Abstract
In Clostridium perfringens, conjugative plasmids encode important virulence factors, such as toxins and resistance determinants. All of these plasmids carry a conjugation locus that consists of 11 genes: intP and tcpA to tcpJ. Three proteins, TcpA, a potential coupling protein, TcpF, a putative ATPase that is similar to ORF15 from Tn916, and TcpH, which contains VirB6-like domains, are essential for conjugation in the prototype conjugative plasmid pCW3. To analyze the functional domains of TcpH, a putative structural component of the mating-pair formation complex and deletion and site-directed mutants were constructed and analyzed. The results showed that the N-terminal 581 residues and the conserved 242VQQPW246 motif were required for conjugative transfer. Bacterial two-hybrid and biochemical studies showed that TcpH interacted with itself and with TcpC. An analysis of the tcpH mutants demonstrated that the region required for these interactions also was localized to the N-terminal 581 residues and that the function of the C-terminal region of TcpH was independent of protein-protein interactions. Finally, immunofluorescence studies showed that TcpH and TcpF were located at both cell poles of donor C. perfringens cells. The results provide evidence that TcpH is located in the cell membrane, where it oligomerizes and interacts with TcpC to form part of the mating-pair formation complex, which is located at the cell poles and is closely associated with TcpF.
Clostridium perfringens is a widely distributed pathogen and a major causative agent of gas gangrene (clostridial myonecrosis), gastrointestinal diseases such as food poisoning and necrotic enteritis in humans, and several enterotoxemic diseases in animals (41, 44, 45, 48). This species has a unique family of conjugative plasmids that carry either a novel tetracycline-resistant operon that has not been found in any other genus or toxin genes that are restricted to C. perfringens (6, 22, 37). The 47-kb plasmid pCW3 confers tetracycline resistance and is the prototype conjugative plasmid in C. perfringens. All known conjugative plasmids from C. perfringens, including plasmids encoding toxins such as enterotoxin and ɛ-toxin, carry variants of the pCW3 tcp transfer locus, which consists of 11 genes, intP and tcpA to tcpJ. (6, 7, 22). Therefore, these plasmids appear to have a similar conjugation mechanism. However, the actual mechanism of conjugation is unknown. Three of these genes, tcpA, tcpF, and tcpH, have been shown to be essential for conjugation in pCW3 (6, 40). The tcpA and tcpF genes encode putative ATPases, and TcpH is a putative integral membrane protein.
The bacterial conjugal machinery typically comprises the relaxosome, a coupling protein, and the mating-pair formation (mpf) apparatus, which is a subset of type IV secretion systems (T4SS) (33). The relaxosome is responsible for DNA processing in the cytoplasm, an event that may be triggered upon the close contact of the donor cell with a suitable recipient cell (31). The mpf apparatus is responsible for DNA translocation into the recipient cell and is linked to the relaxosome complex by the coupling protein (16). The coupling protein, which in the pCW3 system appears to be the TcpA protein (40), links the relaxosome to the mpf apparatus. TcpH is a predicted integral membrane protein that is postulated to be a critical transmembrane component of the pCW3 mpf-like apparatus. TcpH is essential for the conjugative transfer of pCW3, since tcpH mutants are nonconjugative (6). TcpH contains eight predicted transmembrane domains (TMDs) at its N terminus and two putative coiled-coil domains at its C terminus. The first half of TcpH, including the eight predicted TMDs, is most similar to ORF15 from the conjugative transposon Tn916. Within the segment containing TMD5 to TMD8 there is a region that has low-level similarity to a conserved TrbL/VirB6 domain. VirB6 from the Agrobacterium tumefaciens T4SS is an integral membrane protein that is involved in mpf assembly and synthesis, particularly in the stabilization of other mpf-related proteins (27). TrbL from RP4 is involved in the synthesis of the conjugation pilus (39) and also has homology with TraG from the F plasmid, which is a bifunctional protein that is involved in F-pilus synthesis and mating aggregate stabilization (13). It is therefore predicted that TcpH plays a similar role in the pCW3-derived mpf apparatus, with the exception of any involvement in pilus synthesis. TcpH also may have additional functions, since it has a C-terminal cytoplasmic region of 402 amino acids (aa) that is not found within VirB6, TrbL, or any other equivalent protein. PSI-BLAST searches indicate that the cytoplasmic domain of TcpH has distant similarity to DNA translocase proteins such as FtsK and SpoIIIE.
The objective of this study was to analyze the role of TcpH in the conjugal transfer of pCW3. The functional domains of TcpH were determined by performing deletion and site-directed mutagenesis on the cloned tcpH gene and examining the ability of the resultant derivatives to complement a tcpH mutant. In addition, the self interaction of TcpH and an interaction between TcpH and another putative pCW3 transfer protein, TcpC, were demonstrated using the bacterial two-hybrid system and confirmed by biochemical analysis. Finally, to determine the cellular location of TcpH and TcpF, another essential conjugation protein, immunofluorescence studies were performed on C. perfringens using TcpH- and TcpF-specific antibodies. Both TcpH and TcpF were localized at the poles of C. perfringens donor cells. The data support the hypothesis that pCW3 is transferred into a recipient cell through a TcpH-containing mpf apparatus located at the cell poles.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The Escherichia coli strains used in this study were DH5α (Invitrogen), XL1-blue (Stratagene), C43(DE3) (36), and BTH101 (28, 36). E. coli cells were grown in 2× YT medium (35) at 30 or 37°C and supplemented with the following antibiotic(s) when necessary: ampicillin (100 μg/ml), chloramphenicol (30 μg/ml), erythromycin (150 μg/ml), kanamycin (20 μg/ml), and/or streptomycin (100 μg/ml). C. perfringens strains used in this study were cultured at 37°C in fluid thioglycolate medium (FTG) (Difco), trypticase-peptone-glucose (TPG) broth (46), brain heart infusion (BHI) broth/agar (Oxoid), or nutrient agar (43) supplemented with the following antimicrobial agents, when necessary: thiamphenicol (10 μg/ml), erythromycin (50 μg/ml), potassium chlorate (1% [vol/vol]), streptomycin (1 mg/ml), and tetracycline (10 μg/ml). C. perfringens agar cultures were incubated in an atmosphere of 10% H2-10% CO2-80% N2 in an anaerobic jar (Oxoid). The plasmids used in this study are listed in Table S1 in the supplemental material.
Molecular techniques.
E. coli plasmid DNA was extracted from the cell pellet using either a commercial QIAprep Spin Miniprep kit (Qiagen) per the manufacturer's instructions or by alkaline lysis (38). Purified C. perfringens DNA was obtained by sodium dodecyl sulfate (SDS) lysis and cesium chloride-ethidium bromide buoyant density centrifugation (1), and crude C. perfringens DNA was extracted as described previously (6). Sequencing was performed on an Applied Biosystems 3730S capillary sequencer and analyzed using Sequencher version 3.0 (Gene Codes Corporation). The TcpH amino acid sequence was analyzed for homologues and conserved domains using PSI-BLAST (2, 3), for TMDs using TopPred (9), and for secondary-structure predictions using SOSUI (20). The amino acid sequences of TcpH and its homologues were aligned using ClustalW (32).
Construction of deletion and site-directed mutants of tcpH.
Several strategies were adopted to generate TcpH deletion constructs. pJIR3075 was constructed by digesting pJIR2901 using the restriction enzyme XmnI. The digested plasmid was religated, and the shortest construct was selected. The plasmids encoding further C-terminal deletions in TcpH (pJIR3112, pJIR3090, pJIR3113, and pJIR3091) were constructed by cloning corresponding tcpH-specific PCR fragments directly into the C. perfringens-E. coli shuttle vector pJIR750. The plasmid pJIR3115, which encoded a protein containing an internal in-frame deletion of the last four predicted transmembrane segments in TcpH (TcpHΔ311-450), and the plasmids pJIR3134, pJIR3135, and pJIR3145, which contained site-directed mutations in tcpH, were constructed by using splice overlap extension PCR (21) to generate the desired modified PCR product. The resultant fused fragments were inserted separately into pJIR750 (5). All constructs expressing altered or truncated TcpH proteins were confirmed by sequence analysis and tested for their ability to restore conjugation function in the tcpH mutant JIR4885.
Conjugation.
All of the C. perfringens tcpH derivatives were tested for their conjugative ability by being mated on solid media, as described previously (43, 46). The recipient was JIR4394, a streptomycin- and chlorate-resistant derivative of strain 13 (6). The frequency of conjugative transfer is reported as the number of transconjugants per donor cell.
Production of TcpF and TcpH antiserum.
Anti-TcpH serum was generated by immunizing two guinea pigs with a truncated TcpH protein comprising the C-terminal region (TcpH441-832-His6). Anti-TcpF serum was generated by immunizing two rabbits with diphtheria toxoid conjugated to a TcpF-specific peptide of 15 aa residues, RLTPNDEDKGKLDPF, which correspond to residues 543 to 557 of TcpF. The immunization experiments were performed by Antibodies Australia and Chemicon, Australia. The TcpF-specific peptide was synthesized and conjugated to diphtheria toxoid by Mimotopes, Australia.
Bacterial two-hybrid assay.
To test for protein-protein interactions, the genes encoding the potential interacting proteins were cloned separately into the vectors pKT25 and pUT18C (28), and the resultant constructs were used to cotransform strain BTH101. For negative controls, a vector expressing a fusion protein and a corresponding empty vector (pKT25 and pUT18C, respectively) were used for the transformation. The transformants of interest were isolated and used immediately for quantitative β-galactosidase assays (4). Cultures were grown in 2× YT broth supplemented with ampicillin (100 μg/ml) and kanamycin (20 μg/ml) at 30°C for 24 h, cooled to room temperature, and diluted with fresh broth to a turbidity at 600 nm of approximately 0.5. Samples (300 μl) of the diluted cells were transferred into a 96-well microtiter plate, and the turbidity at 620 nm was immediately determined using a Multiskan plate reader (Labsystems). An equal volume of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM 2-mercaptoethanol, pH 7.0) was added to 500 μl of the diluted cells in a microcentrifuge tube. To lyse the cells, two drops of chloroform and one drop of SDS (0.1% [wt/vol]) were added to the cell suspensions, and the mixtures were vortexed for 10 s. The cell lysate (100 μl) was transferred into the 96-well microtiter plate and incubated at 30°C for 15 min. The reaction was started by adding 200 μl of 0.7 mg/ml 2-nitrophenyl-β-d-galactopyranoside (ONPG) (Sigma) in Z buffer. The absorbance was recorded at 414 nm (for O-nitrophenol color change) and 570 nm (for cell debris). β-Galactosidase activity (in Miller units) was calculated as previously described (4) when the absorbance at 414 nm just exceeded 0.6.
Chemical cross-linking assay.
Cross-linking experiments were performed using dimethyl 3,3′-dithiobispropionimidate-2HCl (DTBP; Pierce Biotechnology), a cleavable and bifunctional imidoester cross-linker. To study TcpH-TcpH interaction, pJIR3287 and pJIR3191, which encoded TcpH fused to either His or glutathione S-transferase (GST) tags (17), respectively, were coexpressed in E. coli C43(DE3) cells at 30°C for 3 h after induction with 1 mM of IPTG. Cells were harvested from a 500-ml culture (2× YT broth supplemented with ampicillin [100 μg/ml] and chloramphenicol [30 μg/ml]) and resuspended in 12 ml of phosphate-buffered saline (PBS) before being lysed by passage three times through a prechilled French press. The cell debris was removed by centrifugation at 17,000 × g for 50 min at 4°C, and the resultant clear lysate was separated into 3-ml aliquots. DTBP was added immediately to a final concentration ranging from 0 to 4 mM, inverted gently to mix the suspension, and incubated at room temperature for 1 h to allow cross-linking to occur. The cross-linking reaction was quenched by adding 1 M Tris-HCl to a final concentration of 50 mM, followed by incubation at room temperature for 10 min. The cross-linked samples either were immediately analyzed by immunoblotting or stored at −20°C until required.
Chemical cross-linking assays also were performed on purified maltose binding protein (MBP) and MBP-TcpH. Rosetta Two cells expressing MBP or MBP-TcpH were cultured in autoinduction media overnight at 28°C and then for an additional 24 h at 22°C. The proteins were affinity purified from cell lysates with Amylose resin (New England Biolabs). Solutions of purified proteins (100 μg/ml in 50 mM HEPES, pH 8.2) were cross-linked in serial dilutions of DTBP at 25°C for 60 min. The reactions were quenched with 50 mM Tris-HCl.
Samples were separated by SDS-polyacrylamide gel electrophoresis (PAGE) (30) at 4°C under both reducing and nonreducing conditions. Reducing conditions were achieved by adding 1,4-dithiothreitol (DTT) (Roche Diagnostics, Germany) to a final concentration of 150 mM in 2× sample buffer ([0.5 M Tris-HCl, 0.4% {wt/vol} SDS, pH 6.8], 20% glycerol, 2.5% SDS, and 0.013% bromophenol blue) and then mixing the solution with an equal volume of cross-linked sample. Incubation was at 30°C for 30 min. For nonreducing conditions, no DTT was added and equal volumes of cross-linked samples were mixed with 2× sample buffer and incubated at 53°C for 30 min. The transfer of the proteins onto the nitrocellulose membrane (Schleicher and Schuell) using a mini Trans-Blot electrophoretic transfer cell (Bio-Rad) in transfer buffer (12.5 mM Tris-HCl [pH 8.3], 100 mM glycine, and 10% methanol) was carried out at 200 V for 2 h at 4°C, and immunoblotting was carried out using the following antibodies: Penta-His (Qiagen), anti-GST rabbit polyclonal (Chemicon), anti-MBP rabbit polyclonal (New England BioLabs), anti-TcpH guinea pig polyclonal sera (Antibodies Australia), horseradish peroxidase-conjugated sheep anti-mouse, sheep anti-rabbit, and goat anti-guinea pig (Chemicon).
Far-Western analysis.
Purified His-TcpCΔ1-98 (1 mg/ml in 50 mM HEPES, pH 8.2; kindly provided by Corrine Porter) was labeled with biotin amidohexanoic acid 3-sulfo-N-hydroxysuccinide ester using the BiotinTag Micro biotinylation kit (Sigma). Labeling was performed at 4°C for 4 h, at which time the reaction was stopped by the addition of 50 mM Tris. Biotin-labeled protein then was buffer exchanged using a Microcon MW30 mini spin column (Millipore) to remove unbound biotin. Far-Western analysis was performed on purified MBP and MBP-TcpH proteins that had been separated for 50 min at 200 V on 10% polyacrylamide gels (30) and transferred to a nitrocellulose membrane. The membrane was blocked in 5% skim milk-Tris-buffered saline-Tween solution for 1 h at room temperature and then incubated in the presence of a 1/2,000 dilution of biotin-labeled probe overnight at room temperature. The streptavidin derivative ExtrAvidin peroxidase (Sigma) was used to detect the presence of the biotin-labeled protein.
Cell fractionation.
Crude membrane preparations of C. perfringens were isolated by a modification of a previous method (23). The cells were grown overnight on two BHI agar plates and harvested by resuspension in 10 ml of BHI diluent and centrifugation at 5,000 × g for 5 min at 4°C. The cells were washed once with 10 ml of PBS before being resuspended in 10 ml of PBS supplemented with one tablet of the Complete cocktail protease inhibitor (Roche). The cells were disrupted twice in a precooled French press, and cell debris was removed by centrifugation at 17,000 × g for 50 min at 4°C. To pellet the membrane fraction, 1 ml of the lysate was centrifuged at 300,000 × g for 2 h at 4°C. The membrane pellet was washed once using 1 ml of PBS, and the final membrane fraction was pelleted by centrifugation at 300,000 × g for 30 min at 4°C and resuspended in 200 μl of PBS. To obtain concentrated membrane preparations, two membrane pellets were combined and resuspended in a final volume of 200 μl of PBS.
Immunofluorescence microscopy.
Immunofluorescence staining was performed with slight modifications of a previous method (19). C. perfringens cells from an agar culture grown overnight at 37°C were resuspended in 250 μl of 30 mM sodium phosphate buffer (pH 7.5) and briefly vortexed to disrupt any clumps of bacteria. To fix the cells, paraformaldehyde (BDH) was added to the cell suspension to a final 1.0% (wt/vol) concentration and incubated for 15 min at room temperature, followed by a 35-min incubation on ice. The fixed cells were washed three times in PBS (pH 7.4) at room temperature and resuspended in 100 μl of GTE (50 mM glucose, 20 mM Tris-HCl [pH 7.5], and 10 mM EDTA). A freshly prepared lysozyme solution in GTE was added to a final concentration of 2 mg/ml. A 10-μl aliquot of the cell suspension was immediately spread on a poly-l-lysine-coated coverslip and allowed to dry at room temperature. The coverslip was dipped in −20°C methanol for 5 min and −20°C acetone for 30 s and dried at room temperature. An aliquot of 100 μl of the blocking solution (2% [wt/vol] bovine serum albumin sigma in PBS) was added to the coverslip and incubated at room temperature for 1 h. Coverslips with the fixed cells were rinsed three times in PBS and incubated with 100 μl of primary antibody in PBS for 2 h at room temperature. The anti-TcpH guinea pig serum was used at a 1:10 dilution, and the anti-TcpF rabbit serum was pre-adsorbed with a cell lysate from the tcpF mutant JIR4940 and used undiluted. For double staining, the primary antisera were added and incubated sequentially, with three washes in PBS between the incubation steps. The cells were washed 10 times in PBS before being incubated in the dark with the appropriate fluorescein-conjugated secondary antibody in PBS for 2 h at room temperature. Alexa Fluor 488 goat anti-guinea pig was used at 10 μg/ml, and Alexa Fluor 488 goat anti-rabbit and Alexa Fluor 568 goat anti-rabbit each were used at 4 μg/ml (Molecular Probes). The nuclear stain 4′,6′-diamidino-2-phenylindole (DAPI; Molecular Probes) was coincubated with the secondary antibodies at a final concentration of 25 μg/ml. The cells were washed 10 times in PBS and mounted to a glass slide using fluorescence mounting medium (DAKO). The stained cells usually were observed immediately by fluorescence microscopy using an Olympus BX51 microscope with a ×100 oil immersion objective and 1.35 numerical aperture (Olympus). The stained cells were photographed using an Olympus DP-70 digital camera and merged using DP manager software version 1.1.1.71 (Olympus).
RESULTS
Identification of functional transfer regions of TcpH.
Previously, two independent tcpH mutants were isolated by homologous recombination events that led to double crossovers (6). The mutants were transfer deficient (<1.1 × 10−8 transconjugants per donor cell), and complementation with an intact copy of the tcpH gene partially restored conjugation function in the mutants, providing evidence that tcpH is essential for the conjugal transfer of pCW3. To determine if conserved regions of TcpH were essential for the transfer functions of TcpH, deletion analysis was performed on the wild-type tcpH gene that is located on the complementation plasmid pJIR2901 (6), resulting in a series of successive C-terminal deletions (Fig. 1A). The mutated tcpH genes were sequenced to confirm that no additional mutations were present. These constructs were used to transform the transfer-deficient tcpH mutant JIR4885, which carried pCW3ΔtcpH::ermQ. Genomic DNA was extracted from the various complemented tcpH mutants, and the presence of the correct plasmid construct was confirmed by PCR analysis using tcpH-specific primers (data not shown).
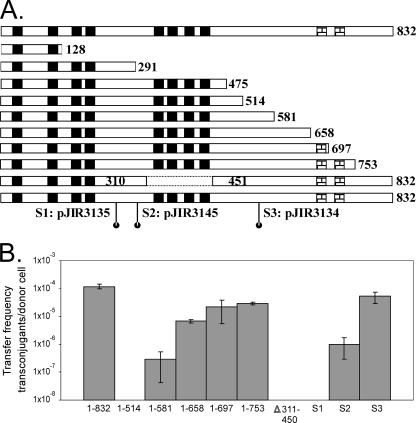
FIG. 1.
Determination of functional regions of TcpH. (A) Diagrammatic presentation of deletions and site-directed substitutions of TcpH. The predicted domains in wild-type TcpH (832 aa) are presented at the top of the diagram. Black boxes represent putative TMDs, and checked boxes represent predicted coiled-coil domains. The numbers represent amino acid sequence positions. All of the plasmids express TcpH proteins with C-terminal truncations, with the exception of pJIR3115, which is the second to last TcpH derivative represented. In pJIR3115, an internal sequence (encoding aa 311 to 450) of tcpH was deleted. Three independent plasmids expressed the following TcpH site-directed substitutions: pJIR3135 (S1; 242VQAAA246), pJIR3145 (S2; 287DAAAK291), and pJIR3134 (S3; 545AAA547). (B) Conjugation frequencies of complemented tcpH mutants. Plasmids expressing the various TcpH derivatives (as indicated below the graph) were used to transform the transfer-deficient tcpH mutant JIR4885, and the resultant transformants were used independently as donors in matings with strain JIR4394 as the recipient. The frequency of conjugative transfer is reported as the number of transconjugants per donor cell. No transfer was observed with the TcpH1-128, TcpH1-291, or TcpH1-475 derivative (data not shown).
Transformation with a plasmid encoding the wild-type TcpH1-832 protein was able to restore transfer to a frequency of (1.2 ± 0.2) × 10−4 transconjugants per donor cell in the tcpH mutant (Fig. 1B). To determine if the predicted coiled-coil domains were essential, the TcpH1-658 derivative without these domains was examined. This derivative was able to complement transfer function in the tcpH mutant to a frequency of (6.7 ± 1.0) × 10−6 transconjugants per donor cell, which was significantly reduced (P < 0.05) compared to that of full-length TcpH1-832. The TcpH1-697 and TcpH1-753 derivatives, which had one or both coiled-coil domains, respectively, further increased the transfer frequency to an order of magnitude higher than that of TcpH1-658 but still significantly (P < 0.05) reduced compared to that of TcpH1-832. These data indicated that the predicted coiled-coil domains were important, but not essential, for the transfer function of TcpH and that the complete C-terminal segment was required for maximal function.
The three most extensive deletion derivatives, TcpH1-128, TcpH1-291, and TcpH1-475, were not able to complement transfer in the conjugation-deficient tcpH mutant (data not shown), indicating that the N-terminal region containing the TMDs was insufficient for conjugative transfer. The TcpH1-514 and TcpH1-581 derivatives then were constructed to determine the minimal region of TcpH required for transfer. Only the TcpH1-581 construct was able to complement transfer, and it did so at a very low frequency (Fig. 1B). It was concluded that the first 581 aa residues, including the eight predicted TMDs, was the minimum-sized region required to restore at least some transfer function to the tcpH mutant and that the C-terminal region (aa 581 to 832) was required for optimal TcpH function.
In addition, there was a TrbL/VirB6-like region within the last four TMDs in TcpH. To determine if this region was essential for transfer, an in-frame deletion of the region encoding aa 311 to 450 was generated. The deletion of the predicted TMD5 to TMD8 region abolished the ability to complement the tcpH mutant (Fig. 1B), indicating that the VirB6-like domain either was involved in facilitating transfer or was required for the structural integrity of TcpH.
Sequence alignments of the pCW3-derived TcpH protein, its homologues derived from the other related pCW3-like plasmids from C. perfringens, and ORF15 from Tn916 revealed three conserved motifs (Fig. 2). To see if these motifs were functional, site-directed mutagenesis was performed on conserved residues from each region, and the resultant constructs were tested for their ability to facilitate transfer in the transfer-deficient tcpH mutant. A 242VQAAA246 mutant, with changes located between TMD4 and TMD5, did not restore conjugation function in the tcpH mutant (Fig. 1B), indicating that the 242VQQPW246 motif was essential for the assembly of a functional conjugation apparatus. Mutations in the conserved 287DEREK291 region resulted in a reduced transfer frequency (P < 0.05) compared to that of wild-type TcpH (Fig. 1B). Much higher transfer frequencies were observed when the 545DTK547 motif was mutated (Fig. 1B). Since the TcpH287DAAAK291 and TcpH545AAA547 derivatives were able to complement transfer, the DEREK and DTK motifs cannot be essential for TcpH function, although the DEREK motif clearly must play some functional role.
FIG. 2.
Amino acid sequence alignment of TcpH homologues. The TcpH amino acid sequence of pCW3 was aligned with sequences of the equivalent regions from Tn916 (14), pMRS4969, pJIR2774, and pJIR26 (GenBank accession numbers DQ338472, DQ338473, and DQ338471, respectively), encoding CPE toxin production, lincomycin resistance, and tetracycline resistance, respectively (6), and pJGS1495 and pJGS1721, which represent plasmids derived from C. perfringens type C and type D strains encoding β- and ɛ-toxins, respectively (40). Highlighted in the boxed regions are the three conserved sequences: VQQPW, DEREK, and DTK. The symbols below the alignment indicate identical residues (*), strongly similar residues (:), and residues with weak similarities (.). The numbers represent amino acid sequence positions in TcpH from pCW3.
TcpH interacts with itself and TcpC.
Protein-protein interactions are important for the formation of a functional mpf apparatus. In this study, a bacterial two-hybrid system was used to detect such interactions. The system is based on the interaction of the T25 and T18 domains of adenylate cyclase from Bordetella pertussis, in which the successful interaction between test proteins results in the increased expression of a lacZ reporter gene (28). Negative controls were BTH101 cells cotransformed with a vector expressing the test protein fused to either the T25 or T18 domains and a corresponding control plasmid (pUT18C or pKT25, respectively). In the transformants coexpressing the chimeric proteins, the detection of β-galactosidase production that was significantly higher than that of the corresponding negative controls provides evidence for interactions between the test proteins.
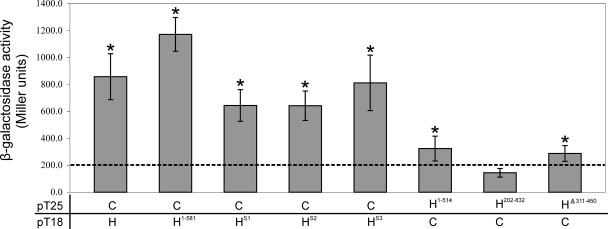
To determine if TcpH interacts with itself, as would be expected for a protein that formed the integral transmembrane component of the mpf complex, BTH101 cells were cotransformed with separate derivatives that encoded the chimeric T25-TcpH and TcpH-T18 proteins and analyzed for β-galactosidase activity (Fig. 3). The results showed that there was significantly higher β-galactosidase activity (P < 0.01) in the transformant coexpressing the T18 and T25 proteins fused to wild-type TcpH than in the negative controls (Fig. 3). These data provided evidence that TcpH monomers interacted with each other to form a homooligomer.
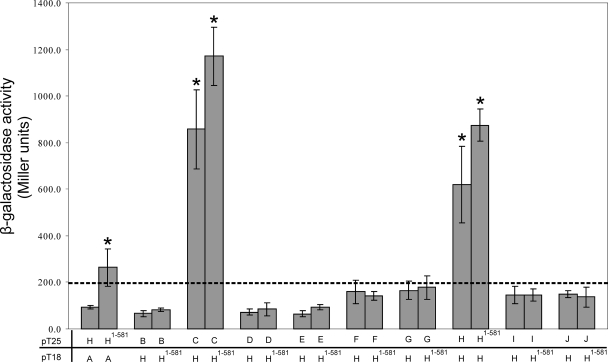
FIG. 3.
Protein-protein interactions between TcpH and other Tcp proteins. β-Galactosidase assays were performed in duplicate on three separate cultures. Samples were derived from E. coli BTH101 cells cotransformed with pUT18C and pKT25 that were fused to DNA sequences encoding TcpH (H) and the other Tcp derivatives (A, B, C, etc.) as shown. As negative controls, BTH101 cells were cotransformed with a vector expressing the test protein fused to either the T25 or T18 domain and a corresponding control plasmid (pUT18C or pKT25, respectively). The dashed line represents the highest level of β-galactosidase activity from the negative controls. Statistical analysis was carried out using the Student's t test after analysis of variance, as indicated by an asterisk (P < 0.05 compared to results for the negative control).
To determine if TcpH interacts with each of the other Tcp proteins, TcpH was used as the bait to search for interacting partners among the remaining nine Tcp proteins that are encoded by pCW3 (TcpA to TcpJ) (Fig. 3). The bacterial two-hybrid assay was carried out with the Tcp proteins fused to the T25 domain, with the exception of TcpA. In addition to its self interaction, TcpH was found to interact with TcpC, since the level of β-galactosidase activity derived from transformants coexpressing TcpC and TcpH was significantly higher than that of the negative controls. The smallest TcpH derivative that was still capable of functional complementation, the TcpH1-581 derivative, was used to screen for potential interacting partners among the other Tcp proteins. TcpH1-581 interacted with TcpC, confirming the results obtained with full-length TcpH. Similarly, TcpH1-581 did not interact with any other Tcp proteins, with one exception. Transformants coexpressing TcpA and TcpH1-581 showed higher levels of β-galactosidase activity (P < 0.05) than the transformants coexpressing TcpA and TcpH, which was not significantly different from results for the negative control.
Although the C terminus of TcpH was not required for self interaction, the two predicted coiled-coil structures could mediate the interaction with other Tcp proteins. This possibility was tested by using the TcpH515-832 derivative as the bait protein in the bacterial two-hybrid system. This derivative did not interact with any of the proteins, including TcpC and full-length TcpH (data not shown). These results suggested that the region of TcpH that is required for protein-protein interactions was located in the first 581 residues.
Delineation of the regions required for TcpH self interaction.
To map the regions of TcpH that are required for its self interaction, the various mutated TcpH derivatives constructed earlier were examined in the bacterial two-hybrid system. Initial studies involved the TcpH1-697 derivative, which contained one predicted coiled-coil domain and resulted in a higher frequency of transfer than TcpH1-581, the shortest derivative capable of restoring transfer in the tcpH mutant. The results showed that both the TcpH1-697 and the TcpH1-581 derivatives were able to self interact, and to interact with full-length TcpH, at levels comparable to those of the wild-type interaction (Fig. 4). These results confirmed that the TcpH region that is involved in these self interactions was located in the N-terminal 581 aa and did not involve the putative coiled-coil domains that are located closer to the C terminus. The β-galactosidase activity derived from transformants coexpressing the TcpH and TcpH1-581 or TcpH1-697 fusions was significantly (P < 0.05) higher than the activity from the transformant coexpressing the wild-type TcpH fusions (Fig. 4), perhaps as a result of the better tolerance of shorter chimeric protein constructs in the bacterial two-hybrid system (28).
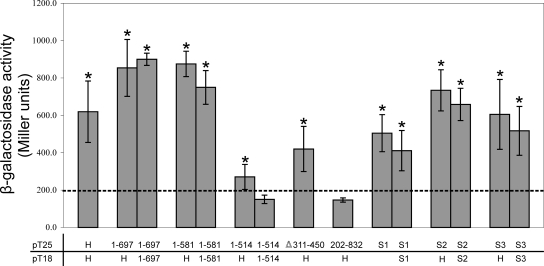
FIG. 4.
Protein-protein interactions between TcpH derivatives in the bacterial two-hybrid system. Bacterial two-hybrid analysis and β-galactosidase assays were performed as described for Fig. 3. The dashed line represents the highest level of β-galactosidase activity from the negative controls. Statistical analysis was carried out using the Student's t test after analysis of variance, as indicated by an asterisk (P < 0.05 compared to results for the negative control).
We then investigated whether the transfer-deficient derivative TcpH1-514 protein still could facilitate protein-protein interactions. β-Galactosidase production resulting from the coexpression of the T18 and T25 fragments fused to TcpH1-514 was not significantly different from that of the negative controls (Fig. 4). However, the transformant coexpressing TcpH and TcpH1-514 produced a slightly higher level of activity (P < 0.05) than the negative controls but significantly lower than that of the TcpH and TcpH1-581 combination (Fig. 4). Another transfer-deficient derivative, TcpHΔ311-450, which did not contain the VirB6-like domain, retained moderate levels of interactions with wild-type TcpH (P < 0.05) (Fig. 4). Based on these results, it was concluded that the region of TcpH composed of aa 514 to 581 was required for both self interaction and conjugative transfer and that the region containing TMD5 to TMD8 may contribute indirectly to TcpH self interactions.
To test whether the region containing TMD1 to TMD4 was involved in protein-protein interactions, we also examined a TcpH202-832 construct in which this region was deleted. The results were not significantly different from those for the negative control (Fig. 4). The absence of interaction could be due to the deletion of binding domains, which may be present in the first 201 aa of TcpH. Alternatively, the deletion of the first 201 aa may affect the stability of the TcpH protein or result in the incorrect folding of the fusion protein.
Similar experiments were carried out to determine whether any of the three conserved motifs that previously were mutagenized contributed to TcpH-TcpH interactions. The results showed that the mutated proteins each were able to self interact and to interact with the wild-type TcpH construct, resulting in β-galactosidase activity that was not significantly different from that observed with the wild-type TcpH fusions (Fig. 4). Since the TcpH242VQAAA246 derivative, which failed to restore conjugation function in the tcpH mutant, was able to self interact, it was concluded that the VQQPW motif, like the VirB6-like region, may be a functional site that is required for transfer but that is not crucial for the formation of TcpH homooligomers.
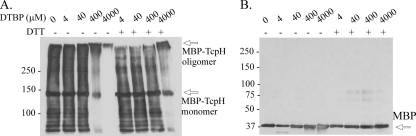
To confirm the TcpH self-interaction at a biochemical level, a chemical cross-linking approach was used. E. coli cells were cotransformed with pJIR3287 and pJIR3191, which expressed His- and GST-tagged TcpH proteins, respectively. The expected size of the His-tagged TcpH protein was approximately 100 kDa, and the addition of the GST domain resulted in a protein with an expected size of approximately 130 kDa. The cell lysates derived from transformants coexpressing these proteins were treated with the imidoester cross-linker DTBP at a final concentration of 4 mM and were examined by immunoblotting using GST- and His-specific antisera. In addition to the monomers (His-TcpH and GST-TcpH), each specific antiserum was able to detect a protein band of a higher molecular size when the lysates were treated with DTBP (Fig. 5), providing evidence that the complex comprised both the His- and GST-tagged TcpH proteins. The same protein band also was detected by TcpH-specific guinea pig antiserum and was dependent upon the concentration of DTBP; it was not present when less than 2 mM DTBP was used (data not shown). The cross-linking reaction mediated by DTBP also could be reversed under reducing conditions, as expected. As shown in Fig. 5, the higher-molecular-size complex protein band was not present when the same samples were separated in the presence of 150 mM DTT. These results indicate that the formation of the higher-molecular-weight complex was specifically mediated by DTBP in a concentration-dependent manner. These data suggested that the His- and GST-tagged TcpH proteins had been cross-linked, providing physical evidence that TcpH forms homooligomers. The results were consistent with the data obtained with the bacterial two-hybrid system.
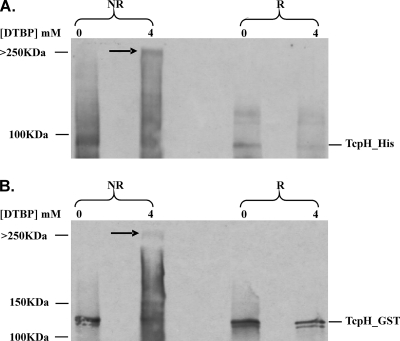
FIG. 5.
Analysis of TcpH interactions by chemical cross-linking and Western blotting. Cell lysates derived from transformants coexpressing His- and GST-tagged TcpH proteins were treated with DTBP (0 or 4 mM, as shown at the top of the gel) and examined by immunoblotting using (A) His-specific antiserum and (B) GST-specific antiserum. R, 150 mM DTT added to produce reducing conditions; NR, nonreducing conditions. The higher-molecular-size protein complex is indicated by the arrow and has a size of >250 kDa.
To further investigate the interaction between TcpH monomers, chemical cross-linking was performed on purified MBP-TcpH fusion proteins. Increasing concentrations of DTBP (4 μM to 4 mM) were added to duplicate samples of MBP-TcpH, which subsequently were separated by SDS-PAGE in the presence and absence of the reducing agent DTT. Under reducing conditions, DTBP-mediated cross-linking is reversed. After electrophoresis, the samples were transferred to nitrocellulose membranes and probed with anti-TcpH-specific antibodies (Fig. 6A). Prior to the addition of DTBP and the reducing agent DTT, both monomeric and oligomeric forms of MBP-TcpH can be seen, indicating that oligomers of TcpH are resistant to SDS and occur independently of the cross-linker. As the amount of DTBP increases, the proportion of oligomeric MBP-TcpH increases, and this effect is reversible upon the addition of DTT at all but the highest concentrations of DTBP. As a control for interactions involving the MBP tag, purified MBP tag also was subjected to chemical cross-linking and detected using anti-MBP antibodies. No cross-linking was observed (Fig. 6B).
FIG. 6.
Formation of MBP-TcpH homooligomers. (A) Duplicate samples of purified MBP-TcpH were exposed to increasing concentrations of the chemical cross-linker DTBP. The resultant products then were separated by SDS-5% PAGE gels in the presence and absence of the reducing agent DTT, as shown. The samples then were transferred to nitrocellulose and probed with anti-TcpH antibodies. (B) MBP also was subjected to chemical cross-linking and separated by SDS-12% PAGE, and the products were probed with anti-MBP antibodies. Protein size standards are indicated (in kilodaltons).
Delineation of regions of TcpH required for interaction with TcpC.
To determine if regions that are essential for TcpH homooligomer formation also were required for the formation of TcpH-TcpC heterooligomers, the various mutant TcpH derivatives were analyzed in the two-hybrid system for their ability to interact with TcpC (Fig. 7). The results indicated that the region between aa 514 and 581, which was essential for TcpH self interaction, also was required for TcpH-TcpC interaction. β-Galactosidase production in transformants coexpressing the TcpC and TcpH1-514 fusions was slightly higher (P < 0.05) than that of the negative controls but significantly reduced compared to that of transformants coexpressing either TcpC and TcpH or TcpC and TcpH1-581.
FIG. 7.
Protein-protein interaction of TcpC and TcpH in the bacterial two-hybrid system. Bacterial two-hybrid analysis and β-galactosidase assays were performed as described for Fig. 3. The dashed line represents the highest level of β-galactosidase activity from the negative controls. Statistical analysis was carried out using the Student's t test after analysis of variance, as indicated by an asterisk (P < 0.05 compared to results for the negative control).
The TcpH202-832 fusion derivative, which lacked TMD1 to TMD4, was not able to interact with the TcpC fusion, whereas the TcpHΔ311-450 protein, which does not contain TMD5 to TMD8, showed limited interaction with TcpC (Fig. 6). These results suggest that the VirB6-like domain of TcpH is involved in interacting with TcpC, although further experiments are required to confirm this hypothesis. Finally, as for the TcpH self interactions, all of the site-directed mutants in the three conserved motifs were able to interact with TcpC (Fig. 7).
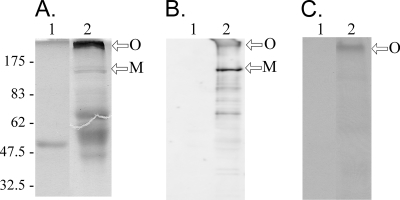
Far-Western analysis was used to confirm the TcpH-TcpC interactions. In these experiments, preparations of purified MBP and MBP-TcpH were separated on SDS-polyacrylamide (10%) gels and either Coomassie blue stained or transferred to a nitrocellulose membrane for Western and Far-Western analysis. The Coomassie gel (Fig. 8A) identified possible oligomeric, monomeric, and breakdown products of MBP-TcpH. These products were confirmed by Western analysis in which the membranes were probed with anti-TcpH-specific antibodies (Fig. 8B). Additional membranes also were probed with biotin-labeled purified His-TcpCΔ1-98, and the interaction with MBP-TcpH was detected with the streptavidin-horseradish peroxidase conjugate ExtrAvidin (Sigma). To control for interactions involving the MBP tag, MBP was analyzed alongside the MBP-TcpH sample. The results (Fig. 8C) indicated that His-TcpCΔ1-98 interacted with a large (>175-kDa) oligomer of TcpH. No reaction was observed with the TcpH monomer alone or with the MBP control. These results were in agreement with those of the interactions observed between TcpH and TcpC in the bacterial two-hybrid system.
FIG. 8.
TcpC interacts with oligomeric MBP-TcpH. Purified MBP (lane 1) and MBP-TcpH (lane 2) were separated on SDS-10% PAGE gels. (A) Coomassie staining of MBP and MBP-TcpH preparations. (B) Western blot of MBP and MBP-TcpH probed with TcpH-specific antibodies. (C) Far-Western blot of membrane preparations containing MBP and MBP-TcpH and then probed with biotin-labeled purified His-TcpCΔ1-98. The protein-protein interaction between His-TcpCΔ1-98 and MBP-TcpH was detected using ExtrAvidin peroxidase (Sigma). Approximate protein sizes are indicated (in kilodaltons). Arrows indicate the MBP-TcpH monomer (M) and oligomer (O).
TcpH is located in the membrane fraction in C. perfringens.
To determine the subcellular location of TcpH, cell lysates derived from the isogenic C. perfringens strains JIR4394 (pCW3 negative) and JIR4195 (pCW3 positive) were separated into cytoplasmic and crude membrane fractions. The fractions were subjected to SDS-12% PAGE and immunoblotting using TcpH-specific guinea pig antiserum (Fig. 9). The native TcpH protein was predicted to be approximately 93 kDa in size, and the TcpH-specific antiserum reacted with a protein band of approximately 100 kDa in both the lysate and concentrated membrane fractions derived from JIR4195, but there was no reaction with the cytoplasmic fraction (Fig. 9). No protein band was observed in the fractions derived from the pCW3-negative strain JIR4394, indicating that the protein band was pCW3 specific. In addition, the TcpH-specific antiserum did not react with a TcpH-specific protein in the membrane fractions derived from the tcpH mutant (JIR4885) (data not shown). This observation indicated that TcpH was, most likely, an integral membrane protein in C. perfringens, which was consistent with the results of our bioinformatic analysis.
FIG. 9.
Localization of TcpH by membrane fractionation in C. perfringens. Western blotting was carried out using TcpH-specific guinea pig antiserum on fractions derived from the pCW3-negative recipient strain JIR4394 (1) and JIR325(pCW3) (2). The membrane fractions were concentrated from two lysate preparations. The various fractions were subjected to SDS-12% PAGE, and the arrow indicates the TcpH-specific band. L, cell lysate; C, cytoplasmic; W, PBS wash; and M, membrane.
TcpH and TcpF are located at the poles in C. perfringens donor cells.
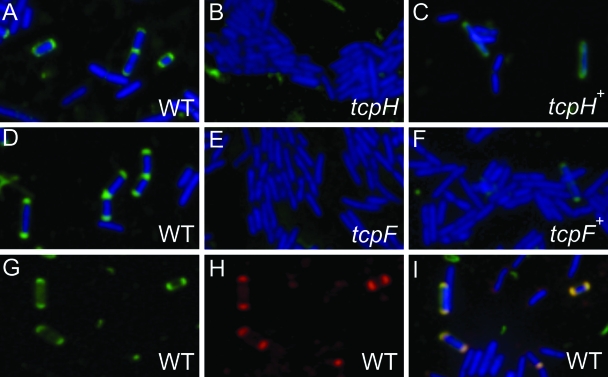
To determine the site(s) of mpf complex formation in C. perfringens donor cells, we examined the subcellular location of TcpH using immunofluorescence microscopy. C. perfringens cells were fixed using a paraformaldehyde solution and permeabilized by treatment with lysozyme. TcpH-specific guinea pig antiserum was used as the primary antibody and fluorescein-conjugated Alexa Fluor 488 goat anti-guinea pig as the secondary antibody. When freshly stained cells were viewed using a fluorescence microscope at ×1,000 magnification, it was clear that TcpH was localized to both cell poles of JIR325(pCW3) cells (approximately 30% of the cells) (Fig. 10A). As expected, the tcpH mutant did not show fluorescent polar foci (Fig. 10B), indicating that the fluorescent signal for JIR325(pCW3) was TcpH specific. As an additional negative control, JIR325(pCW3) cells were treated similarly, except that either the primary or secondary antibodies were omitted. There were no fluorescent polar foci observed in any of these cells (data not shown), again indicating the specificity of the immunofluorescence assay. Finally, the complementation of JIR325(pCW3tcpH) with the wild-type tcpH gene in trans restored the fluorescent polar foci in approximately 20% of the cells (Fig. 10C).
FIG. 10.
Localization of TcpH and TcpF by immunofluorescence microscopy. C. perfringens cells were labeled nonspecifically using the DAPI nuclear stain (blue), specifically using TcpH-specific (A to C) and TcpF-specific (D to F) primary antibodies, and corresponding Alexa Fluor 488(Green)-conjugated anti-guinea pig and anti-rabbit secondary antibodies, respectively. TcpH and TcpF were identified as green fluorescent polar foci. The C. perfringens strains analyzed were JIR325(pCW3) (A, D, and G to I), the tcpH mutant JIR4885 (B), the complemented tcpH mutant JIR4891 (C), the tcpF mutant JIR4940 (E), and the complemented tcpF mutant JIR4942 (F). For the colocalization of TcpF and TcpH by immunofluorescence microscopy, JIR325(pCW3) cells were probed concurrently with (G) TcpH-specific primary antibody and Alexa Fluor 488 (green)-conjugated anti-guinea pig secondary antibody and (H) TcpF-specific primary antibody and Alexa Fluor 568 (red)-conjugated anti-rabbit secondary antibody. The green fluorescent polar foci are TcpH specific, and the red fluorescent polar foci are TcpF specific. (I) Merged image of panels G and H and simultaneously DAPI stained cells.
Since we also had available a TcpF-specific antiserum derived from a rabbit immunized with a fusion construct containing a TcpF-specific peptide (aa 543 to 557) and pre-adsorbed against a cell lysate from JIR325(pCW3tcpF), we carried out similar immunofluorescence studies using this antiserum as the primary antibody. The objective was to determine the subcellular location of TcpF. Fluorescent polar foci were observed in a significant number of JIR325(pCW3) cells (approximately 50%), indicating that, like TcpH, TcpF was located at the cell poles in C. perfringens donor cells (Fig. 10D). The fluorescent polar foci were not observed in the tcpF mutant, as expected, but weak fluorescence was seen when the tcpF mutant was complemented in trans with the wild-type tcpF gene (approximately 1% of cells), providing evidence for the specificity of the reaction (Fig. 10E and F).
To determine if TcpF and TcpH were colocalized, JIR325(pCW3) cells were treated sequentially with TcpF- and TcpH-specific primary antibodies. Both TcpF and TcpH were localized to the cell poles as before (Fig. 10G and H, respectively), and when the images were merged, the fluorescent foci were coincident (Fig. 10I). This colocalization suggests a close association between TcpF and TcpH at the poles of donor cells. Overall, the polar foci observed in C. perfringens were consistent with the polar localization of VirB proteins, including VirB6, in A. tumefaciens (25), indicating that the putative pCW3-mpf complex most likely is assembled at the cell poles.
DISCUSSION
Conjugative plasmids that are very similar to pCW3 and have a closely related tcp region are critical for the dissemination of both antibiotic resistance and toxin genes in pathogenic isolates of C. perfringens (6, 7, 22, 37). The mpf complex is an essential apparatus for bacterial conjugation, and the pCW3-mpf complex most likely is formed by the Tcp proteins. TcpH was predicted to be an essential component of the pCW3-mpf complex, because it was a putative integral membrane protein and tcpH mutants were transfer deficient (6). In this study, the primary functional regions of TcpH were localized to the first 581 aa residues, which included a VirB6-like region. The self interaction of TcpH monomers and an interaction between TcpH and TcpC also was shown by using bacterial two-hybrid and biochemical analyses. This pCW3-mpf complex most likely is located at the poles, since by fluorescence immunomicroscopy both TcpH and TcpF were detected at the poles of C. perfringens donor cells.
Deletion analysis showed that the first 581 aa residues of TcpH were sufficient to restore at least some conjugative transfer to a tcpH mutant. These residues also were shown to be essential for TcpH homooligomer formation and for the formation of TcpH-TcpC oligomers, providing evidence that these protein-protein interactions play an important role in the conjugative transfer of pCW3 DNA. Since the shorter TcpH1-514 derivative was nonfunctional in both the conjugation and protein-protein interaction assays, it was concluded that the region between aa 514 and 581 either was crucial for the proper folding of the TcpH protein or contained active functional sites. By contrast, the TcpHΔ311-450 and the TcpH242VQAAA246 mutants were nonconjugative but still interacted with TcpH and TcpC. These results point out the functional complexity of the large TcpH protein and provide good evidence that different regions of the protein are required for conjugative transfer and for the homo- and heterotypic interactions of TcpH.
Coiled coils comprise two or more right-handed amphipathic α-helices that are wound around one another in a left-handed superhelix, forming the primary structural element in many proteins and also mediating heterooligomeric interactions in others (12, 34). In this study, the two predicted coiled-coil domains that are located in the C-terminal region of TcpH (aa 515 to 832) were shown not to be essential for TcpH-TcpH or TcpH-TcpC interactions. However, the inclusion of the TcpH region from aa 581 to 697 conferred a higher frequency of conjugative transfer in the tcpH mutant, indicating that this region does have a significant functional role in the conjugation process. Further studies are required to establish the role of this unique, presumably cytoplasmic, region of TcpH.
TcpC was able to interact with TcpH and has low-level identity (24%) to ORF13, which is required for the intercellular transfer of Tn916, although its precise function has not been determined (6, 47). Preliminary studies in this laboratory suggest that TcpC is involved in the conjugative transfer of pCW3 (R. Bantwal, T. Bannam, and J. Rood, unpublished results). There is a putative TMD in the N terminus of TcpC, which suggests that it may be membrane associated; results obtained from both the bacterial two-hybrid and biochemical studies suggest that it also is complexed with TcpH. Since no other Tcp proteins were found to interact directly with TcpH in the bacterial two-hybrid assay, TcpC may serve as an intermediate to facilitate further protein-protein interactions within the pCW3-mpf complex. However, a lack of functional complementation in the bacterial two-hybrid system does not necessarily prove that the native proteins do not associate with each other. Steric constraints could be imposed by the three-dimensional structure of the chimeric protein, thereby affecting the resultant adenylate cyclase activity (28). Therefore, there could be other potentially interacting partners of TcpH that need to be identified by other biochemical methods.
The membrane-associated proteins VirB6 to VirB10 are postulated to be the conserved core components of the A. tumefaciens-T4SS complex (8). The VirB6 (295 aa) protein is a polytopic inner membrane protein that forms part of the lumen of the T4SS (24). Similarly, the much larger TcpH protein (832 aa) is postulated to be membrane associated, because it contains eight putative N-terminal TMDs. In this study, the native TcpH protein was found in the membrane fraction of C. perfringens cells encoding pCW3. This result and those from the cross-linking studies are consistent with the hypothesis that TcpH is a membrane-associated protein that forms a complex that acts as a transmembrane channel, through which pCW3 DNA is translocated into the recipient cell. TcpC may act to link the TcpH complex to the likely coupling protein TcpA (40).
The polar localization of the conserved core components of the A. tumefaciens-T4SS complex (VirB6 to VirB10) and the proposed coupling protein VirD4 also indicates that the T4SS complex most likely is assembled at the cell pole (11, 25, 26, 29). Similarly, TcpF and TcpH were localized at the cell poles in C. perfringens. The fluorescent polar foci both were TcpF and TcpH specific, since they were not observed in the corresponding tcpF and tcpH mutants; most importantly, the fluorescent foci at the bacterial poles also were detected, although in lower numbers, in complemented mutant cells. The complementation data also indicate that these genes are not expressed at wild-type levels in the complemented strains, which is consistent with previous conjugation data (6).
In A. tumefaciens, the VirB proteins mostly are localized at one pole, which is consistent with the polar attachment of Agrobacterium cells to the plant (25, 26). However, bipolar staining was observed for TcpH and TcpF in most of the C. perfringens cells examined, suggesting that conjugation can occur at either end of a C. perfringens donor cell. By contrast, the R27-encoded coupling protein (TraG) and the mpf protein (TrhC), which is VirB4 related, form multiple fluorescent foci throughout the cell periphery (15, 18).
ATPases such as VirB4 typically are associated on the cytoplasmic side of the T4SS (10, 42). TcpF contains an ATP binding motif, and comparative sequence analysis showed that it has distant similarity to the VirB4 ATPase encoded by A. tumefaciens. The likely association of TcpF with the mpf complex was supported by the colocalization of TcpF and TcpH. Although TcpH did not interact with TcpF in the bacterial two-hybrid assay, it is possible that the other Tcp proteins function as intermediates to facilitate the interaction between TcpF and the mpf complex.
In summary, this study has determined the TcpH-specific regions essential for its function in conjugation, for homooligomerization, and for its interaction with TcpC. It was shown that the ability to carry out protein-protein interactions does not in itself confer conjugative function, since some transfer-deficient derivatives still were able to interact. It is postulated that TcpH forms a homooligomeric structure that spans the cell membrane and is in close association with another homooligomer, TcpC, as part of the mpf complex that is located at the cell poles. This complex is presumably in close association with the putative ATPase TcpF, facilitating the translocation of pCW3 DNA into the recipient cell.
Supplementary Material
Acknowledgments
We thank Daniel Ladant for kindly providing his bacterial two-hybrid assay system and Steve Morton for photographic assistance.
This research was supported by grants from the Australian Research Council to the ARC Centre of Excellence in Structural and Functional Microbial Genomics and grant AI056177-03 from the U.S. National Institute of Allergy and Infectious Diseases. W.L.T. was the recipient of a Monash postgraduate scholarship. J.A.P. was the recipient of a postgraduate scholarship awarded by the ARC Centre of Excellence and the Department of Microbiology.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 16 May 2008.
REFERENCES
- 1.Abraham, L. J., and J. I. Rood. 1985. Cloning and analysis of the Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid 13155-162. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., and E. V. Koonin. 1998. Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci. 23444-447. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, S. J., C. Daniels, and R. Morona. 1997. PhoP/Q-regulated genes in Salmonella typhi identification of melittin sensitive mutants. Microb. Pathogen. 22165-179. [DOI] [PubMed] [Google Scholar]
- 5.Bannam, T. L., and J. I. Rood. 1993. Clostridium perfringens-Escherichia coli shuttle vectors that carry single antibiotic resistance determinants. Plasmid 29223-235. [DOI] [PubMed] [Google Scholar]
- 6.Bannam, T. L., W. L. Teng, D. Bulach, D. Lyras, and J. I. Rood. 2006. Functional identification of conjugation and replication regions of the tetracycline resistance plasmid pCW3 from Clostridium perfringens. J. Bacteriol. 1884942-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brynestad, S., M. R. Sarker, B. A. McClane, P. E. Granum, and J. I. Rood. 2001. Enterotoxin plasmid from Clostridium perfringens is conjugative. Infect. Immun. 693483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie, P. J., and E. Cascales. 2005. Structural and dynamic properties of bacterial type IV secretion systems. Mol. Membr. Biol. 2251-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comp. Appl. Biosci. 10685-686. [DOI] [PubMed] [Google Scholar]
- 10.Dang, T. A., and P. J. Christie. 1997. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J. Bacteriol. 179453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das, A., and Y. H. Xie. 1998. Construction of transposon Tn3phoA: its application in defining the membrane topology of the Agrobacterium tumefaciens DNA transfer proteins. Mol. Microbiol. 27405-414. [DOI] [PubMed] [Google Scholar]
- 12.Delahay, R. M., and G. Frankel. 2002. Coiled-coil proteins associated with type III secretion systems: a versatile domain revisited. Mol. Microbiol. 45905-916. [DOI] [PubMed] [Google Scholar]
- 13.Firth, N., and R. Skurray. 1992. Characterization of the F plasmid bifunctional conjugation gene, traG. Mol. Gen. Genet. 232145-153. [DOI] [PubMed] [Google Scholar]
- 14.Flannagan, S. E., L. A. Zitzow, Y. A. Su, and D. B. Clewell. 1994. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid 32350-354. [DOI] [PubMed] [Google Scholar]
- 15.Gilmour, M. W., T. D. Lawley, M. M. Rooker, P. J. Newnham, and D. E. Taylor. 2001. Cellular location and temperature-dependent assembly of IncHI1 plasmid R27-encoded TrhC-associated conjugative transfer protein complexes. Mol. Microbiol. 42705-715. [DOI] [PubMed] [Google Scholar]
- 16.Gomis-Rüth, F. X., M. Sola, F. de la Cruz, and M. Coll. 2004. Coupling factors in macromolecular type-IV secretion machineries. Curr. Pharm. Des. 101551-1565. [DOI] [PubMed] [Google Scholar]
- 17.Guan, K., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192262-267. [DOI] [PubMed] [Google Scholar]
- 18.Gunton, J. E., M. W. Gilmour, G. Alonso, and D. E. Taylor. 2005. Subcellular localization and functional domains of the coupling protein, TraG, from IncHI1 plasmid R27. Microbiology 1513549-3561. [DOI] [PubMed] [Google Scholar]
- 19.Harry, E. J., K. Pogliano, and R. Losick. 1995. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 1773386-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14378-379. [DOI] [PubMed] [Google Scholar]
- 21.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 7751-59. [DOI] [PubMed] [Google Scholar]
- 22.Hughes, M. L., R. Poon, V. Adams, S. Sayeed, S. Saputo, F. A. Uzal, B. A. McClane, and J. I. Rood. 2007. Epsilon toxin plasmids of Clostridium perfringens type D are conjugative. J. Bacteriol. 1897531-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivey, D. M., and L. G. Ljungdahl. 1986. Purification and characterization of the F1-ATPase from Clostridium thermoaceticum. J. Bacteriol. 165252-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakubowski, S. J., V. Krishnamoorthy, E. Cascales, and P. J. Christie. 2004. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J. Mol. Biol. 341961-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Judd, P. K., R. B. Kumar, and A. Das. 2005. Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc. Natl. Acad. Sci. USA 10211498-11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Judd, P. K., R. B. Kumar, and A. Das. 2005. The type IV secretion apparatus protein VirB6 of Agrobacterium tumefaciens localizes to a cell pole. Mol. Microbiol. 55115-124. [DOI] [PubMed] [Google Scholar]
- 27.Judd, P. K., D. Mahli, and A. Das. 2005. Molecular characterization of the Agrobacterium tumefaciens DNA transfer protein VirB6. Microbiology 1513483-3492. [DOI] [PubMed] [Google Scholar]
- 28.Karimova, G., A. Ullmann, and D. Ladant. 2000. A bacterial two-hybrid system that exploits a cAMP signaling cascade in Escherichia coli. Methods Enzymol. 32859-73. [DOI] [PubMed] [Google Scholar]
- 29.Kumar, R. B., and A. Das. 2002. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol. Microbiol. 431523-1532. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 31.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 2241-15. [DOI] [PubMed] [Google Scholar]
- 32.Li, K. B. 2003. ClustalW-MPI: ClustalW analysis using distributed and parallel computing. Bioinformatics 191585-1586. [DOI] [PubMed] [Google Scholar]
- 33.Llosa, M., and F. de la Cruz. 2005. Bacterial conjugation: a potential tool for genomic engineering. Res. Microbiol. 1561-6. [DOI] [PubMed] [Google Scholar]
- 34.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21375-382. [PubMed] [Google Scholar]
- 35.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260289-298. [DOI] [PubMed] [Google Scholar]
- 37.Miyamoto, K., D. J. Fisher, J. Li, S. Sayeed, S. Akimoto, and B. A. McClane. 2006. Complete sequencing and diversity analysis of the enterotoxin-encoding plasmids in Clostridium perfringens type A non-food-borne human gastrointestinal disease isolates. J. Bacteriol. 1881585-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morelle, G. 1989. A plasmid extraction procedure on a miniprep scale. Focus 117-8. [Google Scholar]
- 39.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 239623-663. [DOI] [PubMed] [Google Scholar]
- 40.Parsons, J. A., T. L. Bannam, R. J. Devenish, and J. I. Rood. 2007. TcpA, an FtsK/SpoIIIE homolog, is essential for transfer of the conjugative plasmid pCW3 from Clostridium perfringens. J. Bacteriol. 1897782-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petit, L., M. Gibert, and M. R. Popoff. 1999. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 7104-110. [DOI] [PubMed] [Google Scholar]
- 42.Rashkova, S., G. M. Spudich, and P. J. Christie. 1997. Characterization of membrane and protein interaction determinants of the Agrobacterium tumefaciens VirB11 ATPase. J. Bacteriol. 179583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rood, J. I. 1983. Transferable tetracycline resistance in Clostridium perfringens strains of porcine origin. Can. J. Microbiol. 291241-1246. [DOI] [PubMed] [Google Scholar]
- 44.Rood, J. I. 1998. Virulence genes of Clostridium perfringens. Annu. Rev. Microbiol. 52333-360. [DOI] [PubMed] [Google Scholar]
- 45.Rood, J. I., and S. T. Cole. 1991. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol. Rev. 55621-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rood, J. I., V. N. Scott, and C. L. Duncan. 1978. Identification of a transferable resistance plasmid (pCW3) from Clostridium perfringens. Plasmid 1563-570. [DOI] [PubMed] [Google Scholar]
- 47.Senghas, E., J. M. Jones, M. Yamamoto, C. Gawron-Burke, and D. B. Clewell. 1988. Genetic organization of the bacterial conjugative transposon Tn916. J. Bacteriol. 170245-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Songer, J. G. 1997. Clostridial diseases of animals, p. 153-182. In J. I. Rood, B. A. McClane, J. G. Songer, and R. W. Titball (ed.), The clostridia: molecular biology and pathogenesis. Academic Press, London, United Kingdom.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.