Abstract
Reduced coenzyme F420 (F420H2) is an essential intermediate in methanogenesis from CO2. During methanogenesis from H2 and CO2, F420H2 is provided by the action of F420-reducing hydrogenases. However, an alternative pathway has been proposed, where H2-dependent methylenetetrahydromethanopterin dehydrogenase (Hmd) and F420H2-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) together reduce F420 with H2. Here we report the construction of mutants of Methanococcus maripaludis that are defective in each putative pathway. Their analysis demonstrates that either pathway supports growth on H2 and CO2. Furthermore, we show that during growth on formate instead of H2, where F420H2 is a direct product of formate oxidation, H2 production occurs. H2 presumably arises from the oxidation of F420H2, and the analysis of the mutants during growth on formate suggests that this too can occur by either pathway. We designate the alternative pathway for the interconversion of H2 and F420H2 the Hmd-Mtd cycle.
The methanogenic Archaea (methanogens) occupy a variety of anaerobic habitats, where they play essential roles in the conversion of hydrogen and other intermediates to methane (10). The hydrogenotrophic methanogens use hydrogen to reduce CO2 to methane. In addition, some hydrogenotrophs use formate, and a few substitute certain low-molecular-weight alcohols for hydrogen.
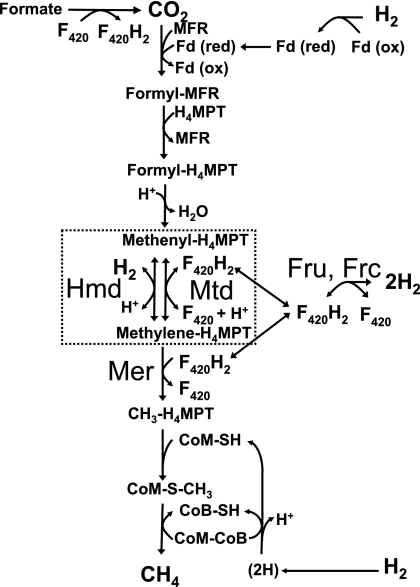
The deazaflavin F420 is an essential coenzyme of methanogenesis. The reduction of CO2 to methane requires reduced F420 (F420H2), since it is the sole electron donor for the step that reduces methylenetetrahydromethanopterin (methylene-H4MPT) (Mer in Fig. 1). In addition, F420H2 is the electron donor for F420H2-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd), one of two enzymes that reduce methenyl-H4MPT. The other enzyme, H2-dependent methylenetetrahydromethanopterin dehydrogenase (Hmd), uses H2 directly. mRNA abundance for mtd increased markedly under hydrogen-limited growth conditions (4), suggesting that Mtd may be more important when H2 is limiting.
FIG. 1.
The hydrogenotrophic methanogenic pathway. See reference 3 for a full description of methanogenesis. The Hmd-Mtd cycle is boxed. Abbreviations: CoB, coenzyme B; CoM, coenzyme M; F420, coenzyme F420; Fd, ferredoxin; Frc, cysteine-containing F420-reducing hydrogenase; Fru, selenocysteine-containing F420-reducing hydrogenase; Mer, methylenetetrahydromethanopterin reductase; MFR, methanofuran.
The F420-reducing hydrogenases (Fru and Frc) reduce F420 with H2. However, an alternative route for this process has been proposed. In Methanothermobacter marburgensis the specific activity of F420-reducing hydrogenase, a Ni-Fe hydrogenase, decreased 20-fold under nickel-limited growth conditions. In contrast, the specific activities of Hmd and Mtd, neither of which requires nickel for activity, increased six- and fourfold, respectively (1). These observations led to the proposal that under nickel-limited conditions, F420 may be reduced by the concerted action of Hmd and Mtd, the former working in the forward direction (with respect to the methanogenic pathway) and the latter in the reverse direction (1, 2). This pathway is boxed in Fig. 1.
Here we report on the properties of mutants of Methanococcus maripaludis that are deficient in Hmd, Mtd, or the F420-reducing hydrogenases. The results demonstrate that neither Hmd nor Mtd is essential, confirming that either enzyme is sufficient for methenyl-H4MPT reduction. The results also indicate that, in vivo, Hmd and Mtd do indeed constitute an alternate pathway for the reduction of F420 with H2, which we designate the Hmd-Mtd cycle. Furthermore, we show that during growth on formate, H2 production occurs, evidently by reversal of either the F420-reducing hydrogenase or the Hmd-Mtd cycle.
MATERIALS AND METHODS
Growth of strains and measurement of H2.
M. maripaludis was grown on H2 and CO2 by standard anaerobic techniques in McCas medium as described elsewhere (6). For growth on formate, McCas medium was modified to contain 200 mM sodium formate and 200 mM MOPS (morpholinepropanesulfonic acid) buffer (pH 7.0), NaCl was decreased to 0.18 M, and the gas atmosphere was 80% N2 and 20% CO2 at a pressure of 15 lb/in2. Cultures (5-ml volume) were inoculated with 0.25 to 0.5 ml of a culture actively growing on formate. Growth was monitored by optical density at 660 nm. The accumulation of H2 in the headspace (20-ml volume) was measured using a Hach CARLE Series 100 AGC gas chromatograph equipped with a Supelco 60/80 mesh molecular sieve 5A column (6 ft by 1/8 in.) and a trace analytical RGD2 reduction gas detector.
Construction of plasmids and strains.
Primers are listed in Table 1, and strains and plasmids are listed in Table 2. PCR products containing the genes hmd, mtd, frcA, and fruA and their flanking regions were generated using the primer pairs hmdcln5for and hmdcln5rev, mtdcln5for and mtdcln5rev, frcAfor2 and frcArev2, and fruAfor and fruArev, respectively. The products were cloned into pCR2.1topo to generate phmdtopo, pmtdtopo, pfrcAtopo, and pfruAtopo. An in-frame deletion of hmd was produced by PCR of phmdtopo using primers hmddel1 and hmddel2, followed by digestion with AscI and ligation to produce phmddeltopo. pmtddeltopo, pfrcAdeltopo, and pfruAdeltopol were generated in the same way using pmtdtopo and the primers mtddel1 and mtddel3, pfrcAtopo and the primers frcdel1 and frcdel2, and pfruAtopo and the primers frudel1 and frudel2, respectively. The in-frame deletion of hmd was amplified from phmddeltopo using the primers hmddelamp1 and hmddelamp3; the resulting fragment was digested with BamHI and ligated into the vector pCRprtneo to produce pCRprtΔhmdneo. pCRprtΔmtdneo was produced in the same way from pmtddeltopo using the primers mtddelamp1 and mtddelamp2 and digesting with BamHI. pCRprtΔfrcneo was produced from pfrcAdeltopo using frcdelamp5 and frcdelamp6 and digesting with XbaI, and pCRprtΔfruneo was produced from pfruAdeltopo using frudelamp5 and frudelamp6 and digesting with XbaI.
TABLE 1.
Primers
| Name | Sequence | Restriction site |
|---|---|---|
| hmdcln5for | GCTGTTGGAATAGACTGCTG | |
| hmdcln5rev | GCCCTTATTACTTCTTTTCC | |
| mtdcln5for | CGTTTCAGCAGGTTCGAAGG | |
| mtdcln5rev | GGGTGTTGCATTAATTGGCG | |
| frcAfor2 | GCACCTCTTTAAAAGCTTT | |
| frcArev2 | AATGAAACAGCGCCATCTAC | |
| fruAfor | CCAGTACTTCAATATCTTTCAC | |
| fruArev | TACTTCTTCTGACAACCGAC | |
| hmddel1 | AGGCGCGCCACTTTCATATCATACACCTCA | AscI |
| hmddel2 | AGGCGCGCCCAATAAAACCTTAAGTATTAC | AscI |
| mtddel1 | AGGCGCGCCCATTATATCACCGAAAGATAT | AscI |
| mtddel2 | GGGCGCGCCAGAATAAATTTGCATCAAAAT | AscI |
| frcdel1 | GGCGCGCCTTACCCATCAGATCACCTATC | AscI |
| frcdel2 | GGCGCGCCAATAAATACTGGTGAATCATGC | AscI |
| frudel1 | GGCGCGCCACTTTATTCACCTCC | AscI |
| frudel2 | GGCGCGCCAATTCTAAATTCCTGAAAAGG | AscI |
| hmddelamp1 | ATGGATCCGGCTTGCTGTTGGAATAGAC | BamHI |
| hmddelamp3 | TTGGATCCGCCCTTATTACTTCTTTTCC | BamHI |
| mtddelamp1 | GAGCTCGGATCCACTAGTAACGGCCGCCAAGTGT | BamHI |
| mtddelamp2 | AGAATTGGATCCCGTTTCAGCAGGTTCGAAGGA | BamHI |
| frcdelamp5 | ATCTAGAGCACCTTCTTTAAAAGCTTT | XbaI |
| frcdelamp6 | CTCTAGAAATGAAACAGCGCCATCTAC | XbaI |
| frudelamp5 | TTCTAGACCAGTACTTCAATATCTTTCAC | XbaI |
| frudelamp6 | CTCTAGATACTTCTTCTGACAACCGAC | XbaI |
TABLE 2.
Strains and plasmids
| Name | Feature(s) | Source or reference |
|---|---|---|
| Plasmids | ||
| pCR2.1topo | Ampr Kanr cloning vector | Invitrogen |
| pCRprtneo | hmv-promoter-hpt fusion + Neor cassette in pCR2.1topo | 6 |
| phmdtopo | hmd plus flanking DNA in pCR2.1topo | This study |
| pmtdtopo | mtd plus flanking DNA in pCR2.1topo | This study |
| pfrcAtopo | frcA plus flanking DNA in pCR2.1topo | This study |
| pfruAtopo | fruA plus flanking DNA in pCR2.1topo | This study |
| phmddeltopo | In-frame deletion of hmd in pCR2.1topo | This study |
| pmtddeltopo | In-frame deletion of mtd in pCR2.1topo | This study |
| pfrcAdeltopo | In-frame deletion of frcA in pCR2.1topo | This study |
| pfruAdeltopo | In-frame deletion of fruA in pCR2.1topo | This study |
| pCRprtΔhmdneo | In-frame deletion of hmd in pCRprtneo | This study |
| pCRprtΔmtdneo | In-frame deletion of mtd in pCRprtneo | This study |
| pCRprtΔfrcneo | In-frame deletion of frcA in pCRprtneo | This study |
| pCRprtΔfruneo | In-frame deletion of fruA in pCRprtneo | This study |
| Strains | ||
| Mm900 | M. maripaludis Δhpt | 6 |
| Mm1097 | Mm900 Δhmd | This study |
| Mm1020 | Mm900 Δmtd | This study |
| Mm1183 | Mm900 ΔfrcA | This study |
| Mm1145 | Mm900 ΔfruA | This study |
| Mm1184 | Mm900 ΔfrcA ΔfruA | This study |
Strains containing markerless in-frame deletions of hmd, mtd, frcA, and fruA were constructed in strain Mm900 as described elsewhere (6) using the plasmids pCRprtΔhmdneo, pCRprtΔmtdneo, pCRprtΔfrcneo, and pCRprtΔfruneo, respectively, to produce strains Mm1097, Mm1020, Mm1183, and Mm1145, respectively. A double mutant of frcA and fruA was constructed by the same procedure from the frcA mutant strain Mm1183 by using pCRprtΔfruneo to produce Mm1184. Deletions were confirmed by Southern analysis. For experiments testing whether hmd deletion mutations could be made, pCRprtΔhmdneo was transformed into recipient strains. The resultant merodiploids were streak purified, allowed to grow overnight without antibiotic selection, and plated on counterselection plates containing 8-azahypoxanthine. Colonies were analyzed by Southern blotting to distinguish strains containing deletions of the hmd gene from those containing the wild-type hmd gene.
RESULTS AND DISCUSSION
F420 reduction during growth on H2.
We used a genetic approach in M. maripaludis to test whether F420-reducing hydrogenase and the Hmd-Mtd cycle constitute two alternative pathways for the reduction of F420 in vivo. M. maripaludis contains genes for Hmd and Mtd and two sets of genes for F420-reducing hydrogenases, fruADGB and frcADGB (5). FruA contains selenocysteine residues, while FrcA contains cysteine residues in corresponding positions, and in the closely related Methanococcus voltae frc expression is repressed in the presence of selenium in the medium (7, 8). We hypothesized that if Hmd and Mtd can provide an alternative pathway for the reduction of F420, then mutants with deletions in fru and frc should be viable in the presence of wild-type hmd and mtd. Conversely, mutants with mutations in either hmd or mtd should be viable in a fru+ frc+ background.
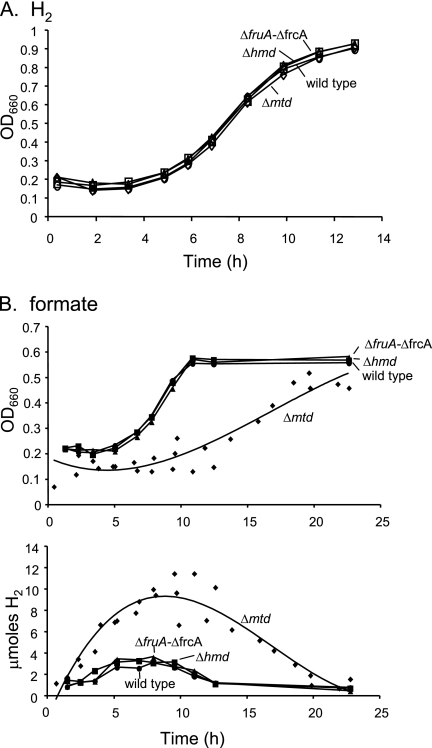
Using H2 and CO2 as growth substrates, we made the following mutants, all containing markerless in-frame deletions: ΔfruA, ΔfrcA, double mutant ΔfruA ΔfrcA, Δmtd, and Δhmd strains. ΔfruA ΔfrcA, Δmtd, and Δhmd strains each grew normally on H2 and CO2 (Fig. 2A). Since F420H2 is essential for methanogenesis, each mutant must retain a pathway for F420 reduction using H2. Hence, the results imply that F420-reducing hydrogenase and the Mtd-Hmd cycle are each sufficient for this function.
FIG. 2.
Growth and H2 production by wild-type and mutant strains of M. maripaludis. (A) Growth on H2; (B) growth and H2 production on formate. For the Δmtd mutant on formate, data from three separate growth experiments are plotted and are represented by a single line. OD660, optical density at 660 nm.
As a formal possibility, a third, unknown pathway for the reduction of F420, different from the F420-reducing hydrogenase and the Mtd-Hmd cycle, could exist. To test this possibility, we attempted to construct an Δhmd mutation in a ΔfruA ΔfrcA background. Following our regular procedure for generation of markerless mutations (6), we introduced Δhmd (containing the N- and C-terminal flanking regions of hmd) on an integrative vector to produce merodiploids of Δhmd and hmd+. We made such merodiploids in the ΔfruA ΔfrcA, ΔfrcA, and fru+ frc+ backgrounds. We then selected for resolution of the merodiploids via a second recombination event and analyzed the resulting strains by Southern blotting. In principle a mixture of wild-type and deletion strains should result, depending on where the second recombination event occurs. We counted the numbers of resulting Δhmd and hmd+ strains in each background. In the fru+ frc+ background six out of eight strains tested contained Δhmd and the remaining two contained hmd+. In the ΔfrcA background, which should express fru and therefore retain active F420-reducing hydrogenase, three strains contained Δhmd and five contained hmd+. In contrast, in the ΔfruA ΔfrcA background all 40 strains tested contained only hmd+. The results indicate that while Hmd can be eliminated in a strain with active F420-reducing hydrogenase, it is essential in a strain lacking F420-reducing hydrogenase. Therefore, no evidence could be found for the existence of a third pathway that would produce F420H2 from H2.
H2 production during growth on formate.
Growth on formate differs from growth on H2 and CO2 because F420H2 is a direct product of formate oxidation (Fig. 1). Neither the F420-reducing hydrogenase nor the Mtd-Hmd cycle should be necessary for the production of F420H2. However, the reversal of either pathway might result in H2 production. We characterized the growth of the ΔfruA ΔfrcA, Δmtd, and Δhmd mutants on formate. The ΔfruA ΔfrcA and Δhmd mutants grew normally, while the Δmtd mutant grew after a lag. For each strain, H2 accumulated in the headspace of the tubes as growth commenced and disappeared when growth ended (Fig. 2B). This observation suggests that H2 is produced from F420H2 and that either the F420-reducing hydrogenase or the Mtd-Hmd cycle can mediate this conversion. H2 accumulated to a substantially higher level in tubes containing cultures of the Δmtd mutant than in tubes containing any of the other strains. In the Δmtd strain, Hmd is the only enzyme for the reduction of methenyl-H4MPT. Therefore, H2 production, which would occur by the action of the F420-reducing hydrogenase, should be essential. Due to the relatively low affinity of Hmd for H2 (9), substantially higher H2 levels accumulate. In contrast, in the other strains Mtd is present and can use F420H2 for the reduction of methenyl-H4MPT. These results indicate that H2 production from F420H2 occurs during growth on formate and that either the F420-reducing hydrogenase or the Mtd-Hmd cycle can carry out this process.
Whether H2 is a necessary intermediate during growth on formate cannot be determined from the present data. The generation of a ΔfruA ΔfrcA Δhmd triple mutant, which is expected to grow in the presence of formate, could resolve this question. Growth of the mutant on formate alone without the addition or generation of H2 would indicate that H2 is not a required intermediate. A requirement for added H2 would indicate that H2 production is required during growth on formate. Efforts to construct such a mutant are under way.
Concluding remarks.
The genetic approach taken here has shown that two alternative pathways, the F420-reducing hydrogenase and the Hmd-Mtd cycle, can function in vivo for the reduction of F420 with H2. Furthermore, during growth on formate the same pathways function in reverse to produce H2 from F420H2. The lack of growth differences between the wild-type and mutant strains on H2 and CO2 (Fig. 2A) suggests that neither pathway for F420 reduction was rate limiting. However, in nature the F420-reducing hydrogenase may constitute the major pathway when sufficient nickel is present, while the Hmd-Mtd cycle may be important when nickel is limiting (1, 2).
Acknowledgments
This work was supported by Department of Energy Basic Research for the Hydrogen Fuel Initiative grant DE-FG02-05ER15709 and grant NCC 2-1273 from the NASA Astrobiology Institute.
We thank William Whitman for helpful comments.
Footnotes
Published ahead of print on 16 May 2008.
REFERENCES
- 1.Afting, C., A. Hochheimer, and R. K. Thauer. 1998. Function of H2-forming methylenetetrahydromethanopterin dehydrogenase from Methanobacterium thermoautotrophicum in coenzyme F420 reduction with H2. Arch. Microbiol. 169206-210. [DOI] [PubMed] [Google Scholar]
- 2.Afting, C., E. Kremmer, C. Brucker, A. Hochheimer, and R. K. Thauer. 2000. Regulation of the synthesis of H2-forming methylenetetrahydromethanopterin dehydrogenase (Hmd) and of HmdII and HmdIII in Methanothermobacter marburgensis. Arch. Microbiol. 174225-232. [DOI] [PubMed] [Google Scholar]
- 3.Deppenmeier, U. 2002. The unique biochemistry of methanogenesis. Prog. Nucleic Acid Res. Mol. Biol. 71223-283. [DOI] [PubMed] [Google Scholar]
- 4.Hendrickson, E. L., A. K. Haydock, B. C. Moore, W. B. Whitman, and J. A. Leigh. 2007. Functionally distinct genes regulated by hydrogen limitation and growth rate in methanogenic Archaea. Proc. Natl. Acad. Sci. USA 1048930-8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hendrickson, E. L., R. Kaul, Y. Zhou, D. Bovee, P. Chapman, J. Chung, E. Conway de Macario, J. A. Dodsworth, W. Gillett, D. E. Graham, M. Hackett, A. K. Haydock, A. Kang, M. L. Land, R. Levy, T. J. Lie, T. A. Major, B. C. Moore, I. Porat, A. Palmeiri, G. Rouse, C. Saenphimmachak, D. Söll, S. Van Dien, T. Wang, W. B. Whitman, Q. Xia, Y. Zhang, F. W. Larimer, M. V. Olson, and J. A. Leigh. 2004. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 1866956-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore, B. C., and J. A. Leigh. 2005. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J. Bacteriol. 187972-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller, S., and A. Klein. 2001. Coordinate positive regulation of genes encoding [NiFe] hydrogenases in Methanococcus voltae. Mol. Genet. Genomics 2651069-1075. [DOI] [PubMed] [Google Scholar]
- 8.Noll, I., S. Muller, and A. Klein. 1999. Transcriptional regulation of genes encoding the selenium-free [NiFe]-hydrogenases in the archaeon Methanococcus voltae involves positive and negative control elements. Genetics 1521335-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thauer, R. K., A. R. Klein, and G. C. Hartmann. 1996. Reactions with molecular hydrogen in microorganisms: evidence for a purely organic hydrogenation catalyst. Chem. Rev. 963031-3042. [DOI] [PubMed] [Google Scholar]
- 10.Zinder, S. H. 1993. Physiological ecology of methanogens, p. 128-206. In J. G. Ferry (ed.), Methanogenesis. Chapman and Hall, London, United Kingdom.