Abstract
Microbes tailor macromolecules and metabolism to overcome specific environmental challenges. Acetic acid bacteria perform the aerobic oxidation of ethanol to acetic acid and are generally resistant to high levels of these two membrane-permeable poisons. The citric acid cycle (CAC) is linked to acetic acid resistance in Acetobacter aceti by several observations, among them the oxidation of acetate to CO2 by highly resistant acetic acid bacteria and the previously unexplained role of A. aceti citrate synthase (AarA) in acetic acid resistance at a low pH. Here we assign specific biochemical roles to the other components of the A. aceti strain 1023 aarABC region. AarC is succinyl-coenzyme A (CoA):acetate CoA-transferase, which replaces succinyl-CoA synthetase in a variant CAC. This new bypass appears to reduce metabolic demand for free CoA, reliance upon nucleotide pools, and the likely effect of variable cytoplasmic pH upon CAC flux. The putative aarB gene is reassigned to SixA, a known activator of CAC flux. Carbon overflow pathways are triggered in many bacteria during metabolic limitation, which typically leads to the production and diffusive loss of acetate. Since acetate overflow is not feasible for A. aceti, a CO2 loss strategy that allows acetic acid removal without substrate-level (de)phosphorylation may instead be employed. All three aar genes, therefore, support flux through a complete but unorthodox CAC that is needed to lower cytoplasmic acetate levels.
Acetic acid bacteria (AAB) are gram-negative, acidophilic α-proteobacteria that oxidize ethanol to acetic acid using membrane-bound alcohol dehydrogenase (ADH) and aldehyde dehydrogenase enzymes. AAB are remarkably resistant to the membrane-permeable toxic compounds ethanol and acetic acid at a low pH. Members of the AAB genus Acetobacter were historically differentiated from those of the genus Gluconobacter by a preference for ethanol and the ability to “overoxidize” acetate to CO2, usually when ethanol is unavailable (5). Acetate oxidation implicates the oxidative decarboxylations performed by citric acid cycle (CAC) dehydrogenases and a complete CAC.
The molecular mechanisms of acetic acid resistance in Acetobacter aceti include adaptation of the cytoplasmic components (9, 14) to internal acidification (32), acetic acid efflux via the AatA acetic acid:proton antiporter (31, 39), and production of acid-inducible proteins identified by proteomic screens, many with undefined biochemical roles (28, 52).
A. aceti strain 1023, an especially acid-resistant and comparatively thermotolerant vinegar factory isolate (41, 42), requires acetic acid resistance genes aarABC for growth on >50 mM acetic acid at a low pH (16). AarA is a NADH-insensitive hexameric form of citrate synthase (15). AarB is predicted to be a basic protein of 154 amino acids with no homologues or known function (16). AarC is required for acetate oxidation and resembles several acyl-coenzyme A (CoA):carboxylate CoA transferases but is not acetyl-CoA synthetase (17).
The apparent requirement of the A. aceti CAC for acetic acid resistance may indicate a direct role in depleting cytoplasmic acetate by way of acetyl-CoA oxidation to CO2 or a general contribution to energy production. A. aceti requires vigorous oxygenation at high acetate levels and low pHs (37), which is consistent with either CAC role. However, a draft genome sequence of A. aceti strain 1023 (T. J. Kappock, S. W. Clifton, and R. K. Wilson, unpublished data) lacks malate dehydrogenase (Mdh) and succinyl-CoA synthetase (SCS) genes.
The goal of this study was to learn if the A. aceti CAC is interrupted. We found that A. aceti contains a complete but modified CAC, enabling direct acetate incorporation by an internal shunt. Mdh and SCS are functionally replaced by malate:quinone oxidoreductase (Mqo) and succinyl-CoA:acetate CoA-transferase (SCACT), respectively. SCACT is encoded by aarC. This biochemical function explains the genetic evidence for the essential roles of aarC in acetic acid resistance, assimilation, and oxidation (16, 17).
MATERIALS AND METHODS
Reagents and general analytical methods.
Chemicals were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher (Houston, TX). Dethiaacetyl-CoA was synthesized as described previously (15). Oligodeoxynucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA) and used without further purification. A. aceti citrate synthase with a hexahistidine affinity tag (CSH6Aa) was purified as described previously (15). Restriction enzymes, DNA-modifying enzymes, and DNA size standards were purchased from New England Biolabs (Beverly, MA). Absorbance measurements were recorded on a Cary Bio 100 UV-visible spectrophotometer (Varian, Palo Alto, CA) or a diode-array spectrophotometer (Agilent Technologies, Santa Clara, CA). Analytical gel filtration, electrospray ionization-mass spectrometry (ESI-MS), and protein quantitation by the method of Bradford were performed as described previously (14).
Bacterial strains, media, and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. A. aceti strains were propagated in Difco yeast extract-peptone-dextrose medium (Becton Dickinson, Franklin Lakes, NJ) supplemented with 2.5% ethanol at 30°C as described previously (14). DNA was isolated and manipulated using standard protocols (2). PCRs were performed using A. aceti genomic DNA (isolated with DNAzol; Molecular Research Center, Cincinnati, OH), appropriate oligodeoxynucleotide primers, and Vent DNA polymerase (New England Biolabs). All clones contained the expected DNA sequences.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source |
|---|---|---|
| Strains | ||
| A. aceti 1023 | Acid-tolerant factory strain | Etsuko Entanib |
| A. aceti 2002 | A. aceti type strain, DSM 2002 | DSMZ |
| E. coli C41 | F′ ompT hsdSB(rB− mB−) gal dcm (DE3) with an uncharacterized mutation | Avidisc |
| Plasmids | ||
| pET23a | T7 promoter expression vector, Apr | Novagen |
| pET23d | T7 promoter expression vector, Apr | Novagen |
| pJK286 | uctB ligated into NdeI and HindIII sites in pET23a | This study |
| pJK357 | aarC ligated into NdeI and XhoI sites in pET23a | This study |
| pJK358 | uctD ligated into NdeI and EcoRI sites in pET23a | This study |
| pJK359 | uctA ligated into NdeI and EcoRI sites in pET23a | This study |
| pJK360 | uctC ligated into NcoI and EcoRI sites in pET23dd | This study |
| pJK385 | Derivative of pJK357 that produces AarCH6e | This study |
HPLC-coupled SCACT assays.
SCACT assays were performed at 25°C in 50 mM potassium phosphate (pH 8.0), 100 mM KCl, the indicated substrates, and AarC with a C-terminal hexahistidine-containing fusion peptide (AarCH6; 5 to 50 ng), which was used to initiate the reaction. Forward-direction reaction mixtures (LCF assays) contained 350 mM potassium acetate, 0.2 mM succinyl-CoA, or varied amounts of one substrate. Reverse-direction reaction mixtures (LCR assays) contained 20 mM succinic acid, 1 mM acetyl-CoA, or varied amounts of one substrate. After 5 min, an aliquot (0.1 ml) of the reaction mixture was transferred into 6.25% trichloroacetic acid (0.4 ml), vortexed briefly, and centrifuged at 16,100 × g for 3 min. The soluble portion was then transferred to an autosampler vial and analyzed by high-performance liquid chromatography (HPLC) using the method described in the supplemental material. No-enzyme controls were processed in the same manner.
Peak areas were referenced to an acetyl-CoA calibration curve (ɛ260 nm = 16.4 mM−1 cm−1) (12), and velocities were determined from the product CoA thioester peak area. Under the fixed-time assay conditions used, the rate of product formation was linear in each direction for at least 10 min. Velocities were averages of three or four determinations unless otherwise noted. Kinetic constants were determined using Prism (GraphPad) to fit equations for Michaelis-Menten kinetics, competitive inhibition, or substrate inhibition (8). A sum-of-squares F test was used to discriminate between alternative kinetic models.
Inactivation experiments.
AarCH6 (0.5 μg) was incubated for 10 min at 25°C in a 0.5-ml mixture containing 50 mM potassium phosphate (pH 8.0), 100 mM KCl, and 100 μM acetyl-CoA. Sodium borohydride was added to 10 mM, followed by incubation for an additional 10 min. An aliquot (5 μl) of the enzyme solution was then immediately added to an otherwise complete LCR assay to measure the residual enzymatic activity. The control reactions lacked sodium borohydride, acetyl-CoA, or both. The remainder of the treated protein solution was flash frozen and analyzed later by ESI-MS.
Nucleotide sequence accession numbers.
The sequences reported in this paper have the following GenBank accession numbers: uctA, DQ668371; uctB-oxc-uctC-duf1275, DQ668372; uctD, DQ668373; aarA-sixA-tyrA-orf1-aarC, DQ631551; and mqo, DQ674275.
RESULTS
Inferred metabolic fates of acetate.
A draft of the 3-Mb A. aceti strain 1023 genome sequence, analyzed as described in the supplemental material, contains all genes needed for six of the eight enzymes of the canonical CAC. No genes for SCS, Mdh, or glyoxylate shunt enzymes were identified. Membrane-bound Mqo, a replacement for Mdh in diverse bacteria (18, 36), is present. The enzymes needed to convert acetate to acetyl-CoA, allowing acetate removal by the CAC, are also present: acetyl-CoA synthetase (Acs) or acetate kinase (AckA) and phosphotransacetylase (Pta).
The genome sequence contains five CoA-transferases, four with unassigned functions (uctA to uctD) and aarC. The new aarC sequence encodes a protein of 505 amino acids with a predicted isoelectric point of 6.16. The originally reported aarC gene (GenBank accession number D13291) has 496 amino acids and a different sequence in residues 1 to 40 (new AarC residues 1 to 54) (16). The only coding region sequence difference is an insertion of a G after nucleotide 178 in the original sequence report (G210 in the revised sequence).
Gene reannotation in the aarB region.
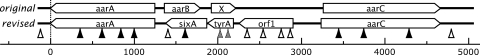
Conditionally essential acetic acid resistance genes were identified in mutant A. aceti 1023 strains that could not survive exposure to 50 mM acetic acid at a low pH but that were able to grow at an equivalent pH generated by the membrane-impermeant mineral acid HCl (16). Insertional inactivation experiments were used to define the aarABC genes and an apparently essential gene, X (Fig. 1) (4).
FIG. 1.
Revised annotation of the aar region in A. aceti strain 1023. The ORFs from the original gene assignment (top) were assigned using a kanamycin cassette insertional inactivation at the restriction sites indicated with vertical triangles (16). Insertion points were characterized as resistant to 50 mM acetic acid (open triangles), sensitive (filled triangles), or apparently essential (hatched triangles). The original assigned functions were as follows: aarA, citrate synthase; aarB, unknown; X, unknown; and aarC, possible CoA-transferase or -hydrolase. As of this writing, neither aarB nor X has significant similarity to other inferred protein sequences. The new assigned functions are as follows: orf1, putative metallophosphoesterase; tyrA, chorismate mutase; and sixA, a phosphoprotein phosphatase that affects the ArcA/ArcB two-component regulator. The revised assignments include a single-base insertion in the aarC region that lengthens the ORF slightly. An insertion at the NsiI site, which does not disrupt acetic acid resistance, would disrupt the aarB gene product after Ala19 or would remove or alter seven or fewer C-terminal amino acids in SixA. The scale bar is in units of bp.
New gene annotations predicted three open reading frames (ORFs) on the opposite strand: orf1, encoding a putative metallophosphoesterase; tyrA, encoding chorismate mutase; and sixA, encoding a transcriptional regulator involved in a phosphohistidine relay system. Gene synteny in the four available AAB genomes supports the new assignments (19, 45; Kappock et al., unpublished; http://genome.jgi-psf.org/finished_microbes/acicr/acicr.home.html). AAB have a colinear orf1-sixA-gltA arrangement (gltA, citrate synthase, is the equivalent of A. aceti aarA), except in Acidiphilium cryptum, which has the putative phosphatase orf1 located elsewhere. In A. aceti and Gluconobacter oxydans, tyrA is inserted between orf1 and sixA, while Granulibacter bethesdensis has two genes inserted between sixA and gltA. In the revised assignment, aarC is divergently transcribed from all other genes in the aar gene region. Transcriptional analyses of the aar genes are in progress.
Enzyme activity assays.
As anticipated from the genome sequence data, six of the eight canonical CAC enzyme activities were detected in A. aceti cell lysates (Table 2). No activity was detected for the glyoxylate shunt enzymes isocitrate lyase and malate synthase (data not shown), SCS, or Mdh, but SCACT and Mqo activities were found. SCACT activity was initially detected as the succinate-dependent decomposition of acetyl-CoA, using a variant of the VisR assay that is described in the supplemental material. Succinate was the only one of 15 carboxylic acids that supported activity.
TABLE 2.
Enzyme activities in A. aceti strains
| Step no. | Enzyme | Activity (nmol product formed/min/mg protein) in A. aceti straina:
|
Assay directionb | pH | |
|---|---|---|---|---|---|
| 2002 | 1023 | ||||
| 1 | Citrate synthase | 1,050 ± 75 | 1,200 ± 60 | F | 8.0 |
| 2 | Aconitase | 5,460 ± 4 | 2,160 ± 150 | H | 8.0 |
| 3,170 ± 230c | |||||
| 3 | Isocitrate dehydrogenase | 530 ± 80 (NAD+) | 1,280 ± 12 (NAD+) | F | 8.0 |
| 520 ± 45 (NADP+) | 590 ± 20 (NADP+) | ||||
| 4 | α-Ketoglutarate dehydrogenase | 98 ± 2 | 28 ± 4 | F | 7.0 |
| 5 | Succinyl-CoA synthetase | NDd | ND | R | 7.2 |
| 5′ | Succinyl-CoA:acetate CoA-transferase | 114 ± 2 | 170 ± 4 | Fe | 8.0 |
| 53 ± 5 | 83 ± 18 | Rf | |||
| 6 | Succinate dehydrogenase | 21 ± 2 | 22 ± 3 | F | 8.0 |
| 7 | Fumarase | 720 ± 115 | 570 ± 110 | R | 7.0 |
| 8 | Malate dehydrogenase | ND | ND | R | 8.0 |
| 8′ | Malate:quinone oxidoreductase | 69 ± 3 | 46 ± 2 | F | 7.5 |
Experimental procedures are described in the supplemental material. Values given are means ± SD of experiments performed in triplicate.
F, biosynthetic; R, nonbiosynthetic; H, citrate→aconitate (half-reaction).
Determined using slow-spin lysate.
ND, activity not detected.
Determined using the VisF assay.
Determined using the VisR assay. In preliminary experiments, 15 carboxylic acids were examined as potential substrates in a variant of the VisR assay (described in the supplemental material). Only succinate supported activity.
Previously published “single-pot” CoA-transferase assays using citrate synthase to quantitate acetyl-CoA were unsuitable for the kinetic characterization. The discontinuous VisF/VisR methods described in the supplemental material were developed for use with complex cell lysates. HPLC-based LCF and LCR assays were used to quantitate individual (acyl-)CoAs, allowing discrimination between acyl-CoA hydrolase and transferase activities. Reaction-quenching conditions that minimized but did not eliminate acyl-CoA decomposition over a timescale of hours were identified. Replicate experiments were performed such that the mean velocity at each concentration was determined with the same average delay. Velocity corrections based on the observed rates of acyl-CoA decomposition would increase the reported kcat values by less than 16%.
Candidate SCACT gene expression and activity screening.
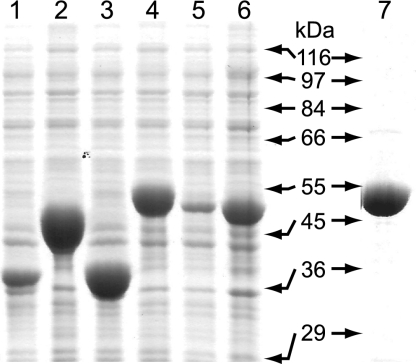
Candidate SCACT genes uctA to uctD and aarC were overexpressed as soluble proteins in Escherichia coli (Fig. 2). Only cells producing UctD or AarC had any activity above that of a vector-only control (VisR assay). The specific activity of SCACT was much higher in the lysate of cells expressing aarC, even though AarC was the least soluble of the five CoA-transferases. Thus, SCACT appears to be produced solely from the aarC gene. We plan to examine aarC-deficient strains to confirm that A. aceti contains only one SCACT gene.
FIG. 2.
Expression of candidate SCACT genes in E. coli strain C41(DE3). Left lanes, cleared cell lysates (centrifugation at 16,000 × g for 10 min) for strains producing an A. aceti CoA-transferase were assayed for the ability to consume acetyl-CoA (VisR assay) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The relative activity of each protein in the same culture volume is expressed as a percentage of the AarC activity (a vector-only control lysate has 0.008% relative activity): UctA (lane 1, 41.5 kDa expected; 0.007%), UctB (lane 2, 47.6 kDa expected; 0.01%), UctC (lane 3, 41.8 kDa expected; 0.01%), UctD (lane 4, 55.5 kDa expected; 0.3%), and AarC (lane 5, 54.8 kDa expected; 100%). Lane 6 shows a comparable amount of total cell protein for cells producing AarC, showing the limited solubility of this protein. A parallel experiment with cells producing AarCH6 (not shown) showed 74% relative activity but also a lower fraction of soluble protein than AarC-producing cells. Right lane, purified AarCH6 (lane 7; 5 μg protein). Size standard positions are indicated.
AarC with a C-terminal hexahistidine-containing fusion peptide (AarCH6) was produced abundantly at 15°C but appeared to be slightly less soluble than the untagged protein (data not shown). The clarified lysates of E. coli cells producing AarCH6 had nearly the same activity as those producing AarC, indicating that the C-terminal fusion peptide does not significantly inhibit enzymatic activity.
Purification and properties of AarCH6.
AarCH6 was purified using an ammonium sulfate fractionation procedure, previously developed for untagged AarC, and an immobilized-metal affinity step that are described in the supplemental material. An 8-liter pJK385/C41(DE3) culture yielded 9.5 mg of purified AarCH6 (Fig. 2).
ESI-MS analysis indicated AarCH6 is a 513-residue protein lacking Met1 (55,847 ± 2 Da observed; 55,847.2 Da expected). Analytical gel filtration gave a single peak with a molecular mass of 160 kDa, consistent with a trimeric or elongated dimeric solution state.
AarCH6 fluorescence emission showed a maximum at 343 nm, which was quenched 8% by the addition of dethiaacetyl-CoA, a nonhydrolyzable analogue of acetyl-CoA that has a methylene group replacing the sulfur atom (30). Fluorescence titrations, performed as described in the supplemental material, were used to determine an apparent Kd (dissociation constant) (Table 3). No fluorescence changes were noted upon the addition of 10 mM succinate to a solution containing 1 μM AarCH6 and 10 μM dethiaacetyl-CoA.
TABLE 3.
Kinetic constants for AarCH6a
| Substrate | Parameter | Value |
|---|---|---|
| Acetateb | kcat (s−1) | 280 ± 37 |
| Km (mM) | 70 ± 20 | |
| kcat/Km (M−1 s−1) | (4 ± 1) × 103 | |
| Ki, acetate (mM) | 1,600 ± 780 | |
| Succinyl-CoAb | kcat (s−1) | 201 ± 8 |
| Km (μM) | 22.1 ± 3.6 | |
| kcat/Km (M−1 s−1) | (9 ± 2) × 106 | |
| Succinatec | kcat (s−1) | 70.9 ± 1.9 |
| Km (mM) | 0.90 ± 0.10 | |
| kcat/Km (M−1 s−1) | (7.9 ± 0.9) × 104 | |
| Acetyl-CoAc | kcat (s−1) | 75.2 ± 1.5 |
| Km (μM) | 22.3 ± 2.0 | |
| kcat/Km (M−1 s−1) | (3.4 ± 0.3) × 106 | |
| Ki, CoA (μM)d | 15.9 ± 2.1 | |
| SCACT reaction | Keqe | 0.137 ± 0.053 |
| Acetoacetatef | kcat (s−1) | 36.5 ± 2.6 |
| Km (mM) | 130 ± 14 | |
| kcat/Km (M−1 s−1) | (2.8 ± 0.4) × 102 | |
| Dethiaacetyl-CoA | Kd (μM)g | 0.75 ± 0.18 |
| Ki (μM)h | 16.6 ± 3.0 |
Determined at 25°C and pH 8.0. Turnover numbers assume one active site per 55.8-kDa subunit, such that 1 s−1 = 1.07 units/mg.
LCF assay. Determined using a fixed concentration of either succinyl-CoA (0.2 mM) or acetate (350 mM).
LCR assay. Determined using a fixed concentration of either acetyl-CoA (1 mM) or succinate (20 mM).
Modified LCR assay. Determined using a variable concentration of acetyl-CoA (5 to 1,200 μM) and fixed concentrations of succinate (20 mM) and CoA (unadjusted or adjusted to a final concentration of 40, 70, or 100 μM). Data were fit to a competitive inhibition model.
Determined using kcat/Km values as (succinyl-CoA × acetate)/(acetyl-CoA × succinate) (7).
Modified LCF assay. Determined using a fixed concentration of succinyl-CoA (0.2 mM) and a variable concentration of lithium acetoacetate.
Determined by fluorescence titration at 20°C and pH 8.0.
Modified LCR assay. Determined using a variable concentration of acetyl-CoA (10 to 300 μM) and fixed concentrations of succinate (20 mM) and dethiaacetyl-CoA (0, 5, 25, and 50 μM). Data were fit to a competitive inhibition model.
Mechanism of AarC.
AarC is predicted to be a class I CoA-transferase from active site sequence alignments that identify Glu435 as a key active site residue (29). Class I CoA-transferases first bind the acyl-CoA substrate, forming a carboxylate product and an enzyme-CoA thioester intermediate (48, 50), which accounts for the observed ping-pong kinetics (22) and susceptibility to borohydride inactivation (23, 25, 50). Sodium borohydride treatment of AarCH6 in the presence of acetyl-CoA caused near-complete loss of enzyme activity (0.7% relative activity), consistent with the reduction of the glutamyl-CoA thioester intermediate to 5-hydroxynorvaline. ESI-MS analysis of the borohydride-treated enzyme complex showed the expected −14-Da change (55,832 ± 2 Da observed; 55,833.2 expected). AarCH6 treated with borohydride in the absence of an acyl-CoA substrate showed partial inactivation (71% activity) relative to a control lacking both acetyl-CoA and sodium borohydride (100% activity; 74 s−1). Partial inactivation may indicate that some of the enzyme is isolated as an AarCH6-CoA adduct.
Steady-state experiments with duplicate LCR assays showed nonintersecting lines in double-reciprocal plots, consistent with ping-pong kinetics and a modified-enzyme mechanism (see Figure S2 in the supplemental material).
Kinetic characterization of AarCH6.
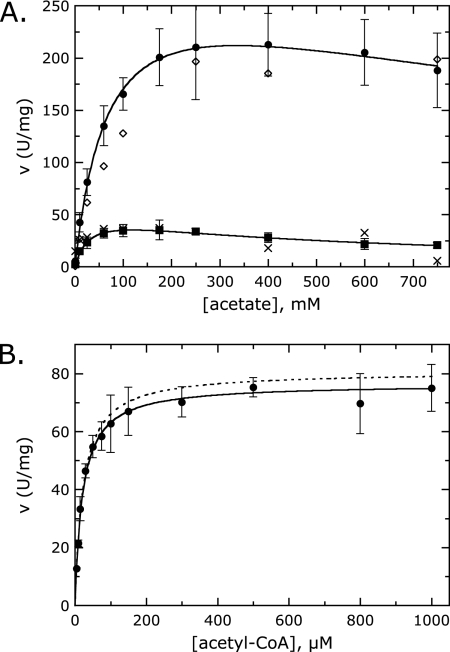
The determination of accurate kinetic parameters for AarCH6 was complicated by the apparent acetate substrate inhibition (Fig. 3A) and the presence of CoA, a common contaminant in commercial acyl-CoAs (∼5% in acetyl-CoA, ∼11% in succinyl-CoA) that hinders substrate saturation. Accurate acid substrate kinetic constants for class I CoA-transferases require the saturation of the enzyme with the acyl-CoA substrate. LCF/LCR assays allowed the simultaneous determination of CoA levels and enzyme activity. Similar kinetic parameters were obtained using both LCF/LCR and VisF/VisR assays (Fig. 3A).
FIG. 3.
Kinetic characterization of AarCH6. (A) Acetate saturation data at pH 8.0 (solid circles, LCF assay; open diamonds, VisF assay) and pH 5.0 (solid squares, LCF assay; crosses, VisF assay). The solid lines are fits to each LCF data set, assuming substrate inhibition. The concentration of succinyl-CoA was 0.2 mM. Fitted parameters are given in the text and Table 3. (B) Acetyl-CoA saturation data at pH 8.0 obtained with the LCR assay (solid circles) at 20 mM succinate. The solid line is a fit to the data accounting for competitive inhibition due to CoA (determined in assay mixtures spiked with CoA). The dotted line is a simulation using the fitted parameters at a CoA concentration of 0.
Kinetic analysis was performed for each substrate at a single, saturating, but minimally inhibitory concentration of the second substrate. LCR assays spiked with CoA showed that CoA is an effective competitive inhibitor versus acetyl-CoA. A plot of velocity versus the concentration of acetyl-CoA is only satisfactorily fit if inhibition due to residual CoA is taken into account (Fig. 3B). The increased scatter in velocities determined with succinyl-CoA (LCF assay) gave fits that were not improved by correcting for CoA inhibition. If CoA is an equally potent competitor for each acyl-CoA substrate, the values given underestimate the succinyl-CoA Km and kcat values by ∼15% but have a negligible effect on Keq (equilibrium constant). The uninhibited kinetic constants obtained by individual substrate analysis (Table 3) are in reasonable agreement with the values obtained by the preliminary double-reciprocal analysis (data not shown).
Acetoacetate is a cosubstrate for several CoA-transferases that convert succinyl-CoA to succinate (10, 24). LCF assays containing acetoacetate instead of acetate showed the formation of acetoacetyl-CoA, identified by comparison to the HPLC retention time of an authentic standard. The kcat/Km for acetoacetate was 7% of that for acetate under the same conditions (Table 3).
pH dependence of acetate kinetic parameters.
Acetate is anticipated to be the true substrate for SCACT. Its mole fraction should decrease as the pH drops below the pKa of acetic acid, 4.76. Kinetic parameters were determined with LCF assays at pH 8.0 (Table 3), pH 6.0 (kcat = 147 s−1; Km = 51 ± 11 mM; Ki = 1,100 ± 310 mM), and pH 5.0 (kcat = 49 s−1; Km = 29 ± 8 mM; Ki = 500 ± 130 mM). The increased substrate inhibition at a lower pH may be due to the increased fraction of acetic acid (Fig. 3A). LCF and VisF velocities were comparable at all pH values tested.
DISCUSSION
Genetic screens have identified two categories of Acetobacter genes affecting acetic acid resistance: (i) those that improve the acetic acid resistance of A. aceti strains when introduced or overexpressed, including the aarABC gene cluster (16) and the aconitase gene (38), and (ii) those that improve the acetic acid resistance of E. coli, including the RecG (51) and AatA (39) genes. The current study was designed to assess the potential role of the CAC and related pathways in acetic acid removal.
The A. aceti CAC converts acetate to CO2, a characteristic capability of Acetobacter strains. A prerequisite is the efficient conversion of acetate to acetyl-CoA, which can be performed by AckA/Pta or Acs. Pta and Acs activities increase in A. aceti during acetate oxidation (49). Sustained acetyl-CoA oxidation and acetate assimilation require a complete CAC. However, SCS was not found in a draft A. aceti genome sequence, and no activity was present in cell lysates.
When grown in the presence of glucose, A. aceti is able to assimilate carbon from acetate (39) or ethanol (46) by way of acetate. Isocitrate lyase and malate synthase activities have been reported in A. aceti cells grown on acetate as the sole carbon source (53), which would be consistent with acetate assimilation by the glyoxylate shunt. However, the genes for glyoxylate shunt enzymes have not been found in any AAB genome (19, 45). Thus, A. aceti must assimilate acetate by a different route. Alternative pathways are present in several other α-proteobacteria that lack isocitrate lyase (1).
Our findings show that A. aceti contains a complete but unorthodox CAC in which the acetic acid resistance protein AarC (SCACT) converts succinyl-CoA and acetate to succinate and acetyl-CoA. This bypass facilitates the metabolic activation and oxidation of acetate and constitutes part of a conditionally essential detoxification pathway that is required for survival at high acetate concentrations at a low pH but is not required for growth at low acetate concentrations, even at the same low pH.
AarC homologues.
SCACT has not to our knowledge been identified in another aerobic α-proteobacterium. Of the four AAB with currently available genome sequences, only G. oxydans has an incomplete CAC (20, 45). The complete genome sequences of the AAB G. bethesdensis CGDNIH1 (19) and A. cryptum contain SCS genes. A. cryptum also contains a close match to AarC (see Table S1 in the supplemental material). Genes resembling aarC are found in numerous proteobacteria, but sequences uniquely associated with SCACT activity have not been identified.
Anaerobic ATP synthesis in trichomonad hydrogenosomes and the distinctive mitochondria of trypanosomes and some helminths require both SCACT and SCS, which convert acetyl-CoA, ADP, and Pi to acetate, CoA, and ATP in the presence of succinate (55, 56). Hydrogenosomal SCACT from Trichomonas vaginalis (55) closely resembles AarC, but the sequence of mitochondrial SCACT from Trypanosoma brucei (47) is similar only in the active site region, (S/T)E(Q/N)GL.
SCACT activity is found in Aspergillus nidulans (6). The recently identified enzyme responsible for this activity (CoaT) has equal activity with acetate, propionate, and acetoacetate in the presence of succinyl-CoA (13). A. nidulans CoaT and similar inferred proteins in six related fungi, all of which contain putative mitochondrial targeting sequences, closely resemble AarC (see Table S1 in the supplemental material) and may function as mitochondrial SCACT enzymes. SCACT is one of a small number of enzyme activities found in both hydrogenosomes and some mitochondria (21). Each organelle is thought to be derived from the α-proteobacteria, a group that includes the AAB. The use of SCACT for a different metabolic purpose may explain the distinctive distribution of this gene.
Comparison with other CAC bypasses.
Helicobacter pylori lacks several typical CAC components (27, 44), including Mdh, which is replaced by Mqo, and SCS, which is replaced by succinyl-CoA:acetoacetate CoA-transferase (10). AarC is also able to support succinyl-CoA:acetoacetate CoA-transferase activity, albeit with a substantially lower kcat/Km than that of the SCACT reaction (Table 3). Given the likelihood that the concentration of acetate is much greater than the concentration of acetoacetate in the A. aceti cytoplasm and that genetic experiments associate aarC with acetate metabolism (16, 17), AarC is likely to function principally as SCACT.
Mycobacterium tuberculosis replaces α-ketoglutarate dehydrogenase and SCS with α-ketoglutarate decarboxylase and succinic semialdehyde dehydrogenase (54). (Activity for these enzymes was not detected in A. aceti lysates [data not shown].) Like the A. aceti SCACT bypass, the succinic semialdehyde bypass avoids a CoA- and nucleotide-dependent step and was detected by the analysis of inconsistent cofactor requirements.
All three bypasses preserve the typical cyclic course of the CAC and thereby differ from the “branched” CAC variants in facultative and obligate anaerobes (11).
Potential advantages of SCACT.
The thiotransesterification reactions performed by CoA-transferases tend to have small |ΔG| values. For SCACT, the slightly unfavorable reaction (Keq = 0.14; ΔG = +1.2 kcal/mol) is mitigated at high acetate levels or when acetyl-CoA is rapidly consumed by the CAC, i.e., during vigorous aeration. The near-thermoneutrality of the SCACT reaction is likely to be maintained in A. aceti as the cytoplasmic pH drops (32), whereas the equilibrium of the substrate-level phosphorylation performed by SCS is likely to depend on pH.
The use of SCACT bypasses a nucleotide requirement in the CAC and the need for initial acetate phosphorylation or adenylation (Fig. 4), decreasing the sensitivity of flux through the CAC to the energy state of the cell. The metabolic demand for free CoA is similarly reduced. Thus, acetic acid detoxification by irreversible oxidation would require only the favorable regeneration of reduced pathway cofactors.
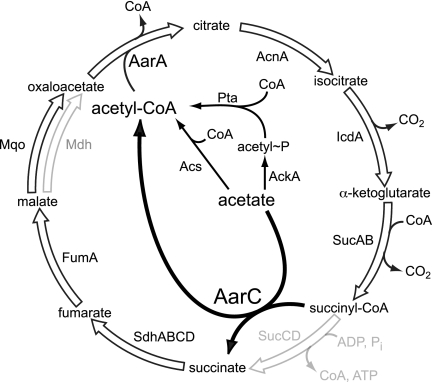
FIG. 4.
An unorthodox A. aceti CAC oxidizes acetate. The gray arrows indicate CAC genes that are not found in a draft A. aceti 1023 genome sequence (Kappock et al., unpublished). Each has a functional replacement. Using periplasmic dehydrogenases, A. aceti produces large quantities of acetic acid from ethanol and must contend with a constant influx of the former. Together with a complete oxidative phosphorylation pathway, this variant CAC functions in the eight-electron oxidation of acetic acid: CH3COOH + 2 O2 → 2 CO2 + 2 H2O. This pathway skips AckA, Acs, and SCS (sucCD), the only steps that would be directly influenced by cytoplasmic nucleotide pools, and reduces the number of enzymes that have free CoA substrates from two to one. The initial electron acceptors, two NAD+, one FAD, and one ubiquinone (presumed to be Q9 in A. aceti), would give a lower energy yield than a canonical CAC but have an additional irreversible step, the Mqo-mediated quinone reduction. Genes for the glyoxylate shunt enzymes isocitrate lyase and malate synthase are not found in A. aceti 1023.
Acetate dissimilation.
Reliance on a complete CAC for acetate removal might explain the vigorous oxygenation requirements of A. aceti cultures, which become critical at high acetic acid levels and low pHs (34, 37). Together with ADH-mediated ethanol oxidation, the CAC supplies energy for many cellular processes, including the maintenance of a proton gradient. Transient O2 deprivation sharply decreases cell viability within minutes, which is not consistent with the energy loss due to a slow collapse of the transmembrane pH gradient. Instead, the evidence presented here indicates that a complete CAC including SCACT supports the eight-electron oxidation of acetate to two CO2 molecules (Fig. 4). Cytoplasmic acetate would rapidly accumulate if cofactor reoxidation slows, which is consistent with the rapid loss of cell viability during inadequate culture aeration at high external acetic acid levels.
In many gram-negative bacteria, NADH limits CAC flux in anaerobic conditions. Citrate synthase (40) and Mdh (57) are susceptible to allosteric and product inhibition, respectively. However, both the acid-tolerant H. pylori and the acidophilic A. aceti contain citrate synthase forms that are insensitive to NADH (15, 44). Both organisms also replace Mdh with Mqo. Ubiquinone reduction by Mqo is more favorable than the Mdh reaction and is largely independent of the ratio of the concentration of NADH to the concentration of NAD+, both of which would tend to favor the production of oxaloacetate. In A. aceti, these adaptations may avoid the suppression of acetate dissimilation during periods of acetic acid stress.
Kinetic characterization.
The kinetic and mechanistic parameters of SCACT delineate its potential effect(s) on metabolic flux. A. aceti survives in >0.5 M acetic acid at a low pH, but aar-disrupted strains are unable to survive in 50 mM external acetic acid (16). Since there is likely no difference in acetate permeation, cytoplasmic acetate removal by oxidation appears to become critical at ∼10−2 M. The observed acetate Km values are in this range (Table 3), consistent with a conditionally essential detoxification role for aarC. Near the Km, SCACT velocity depends linearly on the cytoplasmic concentration of acetate, diminishing as acetate is depleted. At a lower pH, the acetate Km is smaller, which may increase the rate of acetate oxidation during intense acid stress.
An environmentally appropriate overflow pathway?
Acetate secretion by many bacteria is due to the activation of the carbon overflow pathway during anaerobiosis or when the central metabolic pathways are overtaxed (58). NAD+ and CoA are recovered as acetyl-CoA is converted to CoA and acetate, a diffusible compound that leaves the cytoplasm but can be recovered in the stationary phase. In AAB, vigorous periplasmic ethanol oxidation ensures that the acetate concentration gradient is always directed inward, precluding the diffusive loss of excess cytoplasmic acetate. The oxidation of acetate to CO2 by the SCACT-modified A. aceti CAC allows diffusive carbon loss, since CO2 is favored over bicarbonate at a low pH and can traverse bacterial cell membranes (33).
A CAC regulator in the aarA-to-aarC intergenic region.
New assignments in the A. aceti aarA-to-aarC intergenic region explain several previously confusing observations, including the singular gene X and aarB sequences that are apparently unique to A. aceti. Gene X is replaced by tyrA (chorismate mutase), which performs a key step in aromatic amino acid biosynthesis. Disruptants in the X/tyrA region were inviable (16), a finding that might now be attributed to a nutritional deficiency. Additionally, aarB is replaced by sixA. SixA regulates ArcA/ArcB, one of a small number of global regulators in E. coli, which represses the synthesis of aerobic metabolic enzymes during anaerobic conditions (3, 26). An ArcA knockout has increased CAC enzyme activities (43), suggesting that the SixA-dependent suppression of this inhibitory regulator would stimulate flux through the CAC. The reassignment of this conditionally essential sequence region to sixA is congruent with known AarA and AarC functions and suggests an important role for O2 sensing.
Conclusion.
The A. aceti strain 1023 aarC gene product is SCACT. This assignment reconciles previously confusing biochemical and genetic data regarding the important role(s) of aarC in acetic acid resistance and metabolism. SCACT bypasses SCS, which is missing from the genome; allows acetate incorporation without substrate-level phosphorylation; and enables the removal of diffusively trapped cytoplasmic acetate, by acetyl-CoA oxidation, as the readily diffusible CO2.
Supplementary Material
Acknowledgments
This work was supported by a grant to T.J.K. from the National Science Foundation (MCB 0347250). Mass spectrometry provided by the Washington University Mass Spectrometry Resource is supported by the National Institutes of Health (P41RR0954).
We thank Bob Blankenship and Petra Levin for comments on the manuscript, Eric Welsh for advice on annotation strategies, Hong Jiang for dethiaacetyl-CoA, and Courtney Starks for gel filtration analysis.
Footnotes
Published ahead of print on 23 May 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alber, B. E., R. Spanheimer, C. Ebenau-Jehle, and G. Fuchs. 2006. Study of an alternate glyoxylate cycle for acetate assimilation by Rhodobacter sphaeroides. Mol. Microbiol. 61297-309. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. D. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 3.Bauer, C. E., S. Elsen, and T. H. Bird. 1999. Mechanisms for redox control of gene expression. Annu. Rev. Microbiol. 53495-523. [DOI] [PubMed] [Google Scholar]
- 4.Beppu, T. 1994. Genetic organization of Acetobacter for acetic acid fermentation. Antonie van Leeuwenhoek 64121-135. [DOI] [PubMed] [Google Scholar]
- 5.Bergey, D. H., N. R. Krieg, and J. G. Holt. 1984. Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, MD.
- 6.Brock, M., and W. Buckel. 2004. On the mechanism of action of the antifungal agent propionate. Eur. J. Biochem. 2713227-3241. [DOI] [PubMed] [Google Scholar]
- 7.Cleland, W. W. 1982. An analysis of Haldane relationships. Methods Enzymol. 87366-369. [DOI] [PubMed] [Google Scholar]
- 8.Cleland, W. W. 1979. Statistical analysis of enzyme kinetic data. Methods Enzymol. 63103-138. [DOI] [PubMed] [Google Scholar]
- 9.Constantine, C. Z., C. M. Starks, C. P. Mill, A. E. Ransome, S. J. Karpowicz, J. A. Francois, R. A. Goodman, and T. J. Kappock. 2006. Biochemical and structural studies of N5-carboxyaminoimidazole ribonucleotide mutase (PurE) from the acidophilic bacterium Acetobacter aceti. Biochemistry 458193-8208. [DOI] [PubMed] [Google Scholar]
- 10.Corthesy-Theulaz, I. E., G. E. Bergonzelli, H. Henry, D. Bachmann, D. F. Schorderet, A. L. Blum, and L. N. Ornston. 1997. Cloning and characterization of Helicobacter pylori succinyl CoA:acetoacetate CoA-transferase, a novel prokaryotic member of the CoA-transferase family. J. Biol. Chem. 27225659-25667. [DOI] [PubMed] [Google Scholar]
- 11.Cronan, J. E., Jr., and D. Laporte. 1996. Tricarboxylic acid cycle and glyoxylate bypass, p. 206-216. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, M. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 12.Dawson, R. M. C. 1986. Data for biochemical research, 3rd ed. Clarendon Press, Oxford, United Kingdom.
- 13.Fleck, C. B., and M. Brock. 2008. Characterization of an acyl-CoA: carboxylate CoA-transferase from Aspergillus nidulans involved in propionyl-CoA detoxification. Mol. Microbiol. 68642-656. [DOI] [PubMed] [Google Scholar]
- 14.Francois, J. A., and T. J. Kappock. 2007. Alanine racemase from the acidophile Acetobacter aceti. Protein Expr. Purif. 5139-48. [DOI] [PubMed] [Google Scholar]
- 15.Francois, J. A., C. M. Starks, S. Sivanuntakorn, H. Jiang, A. E. Ransome, J.-W. Nam, C. Z. Constantine, and T. J. Kappock. 2006. Structure of a NADH-insensitive hexameric citrate synthase that resists acid inactivation. Biochemistry 4513487-13499. [DOI] [PubMed] [Google Scholar]
- 16.Fukaya, M., H. Takemura, H. Okumura, Y. Kawamura, S. Horinouchi, and T. Beppu. 1990. Cloning of genes responsible for acetic acid resistance in Acetobacter aceti. J. Bacteriol. 1722096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukaya, M., H. Takemura, K. Tayama, H. Okumura, Y. Kawamura, S. Horinouchi, and T. Beppu. 1993. The aarC gene responsible for acetic acid assimilation confers acetic acid resistance on Acetobacter aceti. J. Ferment. Bioeng. 76270-276. [Google Scholar]
- 18.Goldie, A. H., S. Narindrasorasak, and B. D. Sanwal. 1978. An unusual type of regulation of malate oxidase synthesis in Escherichia coli. Biochem. Biophys. Res. Commun. 83421-426. [DOI] [PubMed] [Google Scholar]
- 19.Greenberg, D. E., S. F. Porcella, A. M. Zelazny, K. Virtaneva, D. E. Sturdevant, J. J. Kupko III, K. D. Barbian, A. Babar, D. W. Dorward, and S. M. Holland. 2007. Genome sequence analysis of the emerging human pathogenic acetic acid bacterium Granulibacter bethesdensis. J. Bacteriol. 1898727-8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenfield, S., and G. W. Claus. 1972. Nonfunctional tricarboxylic acid cycle and the mechanism of glutamate biosynthesis in Acetobacter suboxydans. J. Bacteriol. 1121295-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackstein, J. H. P., J. Tjaden, and M. Huynen. 2006. Mitochondria, hydrogenosomes and mitosomes: products of evolutionary tinkering! Curr. Genet. 50225-245. [DOI] [PubMed] [Google Scholar]
- 22.Heider, J. 2001. A new family of CoA-transferases. FEBS Lett. 509345-349. [DOI] [PubMed] [Google Scholar]
- 23.Hersh, L. B., and W. P. Jencks. 1967. Coenzyme A transferase. Isolation and properties of an enzyme-coenzyme A intermediate. J. Biol. Chem. 2423481-3486. [PubMed] [Google Scholar]
- 24.Hersh, L. B., and W. P. Jencks. 1967. Coenzyme A transferase. Kinetics and exchange reactions. J. Biol. Chem. 2423468-3480. [PubMed] [Google Scholar]
- 25.Hersh, L. B., and W. P. Jencks. 1967. Isolation of an enzyme-coenzyme A intermediate from succinyl coenzyme A-acetoacetate coenzyme A transferase. J. Biol. Chem. 242339-340. [PubMed] [Google Scholar]
- 26.Iuchi, S., and E. C. C. Lin. 1988. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl. Acad. Sci. USA 851888-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kather, B., K. Stingl, M. E. van der Rest, K. Altendorf, and D. Molenaar. 2000. Another unusual type of citric acid cycle enzyme in Helicobacter pylori: the malate:quinone oxidoreductase. J. Bacteriol. 1823204-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lasko, D. R., C. Schwerdel, J. E. Bailey, and U. Sauer. 1997. Acetate-specific stress response in acetate-resistant bacteria: an analysis of protein patterns. Biotechnol. Prog. 13519-523. [DOI] [PubMed] [Google Scholar]
- 29.Mack, M., and W. Buckel. 1997. Conversion of glutaconate CoA-transferase from Acidaminococcus fermentans into an acyl-CoA hydrolase by site-directed mutagenesis. FEBS Lett. 405209-212. [DOI] [PubMed] [Google Scholar]
- 30.Martin, D. P., R. T. Bibart, and D. G. Drueckhammer. 1994. Synthesis of novel analogs of acetyl coenzyme A: mimics of enzyme reaction intermediates. J. Am. Chem. Soc. 1164660-4668. [Google Scholar]
- 31.Matsushita, K., T. Inoue, O. Adachi, and H. Toyama. 2005. Acetobacter aceti possesses a proton motive force-dependent efflux system for acetic acid. J. Bacteriol. 1874346-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menzel, U., and G. Gottschalk. 1985. The internal pH of Acetobacterium wieringae and Acetobacter aceti during growth and production of acetic acid. Arch. Microbiol. 14347-51. [Google Scholar]
- 33.Merlin, C., M. Masters, S. McAteer, and A. Coulson. 2003. Why is carbonic anhydrase essential to Escherichia coli? J. Bacteriol. 1856415-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mesa, M. M., I. Caro, and D. Cantero. 1996. Viability reduction of Acetobacter aceti due to the absence of oxygen in submerged cultures. Biotechnol. Prog. 12709-712. [Google Scholar]
- 35.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260289-298. [DOI] [PubMed] [Google Scholar]
- 36.Molenaar, D., M. E. van der Rest, and S. Petrovic. 1998. Biochemical and genetic characterization of the membrane-associated malate dehydrogenase (acceptor) from Corynebacterium glutamicum. Eur. J. Biochem. 254395-403. [DOI] [PubMed] [Google Scholar]
- 37.Muraoka, H., Y. Watabe, and N. Ogasawara. 1982. Effect of oxygen deficiency on acid production and morphology of bacterial cells in submerged acetic fermentation by Acetobacter aceti. J. Ferment. Technol. 60171-180. [Google Scholar]
- 38.Nakano, S., M. Fukaya, and S. Horinouchi. 2004. Enhanced expression of aconitase raises acetic acid resistance in Acetobacter aceti. FEMS Microbiol. Lett. 235315-322. [DOI] [PubMed] [Google Scholar]
- 39.Nakano, S., M. Fukaya, and S. Horinouchi. 2006. Putative ABC transporter responsible for acetic acid resistance in Acetobacter aceti. Appl. Environ. Microbiol. 72497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen, N. T., R. Maurus, D. J. Stokell, A. Ayed, H. W. Duckworth, and G. D. Brayer. 2001. Comparative analysis of folding and substrate binding sites between regulated hexameric type II citrate synthases and unregulated dimeric type I enzymes. Biochemistry 4013177-13187. [DOI] [PubMed] [Google Scholar]
- 41.Ohmori, S., H. Masai, K. Arima, and T. Beppu. 1980. Isolation and identification of acetic acid bacteria for submerged acetic acid fermentation at high temperature. Agric. Biol. Chem. 442901-2906. [Google Scholar]
- 42.Ohmori, S., T. Uozumi, and T. Beppu. 1982. Loss of acetic acid resistance and ethanol oxidizing ability in an Acetobacter strain. Agric. Biol. Chem. 46381-389. [Google Scholar]
- 43.Perrenoud, A., and U. Sauer. 2005. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J. Bacteriol. 1873171-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pitson, S. M., G. L. Mendz, S. Srinivasan, and S. L. Hazell. 1999. The tricarboxylic acid cycle of Helicobacter pylori. Eur. J. Biochem. 260258-267. [DOI] [PubMed] [Google Scholar]
- 45.Prust, C., M. Hoffmeister, H. Liesegang, A. Wiezer, W. F. Fricke, A. Ehrenreich, G. Gottschalk, and U. Deppenmeier. 2005. Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat. Biotechnol. 23195-200. [DOI] [PubMed] [Google Scholar]
- 46.Rao, M. R., and J. L. Stokes. 1953. Utilization of ethanol by acetic acid bacteria. J. Bacteriol. 66634-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rivière, L., S. W. H. van Weelden, P. Glass, P. Vegh, V. Coustou, M. Biran, J. J. van Hellemond, F. Bringaud, A. G. M. Tielens, and M. Boshart. 2004. Acetyl:succinate CoA-transferase in procyclic Trypanosoma brucei: gene identification and role in carbohydrate metabolism. J. Biol. Chem. 27945337-45346. [DOI] [PubMed] [Google Scholar]
- 48.Rochet, J. C., and W. A. Bridger. 1994. Identification of glutamate 344 as the catalytic residue in the active site of pig heart CoA transferase. Protein Sci. 3975-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saeki, A., K. Matsushita, S. Takeno, M. Taniguchi, H. Toyama, G. Theeragool, N. Lotong, and O. Adachi. 1999. Enzymes responsible for acetate oxidation by acetic acid bacteria. Biosci. Biotechnol. Biochem. 632102-2109. [DOI] [PubMed] [Google Scholar]
- 50.Solomon, F., and W. P. Jencks. 1969. Identification of an enzyme-γ-glutamyl coenzyme A intermediate from coenzyme A transferase. J. Biol. Chem. 2441079-1081. [PubMed] [Google Scholar]
- 51.Steiner, P., and U. Sauer. 2003. Overexpression of the ATP-dependent helicase RecG improves resistance to weak organic acids in Escherichia coli. Appl. Microbiol. Biotechnol. 63293-299. [DOI] [PubMed] [Google Scholar]
- 52.Steiner, P., and U. Sauer. 2001. Proteins induced during adaptation of Acetobacter aceti to high acetate concentrations. Appl. Environ. Microbiol. 675474-5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stouthamer, A. H., J. H. van Boom, and A. J. Bastiaanse. 1963. Metabolism of C2 compounds in Acetobacter aceti. Antonie van Leeuwenhoek 29393-406. [DOI] [PubMed] [Google Scholar]
- 54.Tian, J., R. Bryk, M. Itoh, M. Suematsu, and C. Nathan. 2005. Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of α-ketoglutarate decarboxylase. Proc. Natl. Acad. Sci. USA 10210670-10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Grinsven, K. W. A., S. Rosnowsky, S. W. H. van Weelden, S. Putz, M. van der Giezen, W. Martin, J. J. van Hellemond, A. G. M. Tielens, and K. Henze. 2008. Acetate:succinate CoA-transferase in the hydrogenosomes of Trichomonas vaginalis: identification and characterization. J. Biol. Chem. 2831411-1418. [DOI] [PubMed] [Google Scholar]
- 56.Van Hellemond, J. J., F. R. Opperdoes, and A. G. M. Tielens. 1998. Trypanosomatidae produce acetate via a mitochondrial acetate:succinate CoA transferase. Proc. Natl. Acad. Sci. USA 953036-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wise, D. J., C. D. Anderson, and B. M. Anderson. 1997. Purification and kinetic characterization of Haemophilus parasuis malate dehydrogenase. Arch. Biochem. Biophys. 344176-183. [DOI] [PubMed] [Google Scholar]
- 58.Wolfe, A. J. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 6912-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.