Abstract
Lactococcus lactis is a widely used food bacterium mainly characterized for its fermentation metabolism. However, this species undergoes a metabolic shift to respiration when heme is added to an aerobic medium. Respiration results in markedly improved biomass and survival compared to fermentation. Whole-genome microarrays were used to assess changes in L. lactis expression under aerobic and respiratory conditions compared to static growth, i.e., nonaerated. We observed the following. (i) Stress response genes were affected mainly by aerobic fermentation. This result underscores the differences between aerobic fermentation and respiration environments and confirms that respiration growth alleviates oxidative stress. (ii) Functions essential for respiratory metabolism, e.g., genes encoding cytochrome bd oxidase, menaquinone biosynthesis, and heme uptake, are similarly expressed under the three conditions. This indicates that cells are prepared for respiration once O2 and heme become available. (iii) Expression of only 11 genes distinguishes respiration from both aerobic and static fermentation cultures. Among them, the genes comprising the putative ygfCBA operon are strongly induced by heme regardless of respiration, thus identifying the first heme-responsive operon in lactococci. We give experimental evidence that the ygfCBA genes are involved in heme homeostasis.
The lactic acid bacterium Lactococcus lactis is widely used in industrial milk fermentations and is present in the majority of the >18 million tons of cheese produced yearly (41). One main contribution of L. lactis is to produce lactic acid during fermentation, which acidifies and coagulates milk. However, L. lactis can also shift to an energetically favorable respiratory metabolism in an aerobic environment when heme is supplied (14, 21, 22, 48). Addition of this essential cofactor of the terminal cytochrome bd oxidase complex activates an electron transport chain (14, 52). The absence of heme biosynthetic genes in lactococci explains this requirement (21). Respiration growth has been used for large-scale production of lactococcal starter cultures and is applicable to other lactic acid bacteria (17, 43).
Respiratory metabolism has dramatic consequences on lactococcal physiology, with major positive impacts on biomass and long-term survival (14, 22, 28, 48). The biomass of different lactococcal strains is essentially doubled by respiration, compared to fermentation metabolism (14, 17, 43). This is attained since (i) protons are extruded from cells by the respiratory chain, rather than by the H+-ATPase at the expenditure of ATP (5), and (ii) NAD+ can be regenerated by the respiratory chain NADH oxidase, which makes pyruvate metabolism more flexible. As a result, acetate (which generates ATP) and pH-neutral acetoin are synthesized in place of lactate (5, 7, 14).
Respiration growth extends long-term survival of lactococci from a few days to several months (23). This advantage is attributed to lower cytoplasmic O2, which is converted to water via the membrane cytochrome oxidase, and in part, to the less acidic environment that accompanies the metabolic shift (48). Higher stability and yields of L. lactis has led to novel industrial applications in the preparation of bacterial starter cultures (17, 43).
We previously used a proteomic approach to investigate expression changes in L. lactis due to respiration growth in late exponential phase (55). Some 20 proteins displayed altered expression levels or changed migration positions under respiration compared to fermentation growth. The majority of the changes corresponded to proteins involved in general carbon and nitrogen metabolism. In addition, posttranslational changes were observed, including a dramatic decrease in amounts of the oxidized form of major glycolytic enzyme glyceraldehyde 3-P-dehydrogenase, which was attributed to a less O2-stressed cytoplasm under respiration conditions (48, 55). Moreover, a potential regulator of unknown function, YgfC, was specifically expressed in respiration conditions (55).
In the present study, whole-genome transcriptome analysis was performed on mid-exponential-phase lactococci grown under static fermentation, aerobic fermentation, or respiration (aeration plus heme) conditions. We report that transcriptional responses to the aerobic and respiratory states in lactococci are different and that respiring lactococci are under lower oxidative stress. We report that the ygfCBA operon, encoding a putative transport system and regulator, is strongly induced by heme itself and is likely involved in heme tolerance and homeostasis.
MATERIALS AND METHODS
Strains and growth conditions.
L. lactis subsp. lactis CHCC2862, a commonly used cheese-making strain (Chr. Hansen Culture Collection, Hørsholm, Denmark) was used for transcriptomic analyses. Strain MG1363 (20) was used for construction of ygf operon mutants, as described below. Cultures were grown in M17 (Oxoid A/S, Greve, Denmark) supplemented with 0.5% lactose (M17L) for CHCC2862 and with 1% glucose (M17G) for MG1363. Cells from a static stationary preculture were inoculated at 1% into 250 ml of fresh medium preheated at 30°C under (i) static conditions (nonagitated cultures in full bottles), (ii) aerobic conditions (cultures in 2-liter conical flasks with baffles; agitation at 170 rpm), or (iii) respiratory conditions (aerobic plus 8 μM heme). The optical density at 600 nm (OD600) and pH were monitored over time. Sampling was every 0.5 h; the static culture was gently agitated before sampling. Metabolite concentrations were determined by using gas chromatography and high-pressure liquid chromatography (sampling every 1 h) as described previously (12, 50).
Isolation of total RNA.
At an OD600 of 1.0, samples of 0.67 ml were collected in 1.33 ml of RNAprotect according to the manufacturer's protocol (Qiagen, Valencia, CA). Total RNA was isolated by using an RNeasy minikit (Qiagen) except for the following: 200 μl of TE buffer contained 15 mg of lysozyme (L6876, Sigma-Aldrich, Brøndby, Denmark)/ml, 15 μl of proteinase K (20 mg/ml; catalog no. 19131; Qiagen), and 2 μl of mutanolysin (M9901; Sigma-Aldrich) at 25 U/μl. Vortexing was every 10 s at 37°C for 2 min. The quality and concentration of RNA was determined by using the RNA 6000 Nano Kit on the Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA). RNA was eluted with 2 × 55 μl of H2O, and routinely 20 to 50 μg of total RNA was obtained. Only high-quality total RNA, as indicated by two sharp rRNA peaks, was used.
Design of oligonucleotides.
OligoWiz 1.0 (39) was used to design 65- to 75mer oligonucleotides from most of the coding sequences identified in L. lactis subsp. lactis IL1403 (6). Selected coding sequences from phage bIL170 (11), the lactose plasmid (13), the two plasmid-encoded proteinase genes prtM and prtP (10), and the 5S, 16S, and 23S rRNA genes, were also included. All 2208 DNA oligonucleotides were purchased from Bioneer Corp. at a 50-nmol synthesis scale with BioRP purification and in a lyophilized state (Daedeok-gu, Daejeon, South Korea).
Array spotting.
Oligonucleotides were dissolved at 10 μM in 50% dimethyl sulfoxide (D8418; Sigma-Aldrich) and printed in four replicates on UltraGAPS slides (Corning B.V., Schiphol-Rijk, The Netherlands) by using a Genpak Array 21 arrayer (Genetix, Ltd., Hampshire, United Kingdom) with 16 SMP3 pins (TeleChem International, Inc., Sunnyvale, CA), yielding a spot diameter of 100 to 120 μm. Washing was performed between each print cycle for 20 s in water followed by 20 s in 70% ethanol. The relative humidity was kept at 48% ± 5% by placing a water bath at the appropriate temperature below the air inlet of the arrayer. The oligonucleotides were cross-linked to the surface by using a Stratalinker 1800 UV cross-linker set at 70 mJ/cm2 (Stratagene). Printed arrays were stored in aluminum pouches with five arrays each, and silica gel was used to remove humidity. The platform specifications are available at the NCBI Gene Expression Omnibus (GEO) under accession no. GPL5400.
cDNA synthesis and labeling of total RNA.
A total of 10 μg of total RNA was lyophilized in a Speed-Vac. The copying into cDNA and labeling with either Cy3 (reference) or Cy5 (test) was done by using a CyScribe post-labeling kit according to the manufacturer's protocol (Amersham Biosciences, Hillerød, Denmark). One microliter of random nonamer from the kit was used for priming.
Hybridization, washing, and scanning of arrays.
According to the Amersham protocol, the Cy3- and Cy5-labeled cDNA were combined and lyophilized. Hereafter, they were resuspended in 7 μl of nuclease-free water, and 5 μl of hybridization buffer (4×) and 8 μl of formamide was added. This 20-μl solution was applied to the array under a 24-by-24-mm LifterSlip (Erie Scientific Company, Portsmouth, NH) and put in an airtight box containing paper towels soaked in NaCl-saturated water. Arrays were hybridized for 16 to 20 h at 42°C and then washed using the UltraGAPS protocol (Corning B.V.). Arrays were scanned by using a GenePix 4100A personal scanner with GenePix Pro 6.0 software (Axon Instruments, Inc., Union City, CA). The photo multiplier tube sensitivity values of the scanner software were adjusted to around 660 for Cy5 and 520 for Cy3 to obtain only few saturated spots on each array and about equal total signals for each dye.
Preanalysis of arrays.
In GenePix Pro software, a grid with 110-μm spots was superimposed on the array image and manually adjusted to fit small irregularities in spotting. Spots clearly covered by, for example, dust particles were flagged as “bad” (equal to a Flag value of −100). The intensities of all the spots were then calculated by using the “analyze” function. Spots with very weak signals in both channels were also flagged “bad.” These were spots where the signal-to-noise ratio was <1 in both channels or the percentage of feature pixels with intensities more than two standard deviations above the background pixel intensity was <20, i.e., (% > B + 2SD) < 20, in both channels (see the GenePix Pro manual for details). All other spots were considered “found” and had a Flag value of ≥0. All data were exported from GenePix Pro in tab-delimited format.
In further analyses, the variables of feature (spot) intensity minus the background intensity around the spot was used, i.e., (F635 median - B635) = Cy5 and (F532 median - B532) = Cy3. The log2(ratio) was calculated as log2(Cy5/Cy3).
Some spots may have a strong signal in one channel but a very low signal in the other. Due to noise this may result in a negative ratio and thus generate an invalid log2(ratio). This was corrected by using a macro in Excel 2000 (Microsoft Corp., Redmond, WA) that changed the “F635 median - B635” and “F532 median - B532” values of <10 to 10 (arbitrary) for “found” spots. The corrected data were exported from Excel and imported into Acuity 4.0 (Axon), where they were ratio-normalized with the default settings.
In Acuity a data set was created. For each array only genes were included where (i) at least three of the four replicate spots were “found,” (ii) the standard deviation was <0.8 for the log2(ratio) among “found” spots, and (iii) <30% of the pixels in “found” spots, in both channels, were saturated. The mean of the log2(ratio) of replicate spots was used as the working variable in Acuity.
Use of array platform for CHCC2862.
Using comparative genome hybridization, we previously found that all oligonucleotides, except about 130 (mainly prophage genes), designed from L. lactis strain IL1403 gave a hybridization signal for CHCC2862. Moreover, selected genes present in CHCC2862 had ca. 99.8% identity with the corresponding IL1403 genes when sequenced (data not shown). Overall, this shows that the platform is applicable to strain CHCC2862.
Three arrays were produced (test versus reference): (i) aerobic versus static, (ii) respiratory versus static, and (iii) respiratory versus aerobic. We previously showed that twofold differential expression can be detected using this platform, without producing replicate arrays (17, 19, 43). Values of the “respiratory versus aerobic” array should be equivalent to those calculated by subtracting the corresponding log2(ratio) values of “aeration versus static” from those of the “respiration versus static” arrays. It was thus used as a control array to validate results. Only for 47 (2.3%), mostly very lowly expressed genes, did the calculated value not fit within twofold of the control array (data not shown). After preanalysis, data from 2,073 genes remained in the combined data set.
Experiments were performed on two biologically independent samples. Since the growth medium is complex and expression changes during the exponential growth phase may be rapid within short time intervals, some quantitative and qualitative differences were observed. To obtain a robust set of genes affected by the tested conditions, we earmarked only genes whose expression was affected similarly in both experiments. The figures and values are presented from one of the experiments. Array data are available at NCBI GEO under accession no. GSE8182.
qPCR validations.
Probes for qualitative reverse transcription-PCR (qPCR) were in all cases dual-labeled LNA probes from the Human ProbeLibrary (Roche, Diagnostics, Basel, Switzerland). For qPCR expression assays, primer sequences and probes for relevant genes were designed by using ProbeFinder software (Roche). cDNA synthesis was made using the Superscript II first Strand Synthesis kit (Invitrogen, Carlsbad, CA), according to provided protocol, with random hexamer priming. The RNA input was 25 ng of total RNA. qPCR was performed in five technical replicates with one mastermix (Low ROW MasterMix; Eurogentec, Liège, Belgium) on an ABI 7500 qPCR machine. Primers were added at 300 nM, and probes were added at 150 nM. PCR conditions used were: 10 min at 95°C, followed by 40 cycles of 15 s of denaturation at 95°C and 1 min of priming/elongation at 60°C. PCR efficiency was in the range of 85 to 100% and consistent in intra-assay comparisons. The standard deviation between replicates was <0.6 cycle threshold level.
Northern blot experiments.
The expression of ygfC was examined by extracting total RNA (FastRNA Pro Blue Kit; Qbiogene, France) from aerobic or respiration cultures at the early, mid, and late stationary phases. The probe was a DNA fragment containing ygfC, obtained by PCR amplification with the primers 5′-ATTCCGAGAGGGAGTTTTTATCAG-3′ and 5′-CGAGTTTATGAACAACTGATTTTAATACT-3′; the purified fragment was labeled with [32P]dCTP (Ready-To-Go; Amersham). Northern blots were performed as described previously (46).
ygfA-lacLM fusion construction and β-galactosidase assay.
A fragment corresponding to the end of the ygfA gene (527 bp; the terminal gene of the operon) was obtained by PCR amplification with the primers yfgAfor 5′-GCGGCCGCCTTTCCTTACAATAGCAGGAGG-3′ and ygfArev 5′-CCCGGGTTATATTTTTGATAAGAGTCC-3′ (nucleotides not in the genome sequence are set in italics). The PCR product was digested by NotI-XmaI and inserted into cloning vector pBC570.1 (B. Cesselin and P. Gaudu, unpublished data) to generate plasmid pKT1, in which the lacLM gene was just downstream of the ygfA stop triplet. pKT1 was introduced at the ygf locus by single-crossover recombination in MG1363. To determine the β-galactosidase activity, cells were recovered from cultures under specified conditions, adjusted to an OD600 of 1, and then assayed as described by Miller (36).
ygf mutant construction, ygf operon complementation, and heme sensitivity test.
We constructed ygfC and ygfB single-crossover mutants of the L. lactis strain MG1363. For ygfC, a 325-bp internal fragment was amplified with the primers 5′-ATTCCGAGAGGGAGTTTTTATCAG-3′ and 5′-CGAGTTTATGAACAACTGATTTTAATACT-3′; for ygfB, a 578-bp internal fragment was amplified with the primers 5′-GGAATGCCGAGAAAATAATTC-3′ and 5′-AATCCATAGCGCTTAGCG-3′. Fragments were first cloned into pCR2.1-TOPO and sequenced. Plasmids were then digested with EcoRI, and fragments of interest were cloned into EcoRI-restricted pRV300 and established in Escherichia coli (32). Resultant plasmids were prepared and used to obtain SCO integrations by electroporation of MG1363 and selection with erythromycin (2.5 μg/ml). Integrations were confirmed by Southern hybridization.
The three adjacent genes ygfC, ygfB, and ygfA were recovered on a single DNA fragment by PCR using chromosomal DNA extracted from L. lactis strain MG1363. The primers were ygf HindIII (5′-AGAAAGCTTGAAACCTGATACTGACCGTATGGC-3′) and ygf EcoRI (5′-AGAGAATTCCCACAACCTATCTTAGCCTTGGC-3′). The resultant 2.9-kb fragment was digested with HindIII and EcoRI, purified, cloned into HindIII-EcoRI-restricted pG+host8 (34), and established directly in L. lactis MG1363 with tetracycline (5 μg/ml) selection; the resultant plasmid is referred to as pYgfCBA. To determine whether the cloned ygf genes complement the ygfB or ygfC mutant, the mutation was established in the pYgfCBA-containing strain by conjugation (24). Transconjugant selection in both cases was with erythromycin at 3 μg/ml and tetracycline at 5 μg/ml.
The wild-type and the ygfC or ygfB mutant strains, as well as strains containing the chromosomal mutation and the complementing pYgfCBA plasmid, were examined for heme sensitivity as follows: stationary-phase cultures were diluted 1:100 and plated on M17G agar plates. Heme (2 μl of a 20 mM stock solution [13 mg/ml]) was spotted onto the plates, which were incubated overnight at 30°C and photographed. The experiments were repeated four times with comparable results.
RESULTS AND DISCUSSION
Growth characteristics and experimental design.
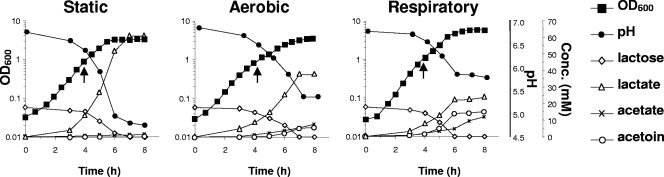
Industrial strain L. lactis subsp. lactis CHCC2862 was grown in rich M17L under static, aerobic, and respiratory conditions (see Materials and Methods). The OD600, pH, sugar, and end product concentrations were monitored during growth (Fig. 1). As previously reported for L. lactis subsp. cremoris MG1363, the OD600 and pH of stationary CHCC2862 cultures were substantially higher for cells grown via respiration versus cells grown in static or aerobic fermentation (OD600 = 5.8 compared to OD600 = 3.2 to 3.5 and about 1 pH unit higher). The end products acetoin and acetate were greatly increased, while lactate production was reduced (Fig. 1). CHCC2862 thus displayed respiration characteristics similar to those of MG1363 (14, 52).
FIG. 1.
Growth and metabolic characteristics of CHCC2862. Growth characteristics for static, aerobic, and respiratory cultures, including measurement of the OD600 (▪), pH (•), lactose consumption (⋄), and production of lactate (▵), acetate (×), and acetoin (○). Vertical arrows indicate an OD of 1.0, the point at which the cells were harvested for transcriptome studies.
For transcriptome studies, cells were harvested from the exponential phase at an OD600 of 1.0 (indicated by arrows in Fig. 1). At this point the pH of all cultures had decreased only slightly, from 6.7 to about 6.4. Microarrays were produced for (i) aerobic versus static, (ii) respiratory versus static, and (iii) respiratory versus aerobic cultures. The data from the “respiratory versus aerobic” array gave information that can be calculated using values from both “respiration versus static” and “aeration versus static” arrays; it was used as a control array to validate results. Below, we discuss genes whose expression differed by >4-fold during aerobic or respiratory growth relative to static conditions in two independently conducted experiments. Since this threshold is high, it is likely that other genes respond at lower levels to the tested growth conditions.
Aerobic fermentation affects the expression of stress response genes.
Expression of at least 20 putative stress response genes was affected mainly under aerobic, nonrespiratory conditions (Table 1) and represented the predominant category of genes with known function (see Table S1 in the supplemental material for a complete list of the categories). Several of the stress response genes encode detoxification enzymes (e.g., AhpCF, GshR, PmsX, Qor, SodA, and YgmK) (37, 45, 51, 53). In line with these values, physiological studies showed that superoxide dismutase gene sodA (∼6-fold induced by aeration compared to 3-fold induced by respiration) is required for survival in aeration but not respiration conditions (14, 51). Genes encoding DNA protection and other stress response functions (e.g., HslB and OsmC) (1, 3, 40, 56) are also more markedly expressed in aerobic cultures. Cytoplasmic NADH oxidase gene noxE is among the most highly induced genes (15-fold) under aerobic growth. NoxE is implicated in the shift from homolactic to mixed acid fermentation (14, 33). It is required in numerous Streptococcaceae for aerobic growth, but respiration chain NADH oxidase activity (5) may compensate for this requirement, as we recently observed in Streptococcus agalactiae (58).
TABLE 1.
Stress response genes are affected by aerobic fermentation but are attenuated in respiration conditionsa
| Genec | ORF description | Ratioc
|
|
|---|---|---|---|
| Aero/ Stat | Resp/ Stat | ||
| ahpC | Alkyl hydroperoxide reductase | 6 | 3 |
| ahpF | Alkyl hydroperoxide reductase | 6 | 2 |
| gshR† | Glutathione reductase | 8 | 4 |
| hslB | HU-like DNA-binding protein | 9 | 4 |
| noxE | Major cytoplasmic NADH oxidase | 15 | 2 |
| osmC† | Osmotically inducible protein | 15 | 7 |
| pmsX | Peptide methionine sulfoxide reductase | 12 | 5 |
| qor | Quinone oxidoreductase | 7 | 2 |
| sodA | Superoxide dismutase | 6 | 3 |
| ybjA† | Peptide methionine sulfoxide reductase | 9 | 4 |
| yfiE† | Organic hydroperoxide resistance protein; OsmC homolog | 11 | 5 |
| yhjA | Similar to B. subtilis stress protein CsbD | 11 | 3 |
| ymgH | Similar to E. faecalis Gls24 stress protein | 4 | 2 |
| ymgK | Aldo-keto reductase involved in detoxification | 14 | 5 |
| ynfC | lactoyl glutathione lyase; serine-rich | 14 | 5 |
| yahB | UspA-like protein | 6 | 2 |
| yjaB | UspA-like protein | 8 | 3 |
| yobA | UspA-like protein | 5 | 2 |
| nifJ† | Pyruvate ferredoxin oxidoreductase | -16 | -8 |
| ypbB | Similar to Mg and Co transporter | -4 | -2 |
Genes related to stress response and showing differences greater than ∼2-fold in aerobic fermentation compared to respiration growth are presented. Signal ratios of >4 of aerobic versus static fermentation were noted. Aero/Stat indicates the ratio of signal expression of aeration versus static (both fermentation) growth conditions. Resp/Stat indicates the ratio of signal expression of respiration versus static growth conditions. Functions are from KEGG annotations of L. lactis strain IL1403 (http://www.genome.jp/kegg/genes.html).
b †, Potential disulfide bonds are present in open reading frames (ORFs).
Negative values indicate downregulation.
Universal stress proteins were described in Escherichia coli as a family of proteins that are induced in response to a wide range of environmental signals, including DNA-damaging agents and respiratory uncouplers (31). Out of six putative universal stress proteins in L. lactis, three were most highly expressed in aeration conditions (yahB, yjaB, and yobA). Among these, the protein encoded by yahB was previously identified as being induced in an O2-sensitive L. lactis thioredoxin reductase-deficient trxB1 mutant (55).
Proteins responding to oxidative stress might undergo conformational changes linked to the formation of disulfide bonds (42). The expression of six genes potentially encoding disulfide bond-containing proteins (http://www.doe-mbi.ucla.edu/∼boconnor/GDAP/query_v2.php? EXP_ID = 1) is altered mainly by aerobic growth (fadA, gshR, nifJ, osmC, ybjA, and yfiE; see Table S1 in the supplemental material). For example, the gene encoding cysteine-rich NifJ (pyruvate-ferredoxin oxidoreductase) is downregulated in aerobic conditions, in keeping with its known O2 sensitivity (29).
Expression of purine de novo synthesis genes was markedly lower, mainly in aeration conditions but also in respiration conditions compared to the static condition (data not shown). However, substantial differences in values were observed between experiments: M17 medium contains purine compounds for growth until around an OD600 of 1.0, as deduced from previous studies of a purine de novo synthesis mutant and time course transcriptome analysis with CHCC2862 at around an OD600 of 1 (C. Garrigues and D. Nilsson, unpublished results). The fluctuation in these values is likely to arise from slight differences in the kinetics of purine utilization and/or M17 batch variation.
The numerous genes that respond to aerobiosis are likely to be involved in the aerobic life of L. lactis. O2 is present under both aerobic and respiratory growth, but differential expression points to a different physiology under the two conditions. The greatly improved growth and survival as found previously for L. lactis during respiratory, compared to aerobic fermentation growth correlates with a markedly lower induction of stress response genes.
L. lactis genes regulated by both aeration and respiration growth.
Relative to static growth, 59 genes responded similarly to aeration and respiration (see Table S2 in the supplemental material), most of which are involved in central metabolism and transport. The mechanism of how these genes respond similarly to aerobic fermentation and respiration conditions remains to be investigated. However, two examples of genes affected in both conditions (see below) suggest that the regulation of some genes might (i) be highly sensitive to O2 or (ii) respond to factors other than O2, such as NAD+ or CO2, which should be present in both conditions.
Pyruvate formate lyase (encoded by pfl), which is highly O2 sensitive, was downregulated by aeration but also by respiration growth (both four- to fivefold; see Table S2 in the supplemental material). Thus, low intracellular O2 levels in respiration cells (48) are likely sufficient to downregulate pfl expression. However, we observed increased amounts of Pfl protein in stationary-phase respiration cultures (B. Cesselin and P. Gaudu, unpublished data); since cydAB genes are induced late in growth (14), increased Pfl might reflect efficient O2 elimination through respiration late in growth.
The arginine catabolic genes arcA, arcB, arcC1, and arcC2 are repressed in aerobic and respiratory compared to static conditions. Enzymes encoded by these genes reportedly contribute to raising internal pH in acidification conditions by liberation of NH3 and are induced by acid stress (8), (47). These genes are reportedly downregulated in the presence of O2 in different bacterial species (e.g., see references 16 and 26). However, since intracellular O2 appears to be lower in respiration than in aeration conditions, other factors might contribute to the repression of arginine catabolic genes (9). One candidate is NAD+, which expectedly reaches increased levels in both aerobic and respiratory growth via NoxE and heme-activated NADH oxidase activity, respectively (4, 5). An alternative signaling molecule common to aeration and respiration conditions is CO2, which might affect the regulation of arginine catabolism (38).
The expression of genes implicated in respiratory chain assembly and respiration activity is constitutively expressed.
An active respiration chain in L. lactis requires an electron donor possibly encoded in L. lactis by noxB (L. Rezaïki, unpublished data), an electron receiver and transmitter (requiring genes of the men operon, plus ispA and ispB (49), and a terminal electron acceptor (encoded by structural genes cydAB and requiring cydCD genes for assembly (14, 44). In addition, L. lactis requires a heme uptake function (fhu operon) (21). Surprisingly, the expression of none of these genes was specifically induced by respiration (Table 2). We conclude that respiration genes are expressed regardless of growth conditions. It is possible that at least some of these genes have functions under nonrespiratory conditions. Nevertheless, their expression under all conditions suggest that cells could activate respiration rapidly once heme and O2 are available. This draws support from early work showing that the respiration chain is immediately activated in fermentation-grown cells by heme addition (52).
TABLE 2.
Lack of specific regulation of genes implicated in respiration chain activitya
| Function and geneb | ORF descriptionc | Ratio
|
|
|---|---|---|---|
| Aero/Stat | Resp/Stat | ||
| Heme synthesis or utilization | |||
| fhuB | Ferrichrome ABC transporter permease protein | 1 | 2 |
| fhuD† | Ferrichrome ABC transporter substrate-binding protein | 1 | 2 |
| fhuG | Ferrichrome ABC transporter permease protein | 1 | 2 |
| fhuR† | fhu operon transcriptional regulator | 1 | 1 |
| hemH† | Ferrochelatase | 1 | 1 |
| hemK | Protoporphyrinogen oxidase | 2 | 1 |
| hemN | O2-independent coproporphyrogen III oxidase | 2 | 2 |
| Electron donor | |||
| noxB† | Putative membrane NADH dehydrogenase | 1 | 1 |
| Electron transfer | |||
| menB† | Naphthoate synthase | 3 | 2 |
| menD | 2-Oxoglutarate decarboxylase/2-succinyl-6- hydroxy-2,4-cyclohexadiene-1-carboxylate synthase | 2 | 2 |
| menE | O-Succinylbenzoic acid-CoA ligase | 2 | 2 |
| menF | Menaquinone-specific isochorismate synthase | 2 | 2 |
| menX | Protein in menaquinone biosynthesis pathway | 2 | 2 |
| yhdB | O-Succinylbenzoate-CoA synthase (menC) | 2 | 2 |
| ybiG | Probable 1,4-dihydroxy-2-naphthoate octaprenyltransferase (menH) | 3 | 3 |
| yljG | Probable SAM-dependent methyltransferase (menA) | 1 | 1 |
| ispA | Geranyltranstransferase: MK tail addition | 1 | 1 |
| ispB | Heptaprenyl diphosphate synthase component II: MK tail addition | 1 | 1 |
| Electron acceptor | |||
| cydA† | Cytochrome bd-I oxidase subunit I | 1 | 1 |
| cydB† | Cytochrome d ubiquinol oxidase subunit II | 1 | 1 |
| cydC† | Cytochrome d ABC transporter ATP-binding and permease protein | 1 | 1 |
| cydD | Cytochrome d ABC transporter ATP-binding and permease protein | 1 | 1 |
Compared to aerobic fermentation, respiration leads to greater production of metabolites acetoin and diacetyl, whereas the amounts of lactate are diminished (Fig. 1) (14, 28). Despite these major changes in pyruvate metabolism the expression of als (encoding acetolactate synthase) and aldBC (encoding acetolactate decarboxylase) was not differentially expressed in respiration conditions; the expression of ackA1A2 (encoding acetate kinase) and pta (encoding phosphate acetyltransferase), which produce acetyl phosphate from acetate or from acetyl-coenzyme A (CoA), respectively, were similarly expressed under all three conditions (see Table S2 in the supplemental material and raw array data). Considering the extensive changes in the end product patterns of respiring lactococci and the lack of changes in expression levels for relevant genes, we suggest that fluxes away from pyruvate might be controlled at the metabolic level, possibly via the redox balance (18, 30).
Eleven L. lactis genes respond mainly to respiration conditions.
Six genes, organized in two and three potential operons, and one monocistronic gene, were induced specifically in respiration. Five genes, including a two-gene cluster, were repressed mainly in respiration (Table 3). Nine of the genes encode putative transport functions. Notably, none of the genes with respiration-specific responses encodes known stress response functions.
TABLE 3.
Genes whose expression responds mainly to respirationa
| Function and geneb | ORF descriptionc | Ratio
|
|
|---|---|---|---|
| Aero/Stat | Resp/Stat | ||
| Metabolism | |||
| adhE | Alcohol dehydrogenase/acetaldehyde dehydrogenase | −23 | −91 |
| frdC | Fumarate reductase flavoprotein subunit; serine-rich domain | −3 | −7 |
| Transport | |||
| amtB† | Ammonium transporter | 1 | 5 |
| glnB† | Nitrogen regulatory protein P-II | 2 | 7 |
| pnuC2 | Nicotinamide mononucleotide transporter | 2 | 5 |
| ygfC† | Transcriptional regulator ygf operon | 1 | 49 |
| ygfB† | ABC transporter permease protein (39% identity YxeA) 29% identity to S. aureus putative heme permease HrtB (54) | 1 | 37 |
| ygfA† | ABC transporter ATP-binding protein (ORF), 59% identity to YxeB and 45% identity to S. aureus heme permease HrtA (54) | 1 | 19 |
| glnQ | Glutamine ABC transporter ATP-binding protein | −4 | −10 |
| mleS§ | Malolactic enzyme | 1 | −4 |
| mleP§ | Malate transporter | 1 | −5 |
As in Table 1, except that the values for Resp/Stat here are at least twofold greater than those for Aero/Stat.
Genes grouped by an associated symbol (†, †, or §) are adjacent and colinear. ygfCBA genes form a putative operon (see the text).
ORF, open reading frame.
The amtB and glnB genes form a putative operon that is induced five- to sevenfold by respiration. These genes encode a channel for ammonium capture that is active in ammonium limitation and feeds into the glutamine synthetase pathway (27). Ammonium, which is generated from the degradation of arginine, may be limiting, since arc genes are repressed in aerobic and respiration conditions (Table 3), and cells undergo a general arginine starvation toward the end of growth (55). The induction of amtB and glnB in respiration cultures may simply reflect a greater demand for nutrients.
The adhE gene codes for alcohol dehydrogenase (AdhE), which uses NADH as a cofactor to mediate the conversion of acetyl-CoA to acetaldehyde and then to ethanol. adhE expression was 91-fold lower in respiration and 23-fold lower in aeration. Repression of adhE in aerobic conditions was previously reported (2). Since NADH oxidase activity is high in both aerobic and respiration conditions, the more repressive effect of respiration conditions on adhE expression might suggest that downregulation is related to limiting the amounts of NADH rather than O2.
The ygfCBA genes form a putative operon in L. lactis strains IL1403 and MG1363 (6, 57) encoding a putative regulator (ygfC), a transporter permease (ygfB), and an ABC transporter ATP-binding protein (ygfA). These genes were specifically induced ∼30-fold in respiration. Respiration-induced expression of the regulator YgfC was previously observed in proteomic studies (55). Further experiments were conducted to characterize these genes (see below).
qPCR validation of microarray data.
Validation of microarray analyses was conducted by qPCR on 34 genes being up-, down-, or unregulated and representing genes of various absolute expression levels. New cultures were grown, and comparisons were made to microarrays of respiration versus static cultures (Table 4). For all genes that were <10-fold differentially expressed on the arrays, microarrays and qPCR gave the same result within 2-fold, with the exception of lowly expressed gene, glpK. In this case, qPCR most likely gives the more accurate result. For the five genes showing >10-fold differential expression, the microarray and qPCR showed the same qualitative result. Overall, the qPCR results validate the results of the array platform.
TABLE 4.
Comparison of microarray versus qPCR data
| Gene | Resp/Stat ratioa
|
Expression levelb (abundance) | |
|---|---|---|---|
| Array | qPCR | ||
| adhE | -128 (−91) | -21 | +++ |
| nifJ | -11 (−8) | -4 | +++ |
| yxcA | -8 (−5) | -4 | + |
| frdC | -7 (−7) | -3 | +++ |
| pfl | -4 (−5) | -3 | +++ |
| galK† | -2 (−2) | 1 | + |
| msmK† | -2 (−3) | 1 | ++ |
| gadR† | -2 (−2) | 2 | + |
| pepM† | 1 (1) | 1 | ++ |
| atpA† | 1 (1) | 1 | ++ |
| rnhA† | 1 (1) | 1 | ++ |
| fhs† | 1 (−2) | 1 | + |
| topA† | 1 (2) | 1 | ++ |
| rpsE† | 1 (1) | 1 | ++++ |
| pflA† | 1 (1) | 1 | ++ |
| yshC† | 1 (1) | 1 | ++ |
| clpC† | 1 (2) | 1 | ++ |
| gyrA† | 1 (1) | 1 | ++ |
| rluD† | 1 (1) | 1 | ++ |
| cydC | 2 (1) | 2 | ++ |
| cydD | 2 (1) | 2 | ++ |
| sodA | 2 (3) | 2 | +++ |
| hemN | 2 (2) | 2 | + |
| fhuG | 2 (2) | 2 | + |
| fhuD | 4 (2) | 4 | ++ |
| lplL | 4 (15) | 6 | +++ |
| pdhA | 4 (7) | 4 | +++ |
| pdhB | 5 (8) | 6 | +++ |
| glpK | 6 (11) | 2 | + |
| amtB | 7 (5) | 7 | ++ |
| pdhD | 7 (4) | 4 | +++ |
| ygfA | 34 (19) | 7 | ++ |
| ygfB | 39 (37) | 11 | +++ |
| ygfC | 45 (49) | 14 | +++ |
Five new arrays were produced for respiratory versus static conditions, including three dye-swap arrays. The average ratio of the respiratory signal divided by the static signal for 34 genes is shown. The genes are sorted from lowest to highest differential expression in respiration conditions. The standard deviation was ≤0.3 for all genes, except for seven genes where it was 0.3 to 0.6. For the 34 genes the average ratio of five qPCR replicates is also shown.
A qualitative measure of the absolute expression level is shown, based on the average spot signal intensity from microarrays (see Materials and Methods). For differentially expressed genes, higher signal intensities were used. Scale: >5,000, ++++ (very high); 1,500 to 5,000, +++ (high); 300 to 1,500, ++ (moderate); <300, + (low).
Evidence that ygfCBA genes are transcribed as an operon whose expression is induced in respiration conditions.
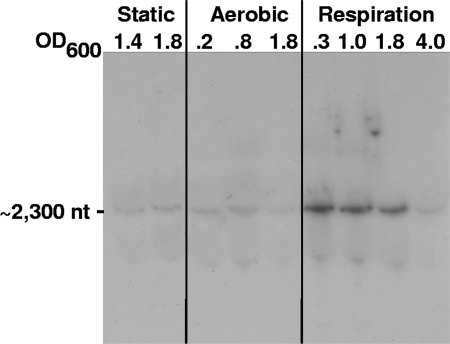
This putative operon organization of ygfCBA genes is conserved in three L. lactis sequenced genomes (IL1403, MG1363, and SK11) (6, 35, 57). The three genes are bounded on either side by putative rho-independent terminators, whereas no terminator is predicted between the genes (data not shown). ygfCBA transcription and expression as a function of time and growth condition was examined by Northern blotting, using a ygfC-specific probe (Fig. 2). A single major transcript of ∼2.3 kb was revealed at the size expected for an mRNA comprising the three genes. Expression was barely detectable in static or fermentation conditions but was strongly induced during respiration growth. Expression in respiration conditions was lower when the cells reached stationary phase. These results confirm that ygfCBA expression is induced under respiration conditions and strongly suggest that these genes constitute an operon.
FIG. 2.
ygfCBA genes are cotranscribed and expressed mainly in exponential growth under respiration-permissive conditions. Cultures of MG1363 were grown under static, aerobic, or respiration conditions. RNA was prepared from cells harvested at the indicated OD600. Northern blotting was performed with a ygfC specific probe. The single major RNA band was estimated at ∼2,300 nucleotides (nt) based on control ladder, as expected for transcription of the three ygf genes as a single operon.
The ygf operon is induced by heme irrespective of respiration.
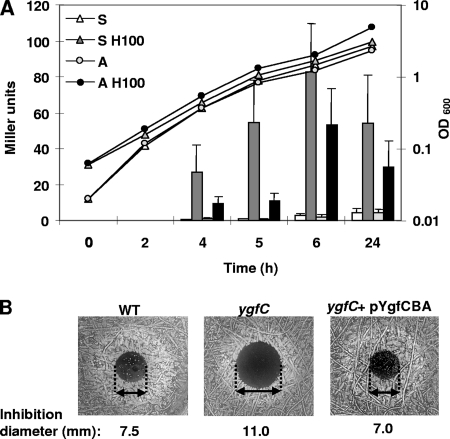
The strong induction of the ygf operon in respiration conditions could be a response to respiration itself or solely to the presence of heme. To distinguish between these possibilities, a fusion of the lacLM reporter (encoding a β-galactosidase) just downstream of ygfA (the last gene of the operon) was introduced at the chromosomal ygf locus; these constructions were done in L. lactis strain MG1363, since it is readily genetically manipulated and has been used for previous respiration studies. The fusion-harboring strain was grown in static and aerobic conditions, with or without 100 μM heme (Fig. 3A). Strong induction of ygf gene expression by heme was observed in respiration conditions and even under static conditions where respiration does not occur. These results confirm the transcriptome results. Furthermore, they show that the ygf operon is induced by heme itself and not by respiration.
FIG. 3.
(A) Induction of the ygf operon by heme. The expression of a chromosomal ygf lacLM fusion in strain MG1363 was monitored under static and aerobic growth without or with 100 μM heme. The β-galactosidase activity (bars, in Miller units, based on the cell OD600 adjusted to 1.0) was examined at different times during growth (line) and corresponds to an average of three independent experiments. A representative growth profile is presented; OD600 values in the three independent experiments differed by <40%. S, static (white bar and white triangle); S H100, static plus hemin (dark gray bar and dark gray triangle); A, aeration (light gray bar and light gray circle); A H100, aeration plus hemin (black bar and black circle). (B) Inactivation of ygf genes results in heme sensitivity. Stationary cultures of L. lactis strain MG1363 and ygfB (results not shown) and ygfC mutants were plated at 1/100 dilutions on M17G agar, and 20, 40, or 100 nmol of heme were pipetted directly onto plates; representative results with 40 nmol of heme are shown. A growth inhibition zone by heme (delimited by arrows) was substantially greater for the ygf mutant strains than for the parental strain and complemented mutants. The ring beyond the inhibition zone corresponds to stimulated growth due to respiration in the presence of heme.
L. lactis ygf genes are involved in heme homeostasis.
The strong induction of the ygfCBA operon led us to investigate possible roles for these genes in lactococci. YgfC was predicted to be a transcriptional regulator of the downstream genes (55). Interestingly, L. lactis genes yxeA and yxeB found elsewhere in the genome encode proteins with relatively high identity to YgfB and YgfA (38 and 59%, respectively). Both sets of proteins showed high identity to Staphylococcus aureus proteins HrtA and HrtB (45 to 46% to HrtA [ATP-binding] and 26 to 29% to HrtB [permease]; see Tables 2 and S2 in the supplemental material). These recently characterized S. aureus genes were found by transcriptomics to be induced by heme (15) and were implicated in heme efflux, since the hrtA mutants were heme sensitive (54). Since only the ygfCBA genes, and not yxeAB, were specifically induced by heme, we investigated whether this operon was involved in heme homeostasis.
ygfB and ygfC mutants were constructed in L. lactis strain MG1363 (genes llmg_0625 and llmg_0626, respectively) (57). Mutant strains were furthermore complemented with a copy of the ygfCBA genes (on plasmid pYgfCBA). Parental, mutant, and complemented strains were examined for heme sensitivity on solid medium in aerobic conditions (Fig. 3B and data not shown). Heme is inhibitory when present at high concentrations but stimulates growth via respiration at lower concentrations. Both these phenomena were visualized on the plates. Both ygfC and ygfB mutants were hypersensitive to heme compared to the parent strain. Heme sensitivity was fully relieved when mutant strains contained the pYgfCBA plasmid. These results lead us to suggest that, like the hrtAB genes in S. aureus, ygfBC genes are likely involved in heme homeostasis. Based on these results, we propose to rename ygfB as hrtA and ygfA as hrtB, in accordance with the names assigned for staphylococci. However, its regulation is likely to be quite different. In S. aureus, hrtAB expression is regulated by an adjacent two-component system. In L. lactis, YgfC is a candidate regulator for the operon. No YgfC homolog is present in S. aureus or in any closely related species: its function is currently under study in our laboratory.
Conclusions.
Respiration growth of L. lactis results in substantial changes in physiology, biomass, and survival. Genome-scale analysis comparing L. lactis expression in static, aerobic, and respiration conditions was used to better understand how each of these conditions affects the bacterium. The findings reported here are being used to further examine the genetic basis for respiration. One main finding is that aerobically grown cells are subject to aerobic stress, as seen by the differential expression of numerous stress response genes. This contrasts with a markedly lower stress response in respiration-grown cells. A confirmed explanation for this is that O2 is efficiently eliminated by respiration chain activity (25). Thus, respiration metabolism can provide the double advantage of being energetically favorable for lactococci and removing O2 for improved survival. We consider it likely that genes whose expression is more affected by respiration than by aeration are likely to be regulated by factors other than O2. NADH oxidases expressed in both of these conditions would lead to lower NADH levels, which evokes the possibility that the redox ratio influences both metabolism and gene expression.
We were initially surprised that genes required for respiration were expressed in the three tested conditions and that relatively few genes were specifically induced. This may indicate that a strategy of “readiness” is incorporated into the genetic program regarding lactococcal respiration, and this may be valuable in the case of rapidly changing environments. We are currently investigating the changes due to respiration at the posttranscriptional level.
The main transcriptional change due to respiration involved strong upregulation of the ygf operon. This operon is induced by heme, rather than by respiration itself. ygf is the first identified heme-inducible operon in lactococci. The role of ygf genes in heme homeostasis is suggested by heme sensitivity of a ygfB mutant and by a similar phenotype for S. aureus hrtAB mutants (54). A novel putative regulator, YgfC, is involved in heme homeostasis, specifically in L. lactis. It is notable that the basal level of heme tolerance of wild-type lactococci appears to be higher than that of staphylococci, i.e., >100 μM versus <10 μM, respectively (Fig. 3) (15, 54). The existence of heme-responsive genes raises questions concerning the natural environments that are home to lactococci. We suggest that dense ecosystems, including heme-producing bacteria, may provide a heme source to trigger L. lactis respiration.
Supplementary Material
Acknowledgments
We thank Maiken Lund Jensen and Karen Fuglede Appel at Chr. Hansen A/S for their excellent experimental work and also Anette Due Kongsbach, Jakob Pedersen, and Marianne Richelieu for the gas chromatography and high-pressure liquid chromatography analysis. We are grateful to Bénédicte Cesselin for expert advice and to Gilles Lamberet, Delphine Lechardeur, Annabelle Fernandez, and Aurélie Derré at the UBLO lab (France) and Eric Johansen (Chr. Hansen) for stimulating discussions and for sharing unpublished results.
Footnotes
Published ahead of print on 16 May 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Almiron, M., A. J. Link, D. Furlong, and R. Kolter. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 62646-2654. [DOI] [PubMed] [Google Scholar]
- 2.Arnau, J., F. Jorgensen, S. M. Madsen, A. Vrang, and H. Israelsen. 1998. Cloning of the Lactococcus lactis adhE gene, encoding a multifunctional alcohol dehydrogenase, by complementation of a fermentative mutant of Escherichia coli. J. Bacteriol. 1803049-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balandina, A., L. Claret, R. Hengge-Aronis, and J. Rouviere-Yaniv. 2001. The Escherichia coli histone-like protein HU regulates rpoS translation. Mol. Microbiol. 391069-1079. [DOI] [PubMed] [Google Scholar]
- 4.Bassit, N., C. Y. Boquien, D. Picque, and G. Corrieu. 1993. Effect of initial oxygen concentration on diacetyl and acetoin production by Lactococcus lactis subsp. lactis biovar diacetylactis. Appl. Environ. Microbiol. 591893-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blank, L. M., B. J. Koebmann, O. Michelsen, L. K. Nielsen, and P. R. Jensen. 2001. Hemin reconstitutes proton extrusion in an H+-ATPase-negative mutant of Lactococcus lactis. J. Bacteriol. 1836707-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooijmans, R. J., B. Poolman, G. K. Schuurman-Wolters, W. M. de Vos, and J. Hugenholtz. 2007. Generation of a membrane potential by Lactococcus lactis through aerobic electron transport. J. Bacteriol. 1895203-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casiano-Colon, A., and R. E. Marquis. 1988. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl. Environ. Microbiol. 541318-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Champomier Verges, M. C., M. Zuniga, F. Morel-Deville, G. Perez-Martinez, M. Zagorec, and S. D. Ehrlich. 1999. Relationships between arginine degradation, pH and survival in Lactobacillus sakei. FEMS Microbiol. Lett. 180297-304. [DOI] [PubMed] [Google Scholar]
- 10.Christensson, C., C. J. Pillidge, L. J. Ward, and P. W. O'Toole. 2001. Nucleotide sequence and characterization of the cell envelope proteinase plasmid in Lactococcus lactis subsp. cremoris HP. J. Appl. Microbiol. 91334-343. [DOI] [PubMed] [Google Scholar]
- 11.Crutz-Le Coq, A. M., B. Cesselin, J. Commissaire, and J. Anba. 2002. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148985-1001. [DOI] [PubMed] [Google Scholar]
- 12.Curic, M., M. de Richelieu, C. M. Henriksen, K. V. Jochumsen, J. Villadsen, and D. Nilsson. 1999. Glucose/citrate cometabolism in Lactococcus lactis subsp. lactis biovar diacetylactis with impaired alpha-acetolactate decarboxylase. Metab. Eng. 1291-298. [DOI] [PubMed] [Google Scholar]
- 13.de Vos, W. M., I. Boerrigter, R. J. van Rooyen, B. Reiche, and W. Hengstenberg. 1990. Characterization of the lactose-specific enzymes of the phosphotransferase system in Lactococcus lactis. J. Biol. Chem. 26522554-22560. [PubMed] [Google Scholar]
- 14.Duwat, P., S. Sourice, B. Cesselin, G. Lamberet, K. Vido, P. Gaudu, Y. Le Loir, F. Violet, P. Loubiere, and A. Gruss. 2001. Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. J. Bacteriol. 1834509-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman, D. B., D. L. Stauff, G. Pishchany, C. W. Whitwell, V. J. Torres, and E. P. Skaar. 2006. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamper, M., A. Zimmermann, and D. Haas. 1991. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J. Bacteriol. 1734742-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrigues, C., E. Johansen, M. B. Pedersen, H. Mollgaard, K. I. Sorensen, P. Gaudu, A. Gruss, and G. Lamberet. 2006. Getting high (OD) on heme. Nat. Rev. Microbiol. 4doi: 10.1038/nrmicro1403-c1. [DOI] [PubMed]
- 18.Garrigues, C., P. Loubiere, N. D. Lindley, and M. Cocaign-Bousquet. 1997. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J. Bacteriol. 1795282-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrigues, C., B. Stuer-Lauridsen, and E. Johansen. 2005. Characterisation of Bifidobacterium animalis subsp. lactis BB-12 and other probiotic bacteria using genomics, transcriptomics, and proteomics. Aus. J. Dairy Tech. 6084-92. [Google Scholar]
- 20.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 1541-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaudu, P., G. Lamberet, S. Poncet, and A. Gruss. 2003. CcpA regulation of aerobic and respiration growth in Lactococcus lactis. Mol. Microbiol. 50183-192. [DOI] [PubMed] [Google Scholar]
- 22.Gaudu, P., K. Vido, B. Cesselin, S. Kulakauskas, J. Tremblay, L. Rezaiki, G. Lamberet, S. Sourice, P. Duwat, and A. Gruss. 2002. Respiration capacity and consequences in Lactococcus lactis. Antonie van Leeuwenhoek 82263-269. [PubMed] [Google Scholar]
- 23.Gaudu, P., Y. Yamamoto, P. Jensen, K. Hammer, and A. Gruss. 2006. Genetics of lactococci, p. 356-368. In V. Fischetti (ed.), gram-positive pathogens, 2nd ed. American Society for Microbiology, Washington DC.
- 24.Godon, J. J., K. Jury, C. A. Shearman, and M. J. Gasson. 1994. The Lactococcus lactis sex-factor aggregation gene cluA. Mol. Microbiol. 12655-663. [DOI] [PubMed] [Google Scholar]
- 25.Goldman, B. S., K. K. Gabbert, and R. G. Kranz. 1996. The temperature-sensitive growth and survival phenotypes of Escherichia coli cydDC and cydAB strains are due to deficiencies in cytochrome bd and are corrected by exogenous catalase and reducing agents. J. Bacteriol. 1786348-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruening, P., M. Fulde, P. Valentin-Weigand, and R. Goethe. 2006. Structure, regulation, and putative function of the arginine deiminase system of Streptococcus suis. J. Bacteriol. 188361-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Javelle, A., G. Thomas, A. M. Marini, R. Kramer, and M. Merrick. 2005. In vivo functional characterization of the Escherichia coli ammonium channel AmtB: evidence for metabolic coupling of AmtB to glutamine synthetase. Biochem. J. 390215-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaneko, T., M. Takahashi, and H. Suzuki. 1990. Acetoin fermentation by citrate-positive Lactococcus lactis subsp. lactis 3022 grown aerobically in the presence of hemin or Cu. Appl. Environ. Microbiol. 562644-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kavanagh, E. P., and S. Hill. 1993. Oxygen inhibition of nitrogenase activity in Klebsiella pneumoniae. J. Gen. Microbiol. 139(Pt. 61307-1314. [DOI] [PubMed] [Google Scholar]
- 30.Koebmann, B. J., H. W. Andersen, C. Solem, and P. R. Jensen. 2002. Experimental determination of control of glycolysis in Lactococcus lactis. Antonie van Leeuwenhoek 82237-248. [PubMed] [Google Scholar]
- 31.Kvint, K., L. Nachin, A. Diez, and T. Nystrom. 2003. The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6140-145. [DOI] [PubMed] [Google Scholar]
- 32.Leloup, L., S. D. Ehrlich, M. Zagorec, and F. Morel-Deville. 1997. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl. Environ. Microbiol. 632117-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez de Felipe, F., M. Kleerebezem, W. M. de Vos, and J. Hugenholtz. 1998. Cofactor engineering: a novel approach to metabolic engineering in Lactococcus lactis by controlled expression of NADH oxidase. J. Bacteriol. 1803804-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 10315611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Moskovitz, J., M. A. Rahman, J. Strassman, S. O. Yancey, S. R. Kushner, N. Brot, and H. Weissbach. 1995. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J. Bacteriol. 177502-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicoloff, H., A. Elagoz, F. Arsene-Ploetze, B. Kammerer, J. Martinussen, and F. Bringel. 2005. Repression of the pyr operon in Lactobacillus plantarum prevents its ability to grow at low carbon dioxide levels. J. Bacteriol. 1872093-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen, H. B., R. Wernersson, and S. Knudsen. 2003. Design of oligonucleotides for microarrays and perspectives for design of multi-transcriptome arrays. Nucleic Acids Res. 313491-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 1835803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Connor, D. 2007. The world market for cheese: 1995-2004. Int. J. Dairy Technol. 6066-67. [Google Scholar]
- 42.Paget, M. S., and M. J. Buttner. 2003. Thiol-based regulatory switches. Annu. Rev. Genet. 3791-121. [DOI] [PubMed] [Google Scholar]
- 43.Pedersen, M. B., S. L. Iversen, K. I. Sorensen, and E. Johansen. 2005. The long and winding road from the research laboratory to industrial applications of lactic acid bacteria. FEMS Microbiol. Rev. 29611-624. [DOI] [PubMed] [Google Scholar]
- 44.Pittman, M. S., H. C. Robinson, and R. K. Poole. 2005. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J. Biol. Chem. 28032254-32261. [DOI] [PubMed] [Google Scholar]
- 45.Poole, L. B. 2005. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch. Biochem. Biophys. 433240-254. [DOI] [PubMed] [Google Scholar]
- 46.Raya, R., J. Bardowski, P. S. Andersen, S. D. Ehrlich, and A. Chopin. 1998. Multiple transcriptional control of the Lactococcus lactis trp operon. J. Bacteriol. 1803174-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raynaud, S., R. Perrin, M. Cocaign-Bousquet, and P. Loubiere. 2005. Metabolic and transcriptomic adaptation of Lactococcus lactis subsp. lactis biovar diacetylactis in response to autoacidification and temperature downshift in skim milk. Appl. Environ. Microbiol. 718016-8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rezaiki, L., B. Cesselin, Y. Yamamoto, K. Vido, E. van West, P. Gaudu, and A. Gruss. 2004. Respiration metabolism reduces oxidative and acid stress to improve long-term survival of Lactococcus lactis. Mol. Microbiol. 531331-1342. [DOI] [PubMed] [Google Scholar]
- 49.Rezaiki, L., G. Lamberet, A. Derre, A. Gruss, and P. Gaudu. 2008. Lactococcus lactis produces short-chain quinones that cross-feed group B Streptococcus to activate respiration growth. Mol. Microbiol. 67947-957. [DOI] [PubMed] [Google Scholar]
- 50.Richelieu, M., U. Houlberg, and J. Nielsen. 1997. Determination of alpha-acetolactic acid and volatile compounds by headspace gas chromatography. J. Dairy Sci. 801918-1925. [Google Scholar]
- 51.Sanders, J. W., K. J. Leenhouts, A. J. Haandrikman, G. Venema, and J. Kok. 1995. Stress response in Lactococcus lactis: cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J. Bacteriol. 1775254-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sijpesteijn, A. K. 1970. Induction of cytochrome formation and stimulation of oxidative dissimilation by hemin in Streptococcus lactis and Leuconostoc mesenteroides. Antonie van Leeuwenhoek 36335-348. [DOI] [PubMed] [Google Scholar]
- 53.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2188-194. [DOI] [PubMed] [Google Scholar]
- 54.Torres, V. J., D. L. Stauff, G. Pishchany, J. S. Bezbradica, L. E. Gordy, J. Iturregui, K. L. Anderson, P. M. Dunman, S. Joyce, and E. P. Skaar. 2007. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1109-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vido, K., D. Le Bars, M. Y. Mistou, P. Anglade, A. Gruss, and P. Gaudu. 2004. Proteome analyses of heme-dependent respiration in Lactococcus lactis: involvement of the proteolytic system. J. Bacteriol. 1861648-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Volker, U., K. K. Andersen, H. Antelmann, K. M. Devine, and M. Hecker. 1998. One of two osmC homologs in Bacillus subtilis is part of the sigmaB-dependent general stress regulon. J. Bacteriol. 1804212-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wegmann, U., M. O'Connell-Motherway, A. Zomer, G. Buist, C. Shearman, C. Canchaya, M. Ventura, A. Goesmann, M. J. Gasson, O. P. Kuipers, D. van Sinderen, and J. Kok. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 1893256-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamamoto, Y., V. Pargade, G. Lamberet, P. Gaudu, F. Thomas, J. Texereau, A. Gruss, P. Trieu-Cuot, and C. Poyart. 2006. The group B Streptococcus NADH oxidase Nox-2 is involved in fatty acid biosynthesis during aerobic growth and contributes to virulence. Mol. Microbiol. 62772-785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.