, as anticipated when the conserved lysine of Walker A motif was replaced. However, neither the substitution of R for K nor A for K eliminated the endogenous ATPase activity. The K115R substitution slightly increased the endogenous rate of ATP hydrolysis, while the SecA2(K115A) variant showed about 60% of the activity of WT SecA2. These results are consistent with those of Mitchell and Oliver (29), where the E. coli SecA(K108R) substitution in the Walker A motif affected biological activity and eliminated effective ATP binding but did not eliminate—and even somewhat increased—the endogenous ATPase activity (29).
The mycobacterial SecA proteins have been shown to be functionally dissimilar from one another in that SecA2 participates only in the export of a specific subset of proteins (3, 4, 14). The data presented here indicate that both M. tuberculosis SecA1 and SecA2 are ATPases. Our data suggest that each M. tuberculosis SecA protein is likely to function in a manner similar to that of E. coli SecA in undergoing cycles of ATP binding and hydrolysis-promoted conformational changes that drive protein export across the cytoplasmic membrane. In addition, we show that the levels of SecA1 and SecA2 in the cell are equivalent. Therefore, the functional differences between SecA1 and SecA2 may be at the level of the specific proteins they recognize for export or the proteins with which they interact to form a translocation complex. For SecA1, we believe the protein works in concert with the membrane-localized SecYEG translocon. The SecA2 protein may also work with SecYEG or with an as-yet unidentified translocation complex to selectively export a subset of proteins. Alternatively, differences in the function of the SecA proteins might be in the regulation of their enzymology. The studies here lay the groundwork for further investigation of the SecA2 export pathway of mycobacteria.
Acknowledgments
We thank James Sacchettini and Arulandu Arockiasamy for the generous gift of the M. tuberculosis SecA1 expression vector, Linda L. Randall for the E. coli SecA expression vector, and Manju Hingorani for the ATP binding assay protocol and for help with the assay.
This work was supported by the University of Connecticut Office of Undergraduate Research (J.M.H.), by NIH grants AI072065 (C.M.T.) and AI054540 (M.B.), by a postdoctoral fellowship from the Heiser Foundation of the New York Community Trust (H.S.G.), and by NIH training grants in infectious disease pathogenesis, AI007151 (H.S.G.), and cell and molecular biology (GM008581).
Footnotes
Published ahead of print on 16 May 2008.
REFERENCES
- 1.Archambaud, C., M. A. Nahori, J. Pizarro-Cerda, P. Cossart, and O. Dussurget. 2006. Control of Listeria superoxide dismutase by phosphorylation. J. Biol. Chem. 28131812-31822. [DOI] [PubMed] [Google Scholar]
- 2.Bensing, B. A., and P. M. Sullam. 2002. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 441081-1094. [DOI] [PubMed] [Google Scholar]
- 3.Braunstein, M., A. M. Brown, S. Kurtz, and W. R. Jacobs, Jr. 2001. Two nonredundant SecA homologues function in mycobacteria. J. Bacteriol. 1836979-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braunstein, M., B. J. Espinosa, J. Chan, J. T. Belisle, and W. R. Jacobs, Jr. 2003. SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 48453-464. [DOI] [PubMed] [Google Scholar]
- 5.Caspers, M., and R. Freudl. 2008. Corynebacterium glutamicum possesses two secA homologous genes that are essential for viability. Arch. Microbiol. [Epub ahead of print.] doi: 10.1007/s00203-008-0351-0. [DOI] [PubMed]
- 6.Chan, K. M., D. Delfert, and K. D. Junger. 1986. A direct colorimetric assay for Ca2+-stimulated ATPase activity. Anal. Biochem. 157375-380. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Q., H. Wu, and P. M. Fives-Taylor. 2004. Investigating the role of secA2 in secretion and glycosylation of a fimbrial adhesin in Streptococcus parasanguis FW213. Mol. Microbiol. 53843-856. [DOI] [PubMed] [Google Scholar]
- 8.de Keyzer, J., C. van der Does, and A. J. Driessen. 2003. The bacterial translocase: a dynamic protein channel complex. Cell. Mol. Life Sci. 602034-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Cruz, J., D. Kressler, and P. Linder. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24192-198. [DOI] [PubMed] [Google Scholar]
- 10.Doyle, S. M., E. H. Braswell, and C. M. Teschke. 2000. SecA folds via a dimeric intermediate. Biochemistry 3911667-11676. [DOI] [PubMed] [Google Scholar]
- 11.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finn, B. E., X. Chen, P. A. Jennings, S. M. Saal'au-Bethell, and C. R. Matthews. 1992. Principles of protein stability. Part 1. Reversible unfolding of proteins: kinetic and thermodynamic analysis, p. 168-189. In A. R. Rees, M. J. E. Sternberg, and R. Wetzel (ed.), Protein engineering: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 13.Franzblau, S. G., and E. B. Harris. 1988. Biophysical optima for metabolism of Mycobacterium leprae. J. Clin. Microbiol. 261124-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbons, H. S., F. Wolschendorf, M. Abshire, M. Niederweis, and M. Braunstein. 2007. Identification of two Mycobacterium smegmatis lipoproteins exported by a SecA2-dependent pathway. J. Bacteriol. 1895090-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein, A., and R. W. Barrett. 1987. Ligand dissociation constants from competition binding assays: errors associated with ligand depletion. Mol. Pharmacol. 31603-609. [PubMed] [Google Scholar]
- 16.Guo, X. V., M. Monteleone, M. Klotzsche, A. Kamionka, W. Hillen, M. Braunstein, S. Ehrt, and D. Schnappinger. 2007. Silencing essential protein secretion in Mycobacterium smegmatis by using tetracycline repressors. J. Bacteriol. 1894614-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunt, J. F., S. Weinkauf, L. Henry, J. J. Fak, P. McNicholas, D. B. Oliver, and J. Deisenhofer. 2002. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science 2972018-2026. [DOI] [PubMed] [Google Scholar]
- 18.Izard, J. W., and D. A. Kendall. 1994. Signal peptides: exquisitely designed transport promoters. Mol. Microbiol. 13765-773. [DOI] [PubMed] [Google Scholar]
- 19.James, B. W., A. Williams, and P. D. Marsh. 2000. The physiology and pathogenicity of Mycobacterium tuberculosis grown under controlled conditions in a defined medium. J. Appl. Microbiol. 88669-677. [DOI] [PubMed] [Google Scholar]
- 20.Klose, M., K.-L. Schimz, J. P. W. van der Wolk, A. J. M. Driessen, and R. Freudl. 1993. Lysine 106 of the putative catalytic ATP-binding site of the Bacillus subtilis SecA protein is required for functional complementation of Escherichia coli secA mutants in vivo. J. Biol. Chem. 2684504-4510. [PubMed] [Google Scholar]
- 21.Kurtz, S., and M. Braunstein. 2005. Protein secretion and export in Mycobacterium tuberculosis. Horizon Bioscience, Norfolk, United Kingdom.
- 22.Kurtz, S., K. P. McKinnon, M. S. Runge, J. P. Ting, and M. Braunstein. 2006. The SecA2 secretion factor of Mycobacterium tuberculosis promotes growth in macrophages and inhibits the host immune response. Infect. Immun. 746855-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanzetta, P. A., L. J. Alvarez, P. S. Reinach, and O. A. Candia. 1979. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 10095-97. [DOI] [PubMed] [Google Scholar]
- 24.Lee, V. T., and O. Schneewind. 2001. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 151725-1752. [DOI] [PubMed] [Google Scholar]
- 25.Lenz, L. L., S. Mohammadi, A. Geissler, and D. A. Portnoy. 2003. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc. Natl. Acad. Sci. USA 10012432-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenz, L. L., and D. A. Portnoy. 2002. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol. Microbiol. 451043-1056. [DOI] [PubMed] [Google Scholar]
- 27.Lill, R., W. Dowhan, and W. Wickner. 1990. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell 60271-280. [DOI] [PubMed] [Google Scholar]
- 28.McCann, J. R., J. A. McDonough, M. S. Pavelka, and M. Braunstein. 2007. Beta-lactamase can function as a reporter of bacterial protein export during Mycobacterium tuberculosis infection of host cells. Microbiology 1533350-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell, C., and D. B. Oliver. 1993. Two distinct ATP-binding domains are needed to promote protein export by Escherichia coli SecA ATPase. Mol. Microbiol. 10483-497. [DOI] [PubMed] [Google Scholar]
- 30.Papanikolau, Y., M. Papadovasilaki, R. B. Ravelli, A. A. McCarthy, S. Cusack, A. Economou, and K. Petratos. 2007. Structure of dimeric SecA, the Escherichia coli preprotein translocase motor. J. Mol. Biol. 3661545-1557. [DOI] [PubMed] [Google Scholar]
- 31.Randall, L. L., J. M. Crane, A. A. Lilly, G. Liu, C. Mao, C. N. Patel, and S. J. Hardy. 2005. Asymmetric binding between SecA and SecB two symmetric proteins: implications for function in export. J. Mol. Biol. 348479-489. [DOI] [PubMed] [Google Scholar]
- 32.Sharma, V., A. Arockiasamy, D. R. Ronning, C. G. Savva, A. Holzenburg, M. Braunstein, W. R. Jacobs, Jr., and J. C. Sacchettini. 2003. Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc. Natl. Acad. Sci. USA 1002243-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swillens, S. 1995. Interpretation of binding curves obtained with high receptor concentrations: practical aid for computer analysis. Mol. Pharmacol. 471197-1203. [PubMed] [Google Scholar]
- 34.Ulbrandt, N. D., E. London, and D. B. Oliver. 1992. Deep penetration of a portion of Escherichia coli SecA protein into model membranes is promoted by anionic phospholipids and by partial unfolding. J. Biol. Chem. 26715184-15192. [PubMed] [Google Scholar]
- 35.van der Wolk, J. P. W., M. Klose, E. Breukink, R. A. Demel, B. de Kruijff, R. Freudl, and A. J. M. Driessen. 1993. Characterization of a Bacillus subtilis SecA mutant protein deficient in translocation ATPase and release from the membrane. Mol. Microbiol. 831-42. [DOI] [PubMed] [Google Scholar]
- 36.Veenendaal, A., C. van der Does, and A. Driessen. 2004. The protein-conducting channel SecYEG. Biochim. Biophys. Acta 169481-95. [DOI] [PubMed] [Google Scholar]
- 37.Vrontou, E., and A. Economou. 2004. Structure and function of SecA, the preprotein translocase nanomotor. Biochim. Biophys. Acta 169467-80. [DOI] [PubMed] [Google Scholar]
- 38.Walker, J. E., A. Eberle, N. J. Gay, M. J. Runswick, and M. Saraste. 1982. Conservation of structure in proton-translocating ATPases of Escherichia coli and mitochondria. Biochem. Soc. Trans. 10203-206. [DOI] [PubMed] [Google Scholar]
- 39.Wang, L., A. Miller, and D. A. Kendall. 2000. Signal peptide determinants of SecA binding and stimulation of ATPase activity. J. Biol. Chem. 27510154-10159. [DOI] [PubMed] [Google Scholar]
- 40.World Health Organization. March 2007, posting date. Fact sheets: tuberculosis, no. 104. WHO, Geneva, Switzerland. http://www.who.int/mediacentre/factsheets/fs104/en.
- 41.Yu, L., H. Yang, Q. Ho, and P. C. Tai. 2006. Expression, purification, and characterization of Pseudomonas aeruginosa SecA. Protein Expr. Purif. 50179-184. [DOI] [PubMed] [Google Scholar]
- 42.Zito, C. R., E. Antony, J. F. Hunt, D. B. Oliver, and M. M. Hingorani. 2005. Role of a conserved glutamate residue in the Escherichia coli SecA ATPase mechanism. J. Biol. Chem. 28014611-14619. [DOI] [PMC free article] [PubMed] [Google Scholar]