Abstract
Proteolytic processes often participate in signal transduction across bacterial membranes. In Salmonella enterica serovar Typhimurium, the transcriptional regulator CadC activates genes of lysine decarboxylase system in response to external acidification and exogenous lysine. However, the signaling mechanism of CadC activation remains unexplored. We report here that CadC is located on the inner membrane under normal growth conditions but rapidly cleaved under acid stress conditions, leading to the induction of target gene transcription. As full-length CadC is degraded, the N-terminal fragment containing the DNA-binding domain accumulates in the inner membrane. Moreover, we show that C-terminal truncations of CadC abolish its degradation, resulting in complete loss of activator function. Together, these observations suggest that site-specific proteolysis at the periplasmic domain of CadC generates a biologically active form of N-terminal DNA-binding domain to promote target gene activation.
The enteric pathogen Salmonella enterica serovar Typhimurium encounters a number of acidic environments during infection of a mammalian host (6, 14, 29). Acid pH often acts as a signal and triggers changes in gene expression profiles (24, 36). Acid-sensing protein CadC is a transcriptional regulator that activates the cadBA operon encoding lysine decarboxylase CadA and lysine-cadaverine antiporter CadB, under conditions of low external pH and exogenous lysine (21, 25, 28, 39). Upon induction of the Cad system, lysine is imported into the cell through CadB and decarboxylated to cadaverine by CadA, contributing to the acid tolerance response (13, 26).
Comparative sequence analysis predicts that S. enterica serovar Typhimurium CadC may be a membrane-spanning protein with an N-terminal cytoplasmic DNA-binding domain and a C-terminal periplasmic domain (19, 39), although there has been no direct demonstration for this. The low pH and lysine signal is believed to be sensed directly or indirectly by CadC and transmitted across the membrane (10, 35). Analysis of the CadC amino acid sequence also predicts it to be a member of the ToxR-like family rather than a member of the two-component system (19, 25), which is a phospho-relay cascade from the membrane-bound autophosphorylating sensor kinase to its corresponding cytosolic response regulator (4, 41). CadC, like the Vibrio cholerae ToxR regulator, has no kinase activity. In addition, although their cytoplasmic domains show sequence similarity to the DNA-binding domains of OmpR-like response regulators of two-component systems, they do not contain the phosphoryl-acceptor domain (2, 10, 39). Thus, the ToxR-like family, including CadC, represents the simplest form of transmembrane signaling system, since the same protein acts as both a signal sensor and response regulator. However, the question of what molecular mechanisms control the activity of CadC, and how CadC reaches its cytoplasmic targets have never been explored.
In a previous study, we reported that transcription of S. enterica serovar Typhimurium cadC is induced by low pH and lysine signal (21). This observation prompted us to examine whether the CadC protein level corresponds to its mRNA level. Surprisingly, we found that the full-length CadC-HA protein is degraded rapidly to undetectable levels in response to acid stress (pH 5.8, 10 mM lysine). This led to the prediction that S. enterica serovar Typhimurium CadC may be a transmembrane protein that undergoes regulated proteolysis by which it is cleaved and activated from a latent state (11). To date, only a few examples of proteolytic transmembrane signaling mechanism have been functionally characterized in bacteria (8, 23, 27, 32, 43). Furthermore, very little is known about this mechanism in bacterial transmembrane activators.
This study was designed to test the hypothesis that the activation of membrane-bound transcriptional regulator CadC is governed by a proteolytic mechanism. We present evidence that regulated proteolysis participates in CadC signal transduction. Our results provide new insights into the mechanism of action of membrane-bound transcriptional activators in bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. enterica serovar Typhimurium UK1 wild-type and ΔcadC strains have been described previously (9, 21). Escherichia coli DH5α was purchased from Gibco-BRL. The plasmids pGEM-T Easy vector (Promega), pACYC184 (NEB), pMW118 (Nippon Gene), and pBAD24 (16) were used for the construction of expression vectors. Cells were routinely cultured at 37°C in LB complex medium or Vogel and Bonner E minimal medium supplemented with 0.4% glucose (22, 38). Lysine decarboxylase broth (0.5% peptone, 0.3% yeast extract, 0.1% dextrose, 0.5% l-lysine, and 0.002% bromcresol purple) was used for the lysine decarboxylase (LDC) assay. When required, ampicillin or chloramphenicol was added to the medium to a final concentration of 60 or 30 μg per ml, respectively. l-lysine was added to a final concentration of 10 mM.
Plasmid construction.
PCR and cloning for plasmid construction were performed by using standard techniques (31). Restriction enzymes, T4 DNA ligase, and Taq polymerase were purchased from TaKaRa. The primers used in the present study are listed in Table 1. pACYC184-HA-CadC was constructed by PCR amplification of the cadC gene and its promoter from S. enterica serovar Typhimurium chromosomal DNA using the two primer pairs, PcadC-F/PcadC-R and ORFcadC-F/ORFcadC-R. pACYC184-CadC-HA was constructed in a similar manner using a pair of primers, PcadC-F/CadCHA-R. For the construction of pBAD24-CadC-HA, the promoterless cadC gene was amplified by PCR using the primers, pBADcadC-F/CadCHA-R. Five C-terminal truncated forms of CadC were generated by PCR using the primer pairs PcadC-F/CadC162-R, PcadC-F/CadC183-R, PcadC-F/CadC200-R, PcadC-F/CadC210-R, and PcadC-F/CadC220-R, respectively. Construct integrity was verified by DNA sequencing.
TABLE 1.
Sequences of primers used in this study
| Primer | Sequence (5′→3′)a | Use |
|---|---|---|
| PcadC-F | CGGGATCCCGGTAGGCTTTATGCAGTT | N-terminal HA tagging |
| PcadC-R | CGGAATTCAGCGTAGTCTGGGACGTCGTATGGGTACATAAAAAGATGCCCTAACG | |
| ORFcadC-F | CGGAATTCCAGCAACCTGTTGTACGCAT | |
| ORFcadC-R | CCAAGCTTTTAATCTTCTGCCAGAAAACTATT | |
| CadCHA-R | CCAAGCTTTTAAGCGTAGTCTGGGACGTCGTATGGGTAATCTTCTGCCAGAAAACTATT | C-terminal HA tagging |
| pBADcadC-F | CGGAATTCATGCAGCAACCTGTTGTA | Arabinose induction |
| CadC162-R | CCAAGCTTTTAGAAAGCGGCAATACGCGG | |
| CadC183-R | CCAAGCTTTTACACCGACATCACGATAAACA | CadC truncation |
| CadC200-R | CCAAGCTTTTATCGTGGGTTAAGTAATAACC | |
| CadC210-R | CCAAGCTTTTAATTGCCGCCTTCAAAACGAA | |
| CadC220-R | CCAAGCTTTTAGGACTCCTGCGAGACCCAGT | |
| CadART-F | AAGAGCCTATTCGTGAACTG | RT-PCR |
| CadART-R | TCATCATCAGATGGGTCAG | |
| 16S RNA-F | AGAGTTTGATCMTGGCTCAG | |
| 16S RNA-R | TACGGYTACCTTGTTACGACTT |
Restriction enzyme recognition sequences are underlined, and the HA sequences are in boldface.
LDC assay.
Strains were grown in Moeller LDC broth (Difco) containing the decarboxylase basal medium supplemented with 0.5% l-lysine and bromcresol purple indicator. One or two colonies were inoculated into fresh culture media. Sterile mineral oil was then added, and the culture was incubated for 36 h at 37°C. If the dextrose is fermented, the medium turns yellow initially, then purple as the decarboxylase reaction elevates pH.
Salmonella subcellular fractionation.
Subcellular fractionation was performed as described previously (20) with slight modifications. Cells were harvested by centrifugation and resuspended in 800 μl of 100 mM Tris-HCl buffer (pH 8.6) containing 500 mM sucrose and 0.5 mM EDTA. Lysozyme (5 μl of a 50-mg/ml stock solution) was added, followed immediately by the addition of 3.2 ml of 50 mM Tris-HCl buffer (pH 8.6) containing 250 mM sucrose, 0.25 mM EDTA, and 2.5 mM MgCl2. After gentle agitation, the suspension was incubated for 15 min on ice. Cells were collected by centrifugation at 7,000 × g for 10 min. Cells resuspended in 4 ml of 20 mM Tris-HCl (pH 8.6) were lysed by sonication and centrifuged at 7,000 × g at 4°C for 10 min to remove unbroken cells. The supernatant was then centrifuged at 132,000 × g at 4°C for 1 h to separate the soluble fraction and the insoluble cell envelopes. To isolate the outer membrane fraction, total-envelope pellets were suspended in 4 ml of 20 mM Tris-HCl (pH 8.6) containing 1% Sarkosyl, followed by incubation for 30 min on ice. The outer membrane fraction was obtained as a pellet after centrifugation at 132,000 × g at 4°C for 1 h. The pellet was resuspended in 4 ml of 20 mM Tris-HCl buffer (pH 8.6).
Immunoblot analysis.
Immunoblotting was performed using standard procedures (31). Samples were resuspended or diluted in sodium dodecyl sulfate (SDS) sample buffer (60 mM Tris-HCl [pH 6.8], 30% glycerol, 2% SDS, 0.1% bromophenol blue, 14.4 mM 2-mercaptoethanol) and boiled for 10 min. Proteins were electrophoresed on a 12 or 14% SDS-polyacrylamide gel and blotted onto nitrocellulose membrane. The blots were probed with mouse monoclonal antibodies against the HA tag (Cell Signaling Technology) or DnaK (Stressgen) or with rabbit polyclonal antibody against OmpW (kindly provided by H. Y. Kang, Pusan National University, Pusan, Korea). Horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit immunoglobulin G (Bio-Rad) was used as the secondary antibody. Proteins were visualized by using enhanced chemiluminescence ECL (Amersham).
RNA preparation and RT-PCR assay.
For reverse transcription-PCR (RT-PCR), cultured cells were collected and incubated with RNA protect bacteria reagent (Qiagen) and disrupted with lysozyme (1 mg/ml). Total RNA was isolated by using the RNeasy minikit (Qiagen) and treated with DNase I. The quantity and purity of RNA was determined by measuring absorbance at 260 and 280 nm. Total RNA (1 μg) was reverse transcribed into cDNA with random primers and amplified by using gene-specific primer sets (Table 1). The reaction was denatured (94°C, 4 min), followed by 18 thermal cycles (94°C for 30 s, 54°C for 30 s, and 72°C for 50 s) and a final extension (72°C for 10 min). Primer pair CadART-F/CadART-R was used to detect the cadA transcript. 16S rRNA was used as a normalization control. Amplified products were separated on a 1.5% agarose gel, stained with ethidium bromide, and visualized.
RESULTS
CadC is located on the inner membrane of S. enterica serovar Typhimurium.
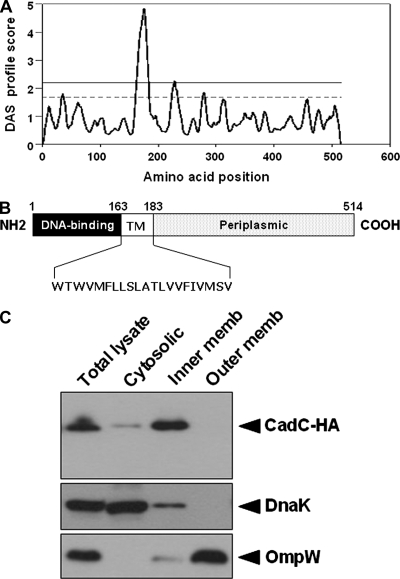
It has not yet been determined experimentally whether CadC is a membrane-spanning protein in S. enterica serovar Typhimurium. Therefore, as a first strategy to identify its exact cellular location, the DAS program (Stockholm University) was used to predict CadC transmembrane regions. CadC was predicted to have one transmembrane domain at amino acid residues 163 to 183 (Fig. 1A). A schematic diagram is shown in Fig. 1B, based on the DAS output and sequence alignment.
FIG. 1.
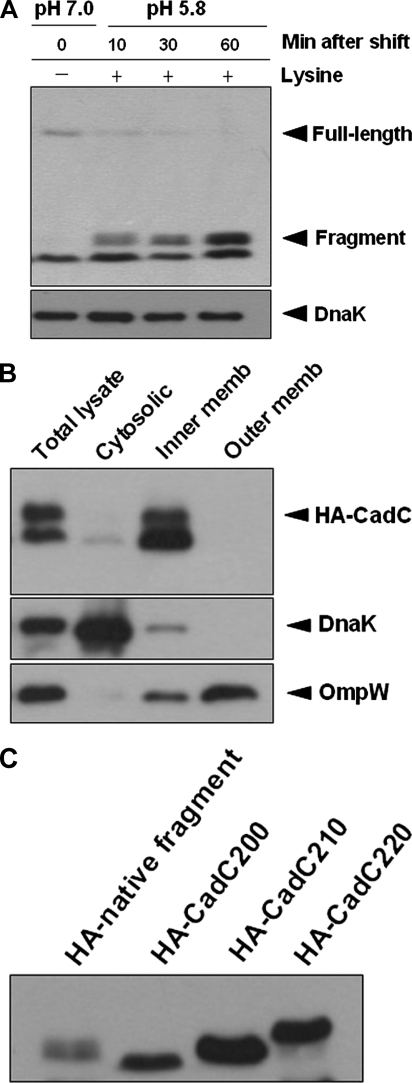
CadC is a membrane-bound protein in S. enterica serovar Typhimurium. (A) Prediction of transmembrane segments by the DAS program. The upper strict cutoff at 2.2 is informative in terms of the number of matching segments. The lower loose cutoff at 1.7 gives the actual location of the transmembrane segment. (B) Schematic representation of CadC based on the DAS output and sequence alignment. The numbers point to relevant amino acid residues. Residues 163 and 183 mark the boundaries of the transmembrane segment (TM). (C) Subcellular localization of HA-tagged CadC. The pACYC184-CadC-HA plasmid was expressed in the ΔcadC strain. Cells were grown in E glucose medium to an OD600 of 0.6, and then the total cell lysates were subjected to fractionation as described in Materials and Methods. The fractions were analyzed by immunoblotting with an anti-HA antibody. DnaK and OmpW were used as controls.
We next performed subcellular fractionation analysis to test the DAS prediction. CadC was tagged at its C terminus with a hemagglutinin (HA) epitope and expressed from its own promoter in the low-copy plasmid pACYC184. Introduction of the resulting plasmid, pACYC184-CadC-HA, into the ΔcadC strain complemented the LDC-negative phenotype, indicating that the HA tag does not affect the activity of CadC (data not shown). Cells grown under normal growth conditions were disrupted by sonication, and then the total cell lysate was fractionated by ultracentrifugation to separate cytosolic and membrane fractions. The inner membrane fraction was released by treatment with the ionic detergent Sarkosyl. The prepared fractions were analyzed by immunoblotting for the presence of CadC protein. DnaK was used as a control since it is known to be always associated with the inner membrane (7). OmpW was used as an outer membrane marker, but the inner membrane fraction by the Sarkosyl method usually contains some outer membrane contamination. As shown in Fig. 1C, the majority of HA-tagged CadC was contained within the inner membrane fraction under nonstress conditions.
CadC is rapidly degraded by low pH and lysine signal.
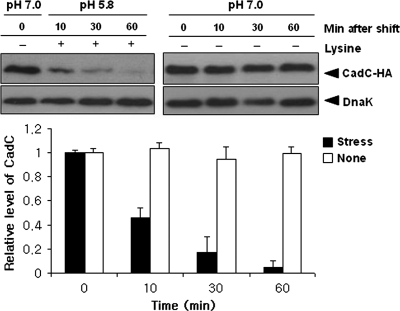
We previously showed that after acid stress (pH 5.8, 10 mM lysine) the levels of cadC mRNA increase rapidly (21). To examine whether the CadC protein level corresponds to its mRNA level, immunoblot analysis was conducted on total protein prepared from the ΔcadC strain harboring pACYC184-CadC-HA. C-terminally HA-tagged CadC (CadC-HA) was expressed under the control of its own promoter to ensure regulation as observed in wild-type. Cells were grown in E glucose medium to an optical density at 600 nm (OD600) of 0.6 and subjected to acid stress. Samples were taken at various time points and then immunoblotted with anti-HA antibodies. As shown in Fig. 2, CadC-HA levels decreased rapidly after acid stress, while they remained unchanged in neutral pH. We obtained the same result using the very-low-copy-number plasmid pMW118, a derivative of pSC101 (data not shown). This striking result raised the possibility that proteolytic cleavage could be a molecular mechanism of acid stress-mediated CadC activation.
FIG. 2.
Acid stress-induced proteolysis of CadC. The ΔcadC strain harboring pACYC184-CadC-HA (a derivative of pACYC184 carrying the cadC-HA gene under the control of the cadC promoter) was grown in E glucose medium to an OD600 of 0.6 and then subjected to acid stress (pH 5.8, 10 mM lysine). Samples were taken at the indicated times, and total proteins were analyzed by immunoblotting with anti-HA and anti-DnaK antibodies. DnaK was used as a loading control. The intensities of these bands were normalized to the band at time zero. Three independent experiments were performed for each strain, and the standard deviation is indicated by error bars.
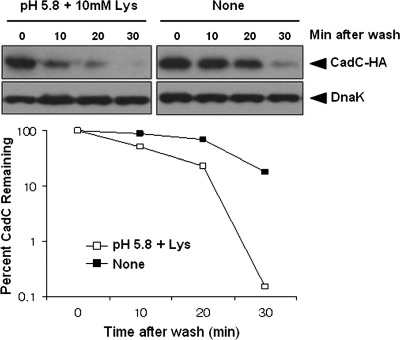
To further confirm CadC degradation, we constructed a pBAD24 derivative (pBAD24-CadC-HA) that expressed CadC-HA under the control of araBAD promoter, which is strictly regulated by the concentration of extracellular arabinose. Cells harboring pBAD24-CadC-HA were grown in E glucose medium to early log phase (OD600 of 0.4), and then arabinose was added to induce CadC expression. After 1 h, the cultures were washed to remove remaining arabinose, and growth was continued under nonstress or acid stress conditions. The results in Fig. 3 show that CadC is somewhat stable through the first 20 min under nonstress conditions, but its stability is strongly decreased under stress conditions. This suggests that CadC undergoes posttranslational proteolysis in response to low pH and lysine signal.
FIG. 3.
Stability of CadC under stress and nonstress conditions. The ΔcadC strain harboring pBAD24-CadC-HA (a derivative of pBAD24 carrying the cadC-HA gene under the control of the inducible araBAD promoter) was grown in E glucose medium to an OD600 of 0.4 and then induced with arabinose (5 mM). After 1 h, the medium was removed and replaced with fresh, arabinose-free medium. Subsequently, the culture was divided, and one portion was subjected to acid stress at time zero. Samples were taken at the indicated times, and total proteins were analyzed by immunoblotting with anti-HA and anti-DnaK antibodies. A representative data set is shown.
Proteolysis of CadC leads to the induction of cadBA operon.
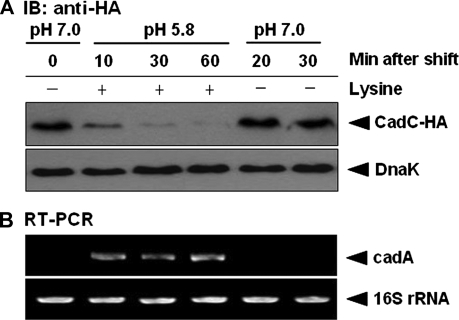
The transcriptional induction of the cadBA operon cannot be explained by the mere disappearance of CadC, since CadC is a transcriptional activator of this operon (21). Thus, we reasoned that CadC degradation would be a major mechanism for target gene regulation. To examine the effects of CadC proteolysis on cadA mRNA synthesis, reverse transcription-PCR (RT-PCR) analysis was conducted on total RNA isolated from the ΔcadC strain harboring pACYC184-CadC-HA. Cells were grown in E glucose medium to an OD600 of 0.6 and exposed to acid stress for 60 min before being shifted back to normal growth conditions. Figure 4A shows that CadC degradation occurs strictly in response to acid stress and is coupled to induction of cadA transcription. After a shift from acid stress conditions to normal growth conditions, CadC returned to basal levels and cadA transcription was precluded (Fig. 4B). These results indicate that the immediate induction of cadA mRNA following stress is correlated with rapid cleavage of CadC.
FIG. 4.
CadC degradation is coupled to transcriptional induction of cadA. (A) Immunoblot (IB) analysis of acid stress-induced CadC degradation in the ΔcadC strain harboring pACYC184-CadC-HA. Cells were grown in E glucose medium to an OD600 of 0.6, followed by acid stress. After 60 min of incubation, the cells were shifted from acid stress conditions to normal growth conditions. DnaK was used as a loading control. (B) RT-PCR analysis of cadA transcriptional induction following acid stress. Total RNA was extracted from the same samples described in panel A. 16S rRNA was used as a loading control.
Full-length CadC disappears and the CadC fragment accumulates in the inner membrane.
To determine whether a fragment containing the cytoplasmic domain of CadC is produced by proteolytic cleavage, a similar analysis was carried out with the N-terminally HA-tagged CadC (HA-CadC). As expected, the N-terminal CadC fragment was shown to accumulate as the full-length CadC disappeared (Fig. 5A). The observed CadC fragment was slightly larger than the expected size of the cytoplasmic domain, implying that it contains the transmembrane domain as well as the cytoplasmic domain. We therefore performed subcellular fractionation analysis after 60 min of acid stress to determine the location of the N-terminal CadC fragment. Figure 5B shows that the CadC fragment is present in the inner membrane fraction only, indicating that the N-terminal cytoplasmic domain is not released from the membrane. Importantly, CadC fragment accumulated to levels greater than those of the preexisting full-length CadC, indicating that CadC protein expression is also induced by acid stress similarly to mRNA expression. To further determine the approximate site of proteolytic cleavage, we constructed three C-terminal truncations of HA-CadC ending at residues 200, 210, and 220. Their electrophoretic mobility was compared to the mobility of the native N-terminal CadC fragment after 60 min of acid stress. As shown in Fig. 5C, HA-CadC210 (residues 1 to 210) nearly comigrated with the native cleavage product, indicating that CadC undergoes proteolytic cleavage in the vicinity of residue 210, which is 27 amino acids distal to the periplasmic end of the predicted transmembrane segment. These results suggest that CadC indeed undergoes proteolytic cleavage to generate a biologically active form of N-terminal DNA-binding domain, which binds to the target gene promoter.
FIG. 5.
N-terminal fragment accumulates in the inner membrane during the degradation of CadC. (A) pACYC184-HA-CadC, encoding a CadC derivative with an N-terminal HA tag (HA-CadC), was expressed under the control of the cadC promoter in the ΔcadC strain. Cells were grown and processed as in Fig. 2. (B) Subcellular localization of the N-terminal CadC fragment after 60 min of acid stress. Cells were grown in E glucose medium at 37°C to an OD600 of 0.6 and subjected to acid stress for 60 min prior to fractionation. Total cell lysates were then fractionated as described in Materials and Methods. (C) Comparison of the electrophoretic migration of the indicated proteins in SDS-polyacrylamide gel. C-terminal truncations HA-CadC200 (residues 1 to 200), HA-CadC210 (residues 1 to 210), and HA-CadC220 (residues 1 to 220) were constructed by inserting a stop codon downstream of the last desired amino acid codon.
C-terminal truncations of CadC abolish its proteolytic activation.
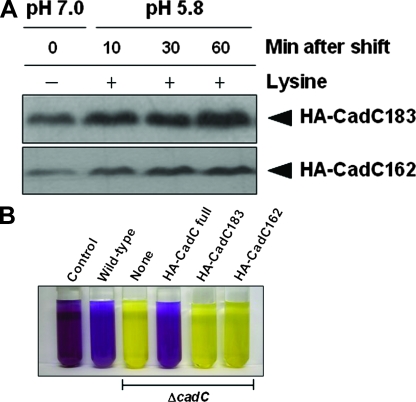
To test the importance of the periplasmic domain for CadC degradation, we constructed two additional C-terminally truncated versions of CadC with an N-terminal HA tag. HA-CadC183 (residues 1 to 183) lacks the entire C-terminal periplasmic domain. HA-CadC162 (residues 1 to 162) lacks both the periplasmic and the transmembrane domains. As shown in Fig. 6A, C-terminal truncations were not degraded and accumulated following acid stress. In addition, HA-CadC183 clearly migrated faster than the native N-terminal CadC fragment in SDS-gels (data not shown), as can be expected from Fig. 5C. To determine whether the periplasmic portion of the native N-terminal CadC fragment is indispensable for its transcriptional activator function, we performed LDC assay using these two C-terminal truncations. In contrast to full-length CadC, both HA-CadC183 and HA-CadC162 cannot complement the LDC-negative phenotype of the cadC knockout strain (Fig. 6B), indicating functional inactivation of these truncated proteins. Together, these findings suggest that site-specific proteolysis at the periplasmic domain of CadC generates a transmembrane signal that switches on the expression of the cadBA operon.
FIG. 6.
Cellular accumulation and functional inactivation of C-terminally truncated CadC. (A) C-terminal truncations of CadC abolish its degradation. HA-CadC183 (residues 1 to 183) and HA-CadC162 (residues 1 to 162) were expressed from pACYC184 in the ΔcadC strain. An HA epitope tag was added to the N terminus. Cells were grown and processed as described in Fig. 2. (B) Effect of C-terminal truncations on CadC activity. LDC assay indicates that both HA-CadC183 and HA-CadC162 have no function in transcriptional activation of the cadBA promoter. Yellow and purple indicate the absence and presence, respectively, of LDC.
DISCUSSION
The central finding of our study is that a membrane-bound transcriptional regulator CadC is activated by proteolytic cleavage in S. enterica serovar Typhimurium. Although the Cad system is relatively well studied in enteric bacteria (21, 25, 28, 39), the molecular mechanisms governing the activation of CadC have thus far remained unknown. The present study implicates a proteolytic mechanism in CadC signal transduction, demonstrating that, upon acid stress, the N-terminal CadC fragment that contains the DNA-binding domain is produced by proteolytic cleavage and thereby functions as a transcriptional activator of cadBA operon.
We have noted that the full-length CadC-HA level does not correlate with its mRNA expression upon acid stress (Fig. 2). To our knowledge, protein levels often correlate poorly or not at all with mRNA expression due to posttranscriptional or posttranslational regulation, and it is thus insufficient to predict protein levels from mRNA transcript levels (17). Therefore, we reasoned that acid stress-induced proteolysis of CadC might be a posttranslational control mechanism for activating membrane-bound CadC because the induction of cadA transcription is accompanied by proteolysis of CadC (Fig. 4).
In recent years, a pivotal role for proteolysis in signal transduction has been reported (11, 37). The data presented here, however, show that S. enterica serovar Typhimurium CadC is activated by a proteolytic mechanism distinct from those described previously (8, 23, 27, 32, 43). Staphylococcal resistance to β-lactam antibiotics is known to be regulated by a transmembrane signal transmitted by sequential proteolytic events. After β-lactam antibiotic binding to the transmembrane sensor-transducer BlaR1, the cytoplasmic transducer domain which contains zinc metalloprotease sequences is proteolytically removed from the membrane and then cleaves the BlaI repressor, allowing transcription of the β-lactamase gene (1, 43). In addition, transmembrane dormant transcription factors can also be cleaved within the transmembrane segment to release cytoplasmic fragments from the membrane. This mechanism, termed regulated intramembrane proteolysis (RIP), is an activation strategy that enables rapid transcriptional responses to an incoming signal (18, 40). RIP was found to function in a wide range of organisms from bacteria to humans and was shown to control diverse cellular functions (5). However, most studies of bacterial RIP have concentrated on the activation of alternative σ factors (8, 27, 32), and very little is known about other bacterial transcriptional activators regulated via the RIP mechanism.
One recently characterized example is TcpP, a membrane-localized virulence activator in V. cholerae. In contrast to the common RIP mechanism that controls transcription in a positive fashion, however, TcpP is degraded by the RIP protease YaeL when it is not needed for virulence gene activation (3, 23). Thus, the biological role of TcpP degradation is fundamentally different from that of CadC degradation. Moreover, we discovered that the N-terminal CadC fragment remains localized to the inner membrane after proteolytic cleavage. In many of the known eukaryotic RIP, the cytoplasmic domain of the membrane-bound transcription factor is liberated from the membrane and enters the nucleus to regulate gene transcription (30, 34, 42). Thus, the proteolytic transmembrane signaling of CadC appears to differ from that in eukaryotic transcription factors. This may be because bacterial chromosomal DNA is highly dynamic and not contained within a cell nucleus (15, 33); therefore, there is no essential requirement for release from the membrane because the cytoplasmic domain is unhindered in its access to target gene promoters.
One possible explanation for our results is that CadC signal transduction may be mediated by proteolysis-induced conformational changes in the cytoplasmic DNA-binding domain. Although HA-CadC183 and HA-CadC162 contain the DNA-binding domain, they have no transcriptional regulatory activity (Fig. 6). Therefore, we can speculate that these C-terminal truncations may remove residues critical for inducing and maintaining the functional conformation of the DNA-binding domain. Importantly, it has been demonstrated that in the bacterial chemosensory system, ligand-binding to the external domain of the transmembrane chemoreceptor induces a conformational change in the cytoplasmic domain (12). Thus, it will be interesting to determine whether proteolytic cleavage at the periplasmic domain of CadC generates a conformational signal that modulates its DNA-binding activity.
In summary, the principal contribution of the present study is the discovery of a novel type of proteolytic transmembrane signaling pathway in S. enterica serovar Typhimurium. Future studies including the identification of the protease(s) responsible for CadC proteolysis will further expand the knowledge of proteolysis-mediated activation of membrane-bound transcriptional regulators in the bacterial cell.
Acknowledgments
We thank Ho Young Kang for the anti-OmpW antibody and all lab members for helpful discussions and suggestions.
This research was supported by grant R01-2006-000-10125-0 from the Korea Science and Engineering Foundation and partly supported by research grant of Chosun University.
Footnotes
Published ahead of print on 16 May 2008.
REFERENCES
- 1.Archer, G. L., and J. M. Bosilevac. 2001. Signaling antibiotic resistance in staphylococci. Science 2911915-1916. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18715-727. [DOI] [PubMed] [Google Scholar]
- 3.Beck, N. A., E. S. Krukonis, and V. J. DiRita. 2004. TcpH influences virulence gene expression in Vibrio cholerae by inhibiting degradation of the transcription activator TcpP. J. Bacteriol. 1868309-8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beier, D., and R. Gross. 2006. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 9143-152. [DOI] [PubMed] [Google Scholar]
- 5.Brown, M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100391-398. [DOI] [PubMed] [Google Scholar]
- 6.Brumell, J. H., and S. Grinstein. 2004. Salmonella redirects phagosomal maturation. Curr. Opin. Microbiol. 778-84. [DOI] [PubMed] [Google Scholar]
- 7.Bukau, B., P. Reilly, J. McCarty, and G. C. Walker. 1993. Immunogold localization of the DnaK heat shock protein in Escherichia coli cells. J. Gen. Microbiol. 13995-99. [DOI] [PubMed] [Google Scholar]
- 8.Chaba, R., I. L. Grigorova, J. M. Flynn, T. A. Baker, and C. A. Gross. 2007. Design principles of the proteolytic cascade governing the σE-mediated envelope stress response in Escherichia coli: keys to graded, buffered, and rapid signal transduction. Genes Dev. 21124-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtiss, R., III, and J. O. Hassan. 1996. Nonrecombinant and recombinant avirulent Salmonella vaccines for poultry. Vet. Immunol. Immunopathol. 54365-372. [DOI] [PubMed] [Google Scholar]
- 10.Dell, C. L., M. N. Neely, and E. R. Olson. 1994. Altered pH and lysine signalling mutants of cadC, a gene encoding a membrane-bound transcriptional activator of the Escherichia coli cadBA operon. Mol. Microbiol. 147-16. [DOI] [PubMed] [Google Scholar]
- 11.Ehrmann, M., and T. Clausen. 2004. Proteolysis as a regulatory mechanism. Annu. Rev. Genet. 38709-724. [DOI] [PubMed] [Google Scholar]
- 12.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster, J. W. 2000. Microbial responses to acid stress, p. 99-115. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, DC.
- 14.Foster, J. W., and M. Spector. 1995. How Salmonella survives against the odds. Annu. Rev. Microbiol. 49145-174. [DOI] [PubMed] [Google Scholar]
- 15.Gitai, Z., M. Thanbichler, and L. Shapiro. 2005. The choreographed dynamics of bacterial chromosomes. Trends Microbiol. 13221-228. [DOI] [PubMed] [Google Scholar]
- 16.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gygi, S. P., Y. Rochon, B. R. Franza, and R. Aebersold. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 191720-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoppe, T., M. Rape, and S. Jentsch. 2001. Membrane-bound transcription factors: regulated release by RIP or RUP. Curr. Opin. Cell Biol. 13344-348. [DOI] [PubMed] [Google Scholar]
- 19.Kim, B. H., H. J. Lee, I. S. Lee, S. H. Bang, J. Kim, and Y. K. Park. 2001. Cloning and sequencing analysis of cadC encoding transcriptional activator CadC from Salmonella typhimurium. J. Microbiol. 39109-115. [Google Scholar]
- 20.Konjufca, V., S. Y. Wanda, M. C. Jenkins, and R. Curtiss III. 2006. A recombinant attenuated Salmonella enterica serovar Typhimurium vaccine encoding Eimeria acervulina antigen offers protection against E. acervulina challenge. Infect. Immun. 746785-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, Y. H., B. H. Kim, J. H. Kim, W. S. Yoon, S. H. Bang, and Y. K. Park. 2007. CadC has a global translational effect during acid adaptation in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1892417-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maloy, S. R., and J. R. Roth. 1983. Regulation of proline utilization in Salmonella typhimurium: characterization of put::Mu d(Ap, lac) operon fusions. J. Bacteriol. 154561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matson, J. S., and V. J. DiRita. 2005. Degradation of the membrane-localized virulence activator TcpP by the YaeL protease in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 10216403-16408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGowan, C. C., A. S. Necheva, M. H. Forsyth, T. L. Cover, and M. J. Blaser. 2003. Promoter analysis of Helicobacter pylori genes with enhanced expression at low pH. Mol. Microbiol. 481225-1239. [DOI] [PubMed] [Google Scholar]
- 25.Merrell, D. S., and A. Camilli. 2000. Regulation of Vibrio cholerae genes required for acid tolerance by a member of the “ToxR-like” family of transcriptional regulators. J. Bacteriol. 1825342-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, Y. K., B. Bearson, S. H. Bang, I. S. Bang, and J. W. Foster. 1996. Internal pH crisis, lysine decarboxylase and the acid tolerance response of Salmonella typhimurium. Mol. Microbiol. 20605-611. [DOI] [PubMed] [Google Scholar]
- 27.Qiu, D., V. M. Eisinger, D. W. Rowen, and H. D. Yu. 2007. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1048107-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhee, J. E., K. S. Kim, and S. H. Choi. 2005. CadC activates pH-dependent expression of the Vibrio vulnificus cadBA operon at a distance through direct binding to an upstream region. J. Bacteriol. 1877870-7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rychlik, I., and P. A. Barrow. 2005. Salmonella stress management and its relevance to behavior during intestinal colonization and infection. FEMS Microbiol. Rev. 291021-1040. [DOI] [PubMed] [Google Scholar]
- 30.Sakai, J., E. A. Duncan, R. B. Rawson, X. Hua, M. S. Brown, and J. L. Goldstein. 1996. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell 851037-1046. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York, NY.
- 32.Schöbel, S., S. Zellmeier, W. Schumann, and T. Wiegert. 2004. The Bacillus subtilis σW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol. Microbiol. 521091-1105. [DOI] [PubMed] [Google Scholar]
- 33.Sherratt, D. J. 2003. Bacterial chromosome dynamics. Science 301780-785. [DOI] [PubMed] [Google Scholar]
- 34.Strooper, B. D., W. Annaert, P. Cupers, P. Saftig, K. Craessaerts, J. S. Mumm, E. H. Schroeter, V. Schrijvers, M. S. Wolfe, W. J. Ray, A. Goate, and R. Kopan. 1999. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398518-522. [DOI] [PubMed] [Google Scholar]
- 35.Tetsch, L., C. Koller, I. Haneburger, and K. Jung. 2008. The membrane-integrated transcriptional activator CadC of Escherichia coli senses lysine indirectly via the interaction with the lysine permease LysP. Mol. Microbiol. 67570-583. [DOI] [PubMed] [Google Scholar]
- 36.Tucker, D. L., N. Tucker, and T. Conway. 2002. Gene expression profiling of the pH response in Escherichia coli. J. Bacteriol. 1846551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urban, S., and M. Freeman. 2002. Intramembrane proteolysis controls diverse signalling pathways throughout evolution. Curr. Opin. Genet. Dev. 12512-518. [DOI] [PubMed] [Google Scholar]
- 38.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 21897-106. [PubMed] [Google Scholar]
- 39.Watson, N., D. S. Dunyak, E. L. Rosey, J. L. Slonczewski, and E. R. Olson. 1992. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J. Bacteriol. 174530-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolfe, M. S., and R. Kopan. 2004. Intramembrane proteolysis: theme and variations. Science 3051119-1123. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto, K., K. Hirao, T. Oshima, H. Aiba, R. Utsumi, and A. Ishihama. 2005. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 2801448-1456. [DOI] [PubMed] [Google Scholar]
- 42.Ye, J., R. B. Rawson, R. Komuro, X. Chen, U. P. Dave, R. Prywes, M. S. Brown, and J. L. Goldstein. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 61355-1364. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, H. Z., C. J. Hackbarth, K. M. Chansky, and H. F. Chambers. 2001. A proteolytic transmembrane signaling pathway and resistance to beta-lactams in staphylococci. Science 2911962-1965. [DOI] [PubMed] [Google Scholar]