Abstract
The Oca family is a novel class of autotransporter-adhesins with highest structural similarity in their C-terminal transmembrane region, which supposedly builds a beta-barrel pore in the outer membrane (OM). The prototype of the Oca family is YadA, an adhesin of Yersinia enterocolitica and Yersinia pseudotuberculosis. YadA forms a homotrimeric lollipop-like structure on the bacterial surface. The C-terminal regions of three YadA monomers form a barrel in the OM and translocate the trimeric N-terminal passenger domain, consisting of stalk, neck, and head region to the exterior. To elucidate the structural and functional role of the C-terminal translocator domain (TLD) and to assess its promiscuous capability with respect to transport of related passenger domains, we constructed chimeric YadA proteins, which consist of the N-terminal YadA passenger domain and C-terminal TLDs of Oca family members UspA1 (Moraxella catarrhalis), EibA (Escherichia coli), and Hia (Haemophilus influenzae). These constructs were expressed in Y. enterocolitica and compared for OM localization, surface exposure, oligomerization, adhesion properties, serum resistance, and mouse virulence. We demonstrate that all chimeric YadA proteins translocated the YadA passenger domain across the OM. Y. enterocolitica strains producing YadA chimeras or wild-type YadA showed comparable binding to collagen and epithelial cells. However, strains producing YadA chimeras were attenuated in serum resistance and mouse virulence. These results demonstrate for the first time that TLDs of Oca proteins of different origin are efficient translocators of the YadA passenger domain and that the cognate TLD of YadA is essential for bacterial survival in human serum and mouse virulence.
Protein secretion in gram-negative bacteria is faced with the serious problem of traversing two different membrane-lipid bilayers. Therefore, several secretory pathways have evolved, which were classified into five different types (9, 11, 20, 29, 51). Recently, even two novel multicomponent secretion systems, type VI and type VII secretion, were introduced (1, 31, 40). While all other types engage a whole machinery of protein exporting helper proteins, the type V secretion mechanism is thought to be less complex, since all necessary information for transport through both membranes is contained in the secreted protein itself, which has also led to the term “autotransporter” protein (20). With over 700 identified proteins to date, the type V secretion family is the largest group, and many of its members are also confirmed virulence factors with effector functions such as adherence, invasion, proteolysis, cytotoxicity, serum resistance, and cell-to-cell spread (14, 37). An N-terminal signal peptide is Sec dependently cleaved during passage through the inner membrane, while the process of translocation through the outer membrane (OM) is still unresolved. The C-terminal region forms a beta-barrel pore before or during integration into the OM and is essential for translocation of the N-terminal passenger domain across the OM (20). However, the detailed process of passenger domain translocation remains unclear (6). Different models have been proposed to approach this issue. According to the hairpin and the threading model, the beta-barrel primarily integrates into the OM, and the passenger domain slides through the pore starting with the C or N terminus (21, 36, 39). The multimeric model suggests that the passenger domain is translocated to the bacterial surface through a central channel built by a multimeric complex of beta-barrel pores (52), while the Omp85 model involves the OM protein Omp85/YaeT in the translocation process (54, 55).
Previously, we could demonstrate that the Yersinia adhesin YadA of Yersinia enterocolitica and Yersinia pseudotuberculosis is the prototype of a novel class of oligomeric autotransporter adhesins (42), which we termed Oca (for oligomeric coiled-coil adhesins), which can be found in alpha-, beta-, and gammaproteobacteria. In addition to YadA, several other family members, such as Hia and Hsf of Haemophilus influenzae, UspA1 and UspA2 of Moraxella catarrhalis, Eib proteins of Escherichia coli, or NadA of Neisseria meningitidis, have been identified in the last few years (8, 12, 15, 23, 45, 48). Conventional autotransporters are monomeric proteins and form their C-terminal translocator domain (TLD) with 12 transmembrane beta-strands from their C-terminal 250 to 300 amino acid (aa) residues, as could demonstrated by the recently resolved crystal structures of NalP and EspP (5, 36). Oca family members form a trimeric 12-stranded beta-barrel pore with their last ∼70 C-terminal amino acids, which means that each of the three monomers contributes four transmembrane antiparallel beta-sheets to the oligomeric pore, roughly comparable with the oligomeric structure of the OM protein TolC from E. coli (25). As demonstrated by C-terminal deletion constructs (50), the assembly of the three TLDs is necessary for translocation of the N-terminal passenger domains to the bacterial surface, which in case of trimeric YadA forms a lollipop-like structure comprising a head, neck, and coiled-coil stalk region and remains uncleaved and covalently attached to the TLDs in contrast to several conventional autotransporters such as the immunoglobulin A (IgA) protease of N. gonorrhoeae (39). It could be shown that the C-terminal 92 aa of YadA efficiently present a FLAG tag on the bacterial surface, thus forming an efficient translocon (42). For the C-terminal 76 aa residues of Hia, comparable results could be obtained (48). Crystal structures of fragments of YadA and Hia N-terminal passenger and C-terminal TLDs, which proved the trimeric architecture of both adhesins, showed that the separate YadA passenger domain also possesses a translocator-independent capacity to oligomerize and that the YadA and Hia C-terminal TLDs form a trimeric beta-barrel, confirming previous biochemical results and secondary structure predictions (24, 26, 34, 56), which finally led to a novel, more restricted term for the Oca family, i.e., trimeric autotransporter adhesins (56). Since it cannot be excluded, however, that in the future tetrameric or other oligomeric variants of autotransporters will be discovered, we suggest using the term Oca as a hypernym.
Certain regions of YadA could be assigned to distinct functional properties associated with virulence. While the ability to bind HEp-2 epithelial cells and extracellular matrix (ECM) proteins such as collagen is localized in an intact YadA head-neck-binding module (19, 42, 43, 49), adherence to neutrophils requires the first 53 aa of its head region (44), and the region necessary for autoagglutination (AA) could not precisely be defined yet, but a hydrophobic region in the head domain seems to be involved (47, 50). Interestingly, the proximally located head-neck region was found to be not directly necessary for YadA-mediated serum resistance (42). The coiled-coil stalk region of YadA, which is formed by a variable number (six to nine) of 15-mer repeats depending on the Yersinia serotype (23), seems to have a spacer function between the YadA head and the bacterial OM, since its length influences the efficiency of type III secretion-mediated Yop translocation (30). Although the stalk region is implicated in YadA-mediated serum resistance, it does not seem to be essential for this phenomenon, since an in-frame deletion of the first four of seven 15-mer repeats of the stalk region of Y. enterocolitica O:8 YadA did not result in a loss of serum resistance (42). Therefore, the exact region for this phenotype could not be defined yet. Furthermore, it is still unknown whether the in vitro-observed phenomenon of serum resistance contributes to in vivo Y. enterocolitica virulence. Although YadA mutants lacking collagen- or neutrophil-binding function were found to be attenuated in the mouse infection model, mutants with full adhesive activity, but without mediation of serum resistance, were not yet available to study (43, 44).
To further elucidate the relative impact of changes in the C-terminal TLD on YadA passenger domain structure and function and to assess the promiscuous capability of Oca TLDs with respect to transport and translocation of related passenger domains, we constructed fusion proteins, which consist of the N-terminal YadA passenger domain and C-terminal TLDs of Oca family members UspA1, EibA, and Hia. These constructs were expressed in Y. enterocolitica and compared for OM localization, surface exposure, oligomerization, resistance to tryptic digestion, adhesion properties, resistance to serum bactericide, and virulence in a Y. enterocolitica mouse infection model. From our results we conclude that C-terminal TLDs of Oca proteins UspA1, EibA, and Hia are efficient translocators of the heterologous YadA passenger domain and maintain its collagen and cell adherence capability but are not sufficient to mediate serum resistance and mouse virulence like the YadA TLD.
(Parts of this study are included in the doctoral thesis of M. Tiller.)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial species and strains used in the present study are listed in Table 1. E. coli, M. catarrhalis, and H. influenzae were grown in Luria-Bertani (LB) medium at 37°C, and Y. enterocolitica was grown at 27°C (18). For the induction of yadA gene expression, overnight cultures cultivated at 27°C in LB were diluted 1:40 in RPMI 1640 medium (Invitrogen, Paisley, United Kingdom) and grown at 37°C for 1.5 h (for binding assays) or 6 h (for immunofluorescence assay [IFA], dot immunoblotting, enzyme-linked immunosorbent assay [ELISA], and OM protein preparations). Antibiotics were used in the following concentrations: ampicillin, 100 μg/ml; kanamycin, 25 μg/ml; nalidixic acid, 60 μg/ml; and spectinomycin, 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| Y. enterocolitica | ||
| WA-314 | Clinical isolate of serotype O:8, carrying virulence plasmid pYVO8 | 18 |
| WA-c | Plasmidless derivative of WA-314 | 18 |
| E. coli | ||
| DH5α | endA1 supE44 hsdR17(rK− mK+) thi-1 recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 (φ80 lacZΔM15) | 17 |
| Sm10λpir | thi-1 thr leu tonA lacY supE recA::RP4-2-TC::Mu-Kan (λpir), Kmr | 28 |
| ECOR-9 | Clinical isolate, EibA+ | 35, 45 |
| H. influenzae NTHi 58670 | Clinical isolate of a nontypeable H. influenzae strain, Hia+ | This study |
| M. catarrhalis O35E | Clinical isolate, UspA1+, UspA2+ | 2 |
| Plasmids | ||
| pUC-A-1 | pUC13 with 5-kb EcoRI-HindIII insert fragment of pYVO8 from WA-314 carrying the yadA gene | 43 |
| pUC-A-YadA-UspA1 | pUC-A1Δ334-422, carrying instead of the yadA linker and anchor region the homologous last 90 aa of UspA1 | This study |
| pUC-A-YadA-UspA1-2 | pUC-A1Δ362-422, carrying instead of the yadA loop and anchor region the homologous last 63 aa of UspA1 | This study |
| pUC-A-YadA-UspA1-3 | pUC-A1Δ369-422, carrying instead of the yadA anchor region the homologous last 56 aa of UspA1 | This study |
| pUC-A-YadA-EibA | pUC-A1Δ334-422, carrying instead of the yadA linker and anchor region the homologous last 87 aa of EibA | This study |
| pUC-A-YadA(D332L, H333E) | pUC-A1Δ334-422, carrying the reinserted yadA linker and anchor region | This study |
| pUC-A-YadA-Hia | pUC-A1Δ334-422, carrying instead of the yadA linker and anchor region the homologous last 90 aa of Hia | This study |
| pGP704 | Mobilizable suicide vector, R6K2 replicon, requires π protein in trans from the λpir-positive strain | 28 |
| pGPS-A-1 | pGP704 + 1.8-kb Spcr cassette in the EcoRI site (=pGPS) carrying the yadA gene as an EcoRI-SphI fragment from pUC-A-1 | 43 |
| pGPS-A-YadA-UspA1 | pGPS carrying the yadA-uspA1 gene as an EcoRI-SphI fragment from pUC-A-YadA-UspA1 | This study |
| pGPS-A-YadA-UspA1-2 | pGPS carrying the yadA-uspA1-2 gene as an EcoRI-SphI fragment from pUC-A-YadA-UspA1-2 | This study |
| pGPS-A-YadA-UspA1-3 | pGPS carrying the yadA-uspA1-3 gene as an EcoRI-SphI fragment from pUC-A-YadA-UspA1-3 | This study |
| pGPS-A-YadA-EibA | pGPS carrying the yadA-eibA gene as an EcoRI-SphI fragment from pUC-A-YadA-EibA | This study |
| pGPS-A-YadA(D332L, H333E) | pGPS carrying the yadA(D332L, H333E) gene as an EcoRI-SphI fragment from pUC-A-YadA(D332L, H333E) | This study |
| pGPS-A-YadA-Hia | pGPS carrying the yadA-hia gene as an EcoRI-SphI fragment from pUC-A-YadA-Hia | This study |
| pYVO8-A-0 | pYVO8, yadA mutant, Kmr cassette inserted in the PstI sites of yadA by allelic exchange | 43 |
| pYVO8-A-1 | pYVO8-A-0 with integrated pGPS-A-1, wild-type yadA | 43 |
| pYVO8-YadA-UspA1 | pYVO8-A-0 with integrated pGPS-A-YadA-UspA1 | This study |
| pYVO8-YadA-UspA1-2 | pYVO8-A-0 with integrated pGPS-A-YadA-UspA1-2 | This study |
| pYVO8-YadA-UspA1-3 | pYVO8-A-0 with integrated pGPS-A-YadA-UspA1-3 | This study |
| pYVO8-YadA-EibA | pYVO8-A-0 with integrated pGPS-A-YadA-EibA | This study |
| pYVO8-YadA(D332L, H333E) | pYVO8-A-0 with integrated pGPS-A-YadA(D332L, H333E) | This study |
| pYVO8-YadA-Hia | pYVO8-A-0 with integrated pGPS-A-YadA-Hia | This study |
Spcr, spectinomycin resistance; Kmr, kanamycin resistance.
DNA manipulations and PCR.
Restriction endonuclease digestion, DNA ligations, transformations, sequencing, and PCR were done according to standard techniques (4). Conjugations were performed as described previously (43). Plasmid DNA preparations and isolation of DNA fragments from agarose gels were done with Macherey-Nagel kits (Macherey-Nagel GmbH & Co.KG, Düren, Germany) as described by the manufacturer.
Sequence alignment.
Amino acid sequence alignment was done with DNAMAN 5.2.9 software (Lynnon BioSoft, Quebec, Canada).
Construction of the YadA-hybrid proteins and expression in Y. enterocolitica O:8.
The pUC-A-1 ClaI-SphI backbone was prepared as described previously (42). YadA-hybrid proteins were generated with three separate PCR fragments (Fig. 1C) —(i) a ClaI-SacI fragment (PCR-1) of the yadA gene encoding the YadA-passenger domain (bp 1 to 993); (ii) a SacI-SacI fragment (PCR-2) encoding the corresponding yadA (for control construct YadA [D332L, H333E]), uspA1, eibA, or hia gene TLD; and (iii) a SacI-SphI fragment (PCR-3) encoding the yadA terminator region directly after the yadA stop codon—were ligated into a ClaI-SphI vector backbone (pUC-A1) and subsequently transformed into E. coli DH5α. The oligonucleotides used in the present study are listed in Table 2.
FIG. 1.
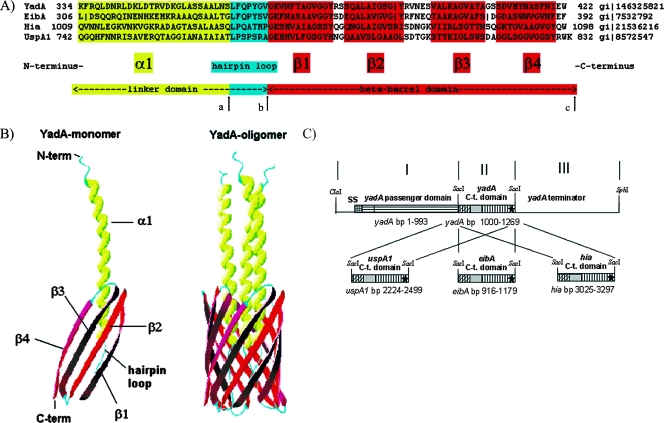
(A) Sequence alignment of the TLD monomers of YadA, EibA, Hia, and UspA1. The TLD monomer consists of two main parts: the linker domain and the beta-barrel domain. The linker domain can be separated into a proximal α1 domain, which traverses the beta-barrel domain, and a distal hairpin loop, which connects the α1 domain to the beta-barrel domain. The beta-barrel domain consists of four transmembrane antiparallel beta-sheets (β1 to β4). In this study, the YadA TLD (aa 334 to 422) was replaced with either the EibA (aa 306 to 392), Hia (aa 1009 to 1098), or UspA1 (aa 742 to 832) TLD region. Arrows mark the replaced amino acids for construction of YadA-UspA1-2 (aa 362 to 422 of YadA replaced by aa 770 to 832 of UspA1) (from arrow a to arrow c) and YadA-UspA1-3 (aa 369 to 422 of YadA replaced by aa 777 to 832 of UspA1) (from arrow b to arrow c). (B) Crystal structure model of the TLD monomer and trimer, e.g., three YadA monomers build the trimeric YadA protein (YadA oligomer). Ribbon diagrams were generated with DeepView PDB. (C) Construction scheme of the yadA chimeras. PCR fragments I, II, and III were ligated into a ClaI-SphI cut pUC-A-1 vector backbone (see Materials and Methods). SS, signaling sequence. The asterisks refer to the corresponding stop codons.
TABLE 2.
Oligonucleotides used in this study
| Primer | Description | Sequence (5′→3′)a |
|---|---|---|
| Construction primers | ||
| PCR-1 | PCR-1:5′ClaI-3′SacI fragments | |
| A-Cla-f | Constant forward primer for PCR-1, the start is 167 bp upstream of yadA start codon | TTTTAAAGATCGATTAGTGCTGT |
| A-993-r | Constant reverse primer for PCR-1, the end is at bp 993 (T331) of yadA | CACGAGCTCTGTGTATTGATTCGATTCACGG |
| PCR-2 | PCR-2:5′SacI-3′SacI fragments | |
| A-1000-f | Forward primer for the yadA(D332L, H333E) insert, the start is at bp 1000 (=K334) of yadA | GGGGAGCTCAAATTCCATCAACTTGACAACC |
| A-1269-r | Reverse primer for yadA(D332L, H333E)-insert, the end is at bp 1269 (yadA stop codon) | ATTGAGCTCTTACCACTCGATATTAAATGATG |
| EibA-916-f | Forward primer for the eibA insert, the start is at bp 916 (L306) of eibA | GTCGAGCTCCTGGACAGCCAGCAGCGCCAG |
| EibA-1179-r | Reverse primer for the eibA insert, the end is at bp 1179 (eibA stop codon) | ATTGAGCTCTTAAAACTCGAAGTTCACACCA |
| UspA1-2224-f | Forward primer for the uspA1 insert, the start is at bp 2224 (Q742) of uspA1 | GACGAGCTCCAGGGTCAGCATTTTAATAATC |
| UspA1-2499-r | Reverse primer for the uspA1 insert, the end is at bp 2499 (uspA1 stop codon) | TATGAGCTCTTATTTCCAGCGGTAACTGCCA |
| Hia-3025-f | Forward primer for the hia insert, the start is at bp 3025 (Q1009) of hia | AACGAGCTCCAAGTCAATAATCTTGAGGGCAA |
| Hia-3297-r | Reverse primer for the hia insert, the end is at bp 3297 (hia stop codon) | ATTGAGCTCTTACCACTGGTAACCAACACC |
| PCR-3 | PCR-3:5′SacI-3′SphI fragments | |
| A-1270-f | Constant forward primer for PCR-2, the start is at bp 1270 of yadA, directly behind the yadA stop codon | CGCGAGCTCTATCATTTAGAAGTTAACAAGTCT |
| A-Sph-r | Constant reverse primer for PCR-3, the end is 569 bp after the yadA stop codon and 30 bp after a SphI site | GTCAATACAGAGATAGAACAGCT |
| PCR mutagenesis | ||
| Mutagenesis primers | ||
| U-2308-f | Forward mutagenesis primer, used for yadA-uspA1-2 together with the primer A-Sph-r; start: bp 2308 of uspA1 | TTACCATCGCCCAGTAGAGCAGGTGAGCAT |
| A-U-1083-r | Reverse mutagenesis primer, used for yadA-uspA1-2 together with the primer A-Cla-f; end: bp 1083 of yadA | TGCTCTACTGGGCGATGGTAAGCTGTTTAAAGC GGCTGAA |
| A-U-2329-f | Forward mutagenesis primer, used for yadA-uspA1-3 together with the primer A-Sph-r; start: bp 2329 of uspA1 | TTGTTCCAGCCATATGGTGTGGGTGAGCATCATGTCTTATTTG |
| A-1104-r | Reverse mutagenesis primer, used for yadA-uspA1-2, together with the primer A-Cla-f; end: bp 1104 of yadA | CACACCATATGGCTGGAACA |
| Standard primers | ||
| A-Cla-f | See PCR-1, here used for yadA-uspA1-2 and yadA-uspA1-3 | |
| A-Sph-r | See PCR-3, here used for yadA-uspA1-2 and yadA-uspA1-3 |
Restriction sites are indicated by an underscore. For the mutagenesis primers, the overlapping part is indicated by an underscore.
In PCR-2 the primers used for the UspA1-insert were UspA1-2224-f and UspA1-2499-r, with M. catarrhalis O35E DNA as a template. We used EibA-916-f and EibA-1179-r for the EibA insert with E. coli ECOR9 DNA, A-1000-f and A-1269-r for the YadA (D332L, H333E) insert with Y. enterocolitica WA-314 DNA, and Hia-3025-f and Hia-3297-r for the Hia insert with H. influenzae NTHi 17035 DNA. Each of these PCR products was ligated with the products of PCR-1 and PCR-3 into the pUC-A-1 ClaI-SphI backbone and transformed into E. coli DH5α.
To replace the yadA beta-barrel domain and hairpin-loop region of the yadA linker domain with the homologous gene sequence of uspA1 or to replace only the beta-barrel domain of wild-type yadA with the corresponding sequence of uspA1, two additional YadA-UspA1 fusion constructs, YadA-UspA1-2 and YadA-UspA1-3, were constructed by using a PCR mutagenesis strategy (22). Briefly, two PCR products with overlapping ends were synthesized, which were then added into a second PCR where, with the help of an appropriate annealing temperature (in this case 60°C), overlap of the two PCR products happens, and a new PCR product is synthesized by external 5′ and 3′ primers. For the synthesis of YadA-UspA1-2, the primers A-Cla-f and A-U-1083-r with Y. enterocolitica WA-314 DNA and the primers U-2308-f and A-Sph-r with M. catarrhalis O35E DNA were used for the construction of PCR products 1 and 2, respectively, and for the synthesis of YadA-UspA1-3, the primers A-Cla-f and A-1104-r with Y. enterocolitica WA-314 DNA and the primers A-U-2329-f and A-Sph-r with M. catarrhalis O35E DNA were used. In the second PCR, the appropriate PCR products 1 and 2 were used as overlapping template DNA and with the primers A-Cla-f and A-Sph-r the fusion constructs were synthesized, digested with ClaI and SphI, and subsequently ligated in the pUC-A-1 ClaI-SphI backbone as described before.
For subsequent expression studies in Y. enterocolitica, all yadA hybrid gene constructs were cloned into the mobilizable suicide vector pGP704 with an inserted streptomycin-resistant spectinomycin-resistant Ω fragment (28) in E. coli SM10λpir for mobilization into Y. enterocolitica strains WA-314 harboring a virulence plasmid (pYVO-A-0) with a deleted yadA gene, as described previously (42). All of the constructs listed in Table 1 were verified by restriction enzyme analysis of plasmid preparations, PCR, and sequencing.
IFA.
To investigate the surface exposure of the YadA chimeras, Yersinia strains were grown at 37°C for 6 h, harvested by centrifugation, and washed with phosphate-buffered saline (PBS). Bacteria were treated with a monoclonal antibody (MAb; 8D1) specific for the lower stalk region of YadA (aa 290 to 330) at 37°C for 30 min and subsequently washed three times with PBS. Surface-bound MAb 8D1 was detected with fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin (Sigma-Aldrich, Munich, Germany) diluted 1:50 in PBS as described previously (42). Glass slides were coated with the unfixed bacteria, which were then visualized by fluorescence microscopy.
ELISA.
To quantitatively compare the surface expression of YadA and YadA hybrid molecules, an ELISA of whole, unfixed bacteria was performed. Yersinia strains were grown at 37°C for 6 h, harvested by centrifugation, and diluted to an optical density at 600 nm (OD600) of 0.2. After two washes with PBS, the bacteria were incubated with MAb 8D1 (1:1,000 in PBS) at 37°C for 1 h, washed again twice, and incubated with secondary antibody anti-mouse IgG-peroxidase conjugate (Sigma-Aldrich) diluted 1:1,000 in PBS at 37°C for 1 h. After two further washing steps, samples were resuspended in 50 μl of PBS and transferred into a Microlon 600 96-well plate (Greiner, Frickenhausen, Germany). For development, 50 μl of orthophenylene-diamine (OPD) solution (1 OPD tablet dissolved in 3 ml of H2O and 1.25 μl of H2O2; Dako, Glostrup, Denmark) was added to each sample, and the plate was incubated for 15 min at room temperature. The reaction was stopped with 0.5 N H2SO4, and extinction was measured at OD492.
Immunoblotting.
OM preparations of YadA were performed as described elsewhere (23). OM samples were resuspended in electrophoresis buffer (1% sodium dodecyl sulfate [SDS] and 0.25% 2-mercaptoethanol), either boiled for 10 min or incubated at 37°C for 60 min, and separated by discontinuous SDS-polyacrylamide gel electrophoresis (PAGE) using 11% polyacrylamide. To check for release of YadA or YadA hybrid protein into the culture broth, supernatants (100 ml each) from bacterial overnight cultures were collected after centrifugation and sterile filtered through a membrane (pore size, 0.2 um; Millipore, Billerica, MA). After the addition of a 0.1 volume (vol) trichloroacetic acid (Roth, Karlsruhe, Germany), the samples were vortex mixed and incubated on ice for 2 h for precipitation, followed by a 30-min centrifugation step (20,800 × g, 4°C). The supernatant was discarded, and the precipitate was resuspended in 1 ml of PBS. Then, a 0.4 volume of acetone (−20°C; Merck, Darmstadt, Germany) was added, followed by a further vortexing and a subsequent 1-h incubation step on ice for acetone precipitation. The samples were centrifuged again (20,800 × g, 4°C, 30 min); the pellets were then resuspended in 1 ml of acetone (−20°C) and centrifuged again at 20,800 × g at 4°C for 3 min and, after removal of the supernatant, resuspended in 50 mM Tris (pH 8.0) and stored at −20°C. The OM and supernatant samples were then transferred to nitrocellulose sheets (BA85; Whatman, Plc., London, United Kingdom) by electrophoresis and blocked with 3% bovine serum albumin (fraction V) in PBS-0.5% Tween overnight at 4°C. Immunostaining of YadA was done with MAb 8D1. Antigen-antibody complexes were detected with an anti-mouse IgG-alkaline phosphatase conjugate (Sigma-Aldrich) diluted 1:5,000 in PBS-0.5% Tween for the MAb, followed by development with indoxylphosphate-tetrazolium (Sigma-Aldrich) as described previously (43).
Protease accessibility assay.
After the stimulation of bacterial YadA production by growth at 37°C for 6 h in LB medium, 107 bacterial cells were washed with PBS and incubated with protease trypsin (Invitrogen) at various concentrations (ranging in 10-fold increments from 0.25 to 250 μg/ml, diluted in PBS) for 1 h at 37°C. After incubation the cells were pelleted, and digestion was stopped by adding 20 μl of electrophoresis buffer (2% SDS, 10% glycerol, 5% 2-mercaptoethanol, 0.001% bromophenol blue). The samples were incubated at 37°C for 60 min and separated by SDS-PAGE as described below. The bacterial cell number was verified by counting the CFU of serial dilutions of the samples.
Binding assays and AA.
Binding to immobilized collagen and the assay for AA were done as described previously (43). Briefly, type I collagen (Sigma-Aldrich) was allowed to react with Microlon 600 96-well plates (Greiner) in a 50-μl volume with concentrations of 2 or 20 μg/ml in PBS for 1 h at 37°C. Nonspecific binding sites were blocked by incubation with 200 μl of coating buffer (PBS, 0.5% bovine serum albumin) for 1 h at 37°C. After five washes with PBS-0.1% Tween 20 the wells were incubated with bacteria (OD600 = 0.5) in PBS-0.1% sodium azide for 1 h at 37°C. The bacteria were then washed again five times with PBS-0.1% Tween 20. The binding reaction was verified by immunostaining with polyclonal 1:10,000-diluted rabbit anti-WA-c antiserum overnight at 4°C and incubation with alkaline phosphatase-conjugated goat anti-rabbit IgG (Sigma-Aldrich), as described previously (27). Then, 1 mg of p-nitrophenyl phosphate per ml in H2O was added as a substrate at 37°C. The reaction was stopped with 0.5 M H2SO4. The absorbance at 405 nm was determined.
For studying cell adherence, monolayers of HEp-2 cells grown on RPMI 1640 medium were incubated with 5 × 107 bacteria (OD600 = 0.3) per ml (multiplicity of infection of 100) for 30 min at 37°C in six-well plates for the plating assay. For microscopic counting of cell-associated bacteria, monolayers were grown on glass slide insets. To reduce bacterial AA and obtain comparable bacterial input values, bacteria were subjected to shearing forces by pushing them through a 27-gauge needle (Braun, Melsungen, Germany) before determination of the OD600. For the removal of loosely attached bacteria, wells were washed three times with prewarmed Dulbecco PBS (Invitrogen) after the first 30 min of incubation. Fresh prewarmed RPMI 1640 medium was then added; plates were reincubated for an additional 30 min and washed again as described above. For the plating assay, the cells were removed from the wells with a cell lysis buffer (0.2% Triton X-100, 0.025% trypsin), and serial dilutions of the lysate were plated out on LB-agar plates for determination of the bacterial CFU. For microscopic counting glass slides were fixed with methanol and stained with Giemsa solution, and cell-associated yersiniae were visualized microscopically. Cell-associated bacteria of 30 randomly selected cells were counted, and the average number per cell ± the standard deviation was determined. The statistical significance was determined by using the Student t test. Each experiment was repeated at least three times.
AA experiments were performed as described previously by Skurnik et al. (47). Briefly, overnight cultures of yersiniae were diluted to an OD600 of 0.1 in RPMI 1640 and grown for 7 h in glass tubes without shaking at 37°C. The AA phenotype was detected as the sedimentation of bacterial clumps and clearance of the medium.
Serum resistance test.
A serum resistance test was performed as described previously (43). Briefly, bacteria were grown overnight in RPMI 1640 medium at 37°C, pelleted by centrifugation, washed in PBS-MgCl2 (5 mM), and then incubated at 37°C in 50% human serum pooled from healthy blood donors (laboratory personnel). Surviving bacteria were defined as the CFU determined after plating out serial dilutions on LB-agar after 0 and 90 min. The CFU counts of the bacterial inputs at time zero were defined as 100% survival. The statistical significance was determined by using the Student t test.
Mouse virulence test.
Virulence tests were carried out as described previously (43). Bacteria were grown for 18 h at 27°C and diluted to the appropriate infectious dose. Groups of five BALB/c mice (6 to 8 weeks old, female) were infected with Y. enterocolitica WA derivatives, each harboring the pYV virulence plasmid with a different yadA hybrid gene. For intravenous or intraperitoneal infection, 5 × 104 bacteria were injected into the tail vein (intravenously) or into the abdomen (intraperitoneally), respectively. For peroral infection, 109 bacteria were administered intragastrically (animal licensing committee permission no. 209.1/211-2531-105/03). At the indicated day after infection, mice were sacrificed, the small intestine was washed with 5 ml of ice-cold PBS, and the Peyer's patches, spleens, and livers were homogenized in 1 ml of PBS. The quantity of yersiniae in the intestinal content and organs was determined by plating out 0.1 ml of serial dilutions of the homogenates on LB-agar plates supplemented with the appropriate antibiotics [nalidixic acid-kanamycin for WA(pYV-O8-A0) and nalidixic acid-spectinomycin for all other strains] and counting the CFU. Five colonies from every experiment were tested for the presence of the corresponding plasmid.
RESULTS
Construction of yadA hybrid genes in Y. enterocolitica serotype O:8.
The most homologous region between Oca family members is localized in their C-terminal TLD, as could be shown by sequence alignment and crystal structure studies for YadA and Hia (26, 56). This TLD can be separated into a proximal linker domain, which consists of three N-terminal heptamer repeats and which forms a left-handed alpha-helical coiled-coil, followed by a small C-terminal hairpin-loop, and the distal beta-barrel domain of four antiparallel transmembrane beta-sheets (Fig. 1A and B). To replace the codons of the predicted TLD of the yadA gene (i.e., codons 334 to 422) with the corresponding TLDs of either the uspA1 gene (codons 742 to 832), the eibA gene (codons 306 to 392), or the hia gene (codons 1009 to 1098), a PCR-based strategy was used (Fig. 1C). The introduction of a SacI-site between the fragments PCR-1 and PCR-2 resulted in two codon changes leading to two amino acid substitutions in YadA(D332L, H333E). Therefore, to rule out an impact of this exchange on YadA function a corresponding mutant yadA(D332L, H333E) gene was also constructed. For expression studies and further functional analysis, the chimeric yadA genes were transferred into Y. enterocolitica WA(pYVO8-A-0), harboring the virulence plasmid pYVO8-A-0 (deleted yadA gene) and integrated into pYV by using the λpir suicide vector pGP704 (see Table 1).
The YadA passenger domain is translocated across the OM and forms an oligomer on the bacterial surface by UspA1, EibA, and Hia TLDs.
Previously, we have shown that the C-terminal 91 aa of YadA (aa 332 to 422), namely, the linker and membrane anchoring region (comprising the TLD), are involved in translocation and oligomerization of its passenger domain (YadA, aa 26 to 331) (26). To test the hypothesis that the homologous TLDs of related Oca family members UspA1, EibA, and Hia are also capable of mediating translocation and oligomerization of the YadA passenger domain, we constructed YadA-UspA1, YadA-EibA, and YadA-Hia fusion proteins. Production of the YadA hybrid proteins was verified in whole-cell lysates of mid-logarithmic-phase cultures by Western blotting with MAb 8D1 (data not shown).
To test whether these YadA hybrids were also localized on the bacterial surface in comparable amounts, an IFA, an ELISA of whole, unfixed Yersinia cells, and Western blotting of Yersinia OM preparations were performed with MAb 8D1 (Fig. 2 and Fig. 3A, IFA results not shown).
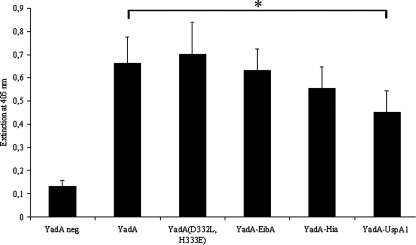
FIG. 2.
Comparison of surface expression of wild-type YadA, YadA(D332L, H333E), YadA-EibA, YadA-Hia, and YadA-UspA1 on whole, unfixed Yersinia WA(pYV-O8) cells by an ELISA with MAb 8D1. The asterisk indicates the highly significantly (P < 0.01) diminished surface expression of YadA-UspA1 compared to wild-type YadA. YadA neg., YadA-negative strain WA(pYVO8-A-0); YadA, strain carrying wild-type YadA WA(pYVO8-A-1).
FIG. 3.
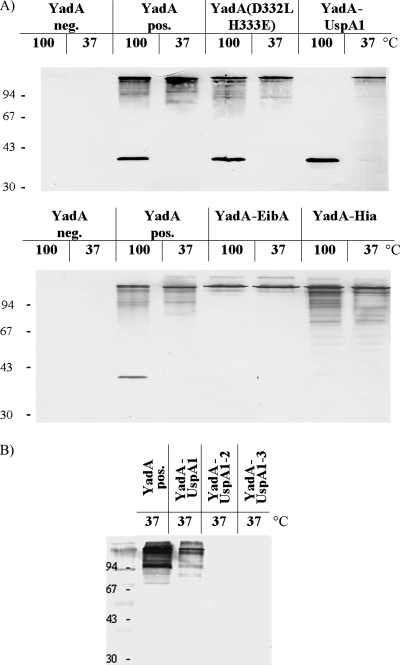
Expression, OM localization, and stability of high-molecular-weight complexes of controls YadA negative, YadA positive, and YadA(D332L, H333E) and of YadA-hybrid proteins YadA-UspA1, YadA-EibA, and YadA-Hia (A) and of three different YadA-UspA1 hybrid proteins: YadA-UspA1, YadA-UspA1-2, and YadA-UspA1-3 (B). OM fractions were prepared from Yersinia WA strains harboring the gene for the corresponding YadA-hybrid protein on their pYV virulence plasmids. Strains were grown at 37°C for 6 h before OM preparation, and 8 μg of each sample was solubilized in sample buffer either for 60 min at 37°C or for 10 min at 100°C, separated by SDS-PAGE, transferred to nitrocellulose sheets, and probed with MAb 8D1. The positions of molecular size markers are shown on the left in kilodaltons.
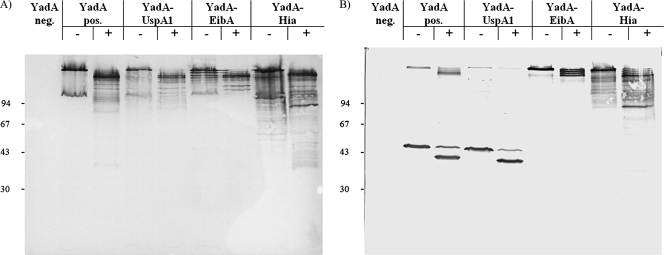
The detection of surface exposure of YadA(D332L, H333E), YadA-EibA, and YadA-Hia by IFA and ELISA using the respective WA strains as antigen and anti-YadA MAb 8D1 as the primary antibody was positive. However, the WA strain expressing the yadA-uspA1 gene showed significantly lower ELISA values than did the other strains (Fig. 2), suggesting a lower amount of surface exposure of YadA-UspA1. This could be confirmed by the immunoblotting of OM preparations using MAb 8D1 (Fig. 3A). Alternatively, it is also conceivable that changes in the YadA stalk structure due to the hybrid anchor domain of UspA1 lead to decreased binding of the MAb 8D1-recognized epitope within the stalk. At 37°C, wild-type YadA characteristically forms oligomers in SDS-PAGE, which disintegrate partially to the YadA monomer after the sample is boiled for 10 min at 100°C or completely after treatment with 8 M urea. The YadA(D332L, H333E) control construct showed disintegration behavior identical to that of wild-type YadA. Interestingly, neither boiling (Fig. 3A) nor treatment with 8 M urea (data not shown) led to disintegration of the oligomeric YadA-EibA and YadA-Hia bands, whereas YadA-UspA1 completely disintegrated to its monomeric form at 100°C. This implies that the TLDs of UspA1, EibA, and Hia determine the oligomeric stability of the YadA hybrid protein. To gain further insights into the structural features and the folding state of the YadA hybrid proteins, we checked protease access to YadA hybrids by treatment of yersiniae with trypsin (ranging from 0.25 to 250 μg/ml). A total of 250 μg of trypsin/ml resulted in complete proteolysis of the YadA passenger domain (negative MAb 8D1 immunoblot [data not shown]), whereas 2.5 μg of trypsin/ml led to truncated YadA and YadA hybrid proteins (Fig. 4, see the double bands of monomeric YadA and YadA-UspA1 after boiling [Fig. 4B]). This approach demonstrates that, in their oligomeric form, all YadA hybrids showed a major truncation approximately comparable to that of wild-type YadA, although there were slight differences in the trypsin cleavage patterns of YadA-EibA and YadA-Hia and thus in trypsin sensitivity.
FIG. 4.
Protease sensitivity assay of Y. enterocolitica WA-314 strains carrying YadA and YadA hybrid constructs. Strains were grown at 37°C for 6 h and 107 bacteria were submitted to a 60 min tryptic digest with 2.5 μg of trypsin/ml, subsequently solubilized in sample buffer for 60 min at 37°C (A) or 100°C (B), separated by SDS-PAGE, and probed with MAb 8D1. YadA neg., YadA-negative strain WA(pYVO8-A-0); YadA pos., strain carrying wild-type YadA WA(pYVO8-A-1); +, trypsin added; −, no trypsin added. The positions of molecular size markers are shown on the left in kilodaltons.
In summary, it could be established that the TLDs of EibA, Hia, and UspA1 were capable of translocating efficiently the heterologous passenger domain of YadA to the bacterial surface. However, the heat stability of the trimeric form of the YadA hybrids was significantly different, with YadA-EibA and YadA-Hia forming more stable and with YadA-UspA1 forming less stable trimers than the wild-type YadA.
Linker and membrane anchor domain of UspA1 form a coherent oligomerizing translocation module.
Next, we studied the impact of the linker domain and its hairpin-loop region on the autotransport capacity of the TLD of UspA1 by replacing these central luminal domains with those of the YadA TLD. YadA-UspA1-3 was constructed by replacing the beta-barrel domain of YadA, i.e., the last 54 aa, with the beta-barrel domain of UspA1. An additional construct YadA-UspA1-2 with replaced beta-barrel domain and 7-aa hairpin-loop of YadA with the corresponding sequence of UspA1 (i.e., the last 61 aa of YadA) was also generated to determine whether the cognate proximal UspA1 coiled-coil region of the linker domain is necessary for the coherence of the UspA1 TLD module. After these constructs were introduced into Yersinia and their expression was induced at 37°C, we could not detect YadA-UspA1-2 or YadA-UspA1-3 in the OM (Fig. 3B). In addition, we checked culture supernatants for released YadA or YadA hybrids and could only detect tiny amounts from 100 ml of culture broth by immunoblotting in all samples, indicating similar stable membrane insertion (data not shown). From this we conclude that the TLD requires for autotransport function the beta-barrel domain together with its cognate hairpin-loop and coiled-coil linker domain, suggesting a close adaptation of these three domains.
YadA TLD-hybrid proteins show comparable adhesive capability as wild-type YadA.
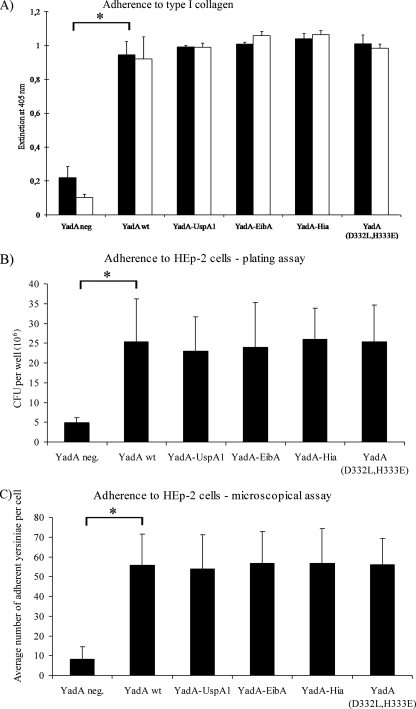
The YadA passenger domain contains an N-terminal binding module for ECM proteins, mammalian cells, and bacterial AA (43, 49, 50). The phenotype AA has been defined as the formation of bacterial aggregates or clumps, which are formed by YadA producing yersiniae during growth in cell culture medium at 37°C (47). As shown previously by electron microscopy, this process is mediated by surface-exposed YadA molecules, which lead to zipper-like interbacterial interactions and require a high YadA surface density and probably lollipop-like structures (16, 23, 50). The exchange of only two amino acid residues (H156Y and H159Y) in this module could be shown to abolish collagen and epithelial cell adherence, which underlines the sensitivity of this module for structural changes (43). In addition, Cotter et al. recently showed that the adhesive capability of this region requires a trimerized passenger domain (13). Therefore, the AA, collagen type I, and HEp-2 cell adherence of Y. enterocolitica WA-314 producing either YadA-UspA1, YadA-EibA, YadA-Hia, YadA(D332L, H333E), or wild-type YadA were compared. All of these strains showed a highly significantly (P < 0.01) enhanced binding to immobilized collagen type I or HEp-2 cells compared to the YadA-negative strain (Fig. 5). Although these results suggest an exposed and functional YadA head domain, the AA test showed that the YadA-UspA1-producing strain was AA negative in contrast to the other strains (data not shown). In summary, the TLD domains do not affect collagen or cell binding of YadA chimera strains. However, these adhesive capabilities could be separated from the AA trait, suggesting differences in the head structure between YadA-UspA1 and the other chimeric YadAs or wild-type YadA.
FIG. 5.
(A) Type I collagen adherence of Y. enterocolitica strains producing YadA or the YadA hybrid proteins as indicated. Collagen coating concentrations: ▪, 20 μg/ml; □, 2 μg/ml. Yersinia adherence was determined by a YadA-specific immunoassay. (B and C) HEp-2 cell adherence of Y. enterocolitica strains producing YadA or the YadA hybrid proteins, determined by CFU per well (values are the averages of triplicate samples, with the ranges indicated, and reflect similar results from several experiments) (B) or by microscopic counting of cell-associated yersiniae (average number of yersiniae per cell obtained from 30 randomly selected cells, with the ranges indicated) (C) (see Materials and Methods). Highly significant differences in these experiments are indicated by an asterisk for the difference between the YadA negative strain and the YadA wild-type strain, which are also representative for the differences between the YadA-negative strain and each of the YadA hybrid-producing strains (P < 0.01).
The YadA TLD contributes to serum resistance.
Another important virulence-associated function of YadA is its ability to mediate resistance to the bactericidal activity of human serum (7, 38, 42). Previously, we could demonstrate that serum resistance is not strictly dependent on the head or neck region of YadA, since deletions of either or both of these two regions did not result in serum-mediated killing of yersiniae. In addition, while the stalk region is thought to be an important contributor to serum resistance, it is not essential for this phenomenon, since an in-frame deletion of the first four 15-mer repeats of the stalk region yielded a serum-resistant phenotype (42). Therefore, we hypothesized that the TLD of YadA could contribute significantly to bacterial survival in human serum. We therefore tested the Yersinia strains carrying the different YadA anchor-hybrid genes for survival in 50% human serum. The results show that the exchange of the YadA TLD with either that of EibA, Hia, or UspA1 leads to a significant loss of Yersinia survival in human serum (Table 3). YadA-EibA or YadA-Hia producing yersiniae appeared to be moderately serum resistant (no significant difference between both strains [P < 0.1]), whereas the YadA-UspA1 Yersinia strain was as serum sensitive as the YadA minus strain (no significant difference [P < 0.9]). As expected, the YadA(D332L, H333E) control strain was comparable in serum resistance to wild-type YadA strain WA(pYVO8-A-1) (no significant difference [P < 0.6]). These results demonstrated that the TLDs are directly or indirectly involved in serum resistance.
TABLE 3.
Survival of WA strains in 50% pooled normal human serum at 37°C
| Strain | Mean % survival ± SD after 90 min of incubationa | P |
|---|---|---|
| YadA, negative | 3 ± 22 | <0.9, <0.005, <0.05, <0.2 |
| YadA, wild type | 229 ± 67 | <0.6, <0.005 |
| YadA(D332L, H333E) | 251 ± 30 | <0.6 |
| YadA-EibA | 84 ± 47 | <0.1, <0.05 |
| YadA-Hia | 28 ± 24 | <0.1, <0.2 |
| YadA-UspA1 | 1 ± 13 | <0.9 |
A 100% value corresponds to the initial bacterial input determined by CFU.
The YadA TLD is necessary for Y. enterocolitica virulence in oral, intraperitoneal, and intravenous BALB/c mouse infection models.
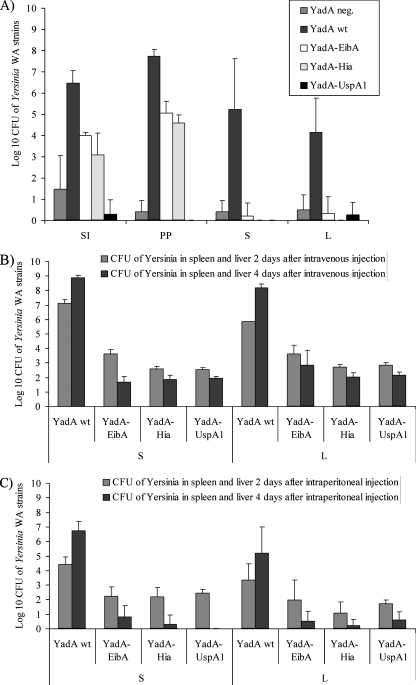
To elucidate the importance of the YadA TLD to Y. enterocolitica virulence in a BALB/c mouse model, we infected groups of five mice orally, intraperitoneally, and intravenously with WA-strains producing the different YadA-hybrid proteins and compared them to WA(pYVO8-A1) and WA(pYVO8-A0) strains. Five days after peroral infection with 109 bacteria and 2 and 4 days after intraperitoneal and intravenous infection with 5 × 104 bacteria, respectively, mice were sacrificed, and the numbers of CFU in the organs were determined. For all three routes of infection, the WA strains producing YadA hybrid proteins showed a highly significant attenuation (Fig. 6). After peroral infection, in comparison to the control WA(pYVO8-A1) strain, only ca. 0.1% of the WA(pYVO8-YadA-EibA) and WA(pYVO8-YadA-Hia) strains could still be detected in the small intestine and Peyer's patches, whereas WA(pYVO8-YadA-UspA1) was completely undetectable. With the peroral route of infection almost no yersiniae producing YadA-hybrid proteins could be detected in the spleen or liver. In the intraperitoneal and intravenous infection experiments WA(pYVO8-A1) efficiently colonized the spleen and liver with a 100-fold increase of CFU from day 2 to day 4, while yersiniae producing YadA-hybrid proteins showed a considerably reduced colonization of the spleen and liver at day 2, which was even more diminished after 4 days, resembling a progressive clearance of infection. In all infection experiments the strain harboring the construct control YadA(D332L, H333E) yielded CFU counts comparable to those obtained with strain WA(pYVO8-A1) (data not shown). The presence of the recombinant plasmids during mouse infection was checked by PCR. From each strain five colonies were reisolated from mouse organs and found to be pYV positive in all cases.
FIG. 6.
Virulence of Y. enterocolitica serotype O:8 strains WA(pYVO8-A-0) (YadA-negative virulence plasmid carrying strain), WA(pYVO8-A-1) (YadA wild type), WA(pYVO8-YadA-UspA1), WA(pYVO8-YadA-EibA), and WA(pYVO8-YadA-Hia) in orally (A), intravenously (B), and intraperitoneally (C) infected groups of five BALB/c mice infected with 1 × 109 CFU (A) or 5 × 104 CFU (B and C) of bacteria. After 5 days (A) or 2 and 4 days (B and C), the CFU of bacteria in the small intestine (SI), Peyer's patches (PP), spleen (S), and liver (L) (A) or in the spleen (S) and liver (L) (B and C) were determined. The data are means ± the standard deviations. wt, wild type.
DISCUSSION
Recently, crystallization and X-ray structure determination experiments of both the YadA and Hia C-terminal regions revealed a trimeric beta-barrel through which the coiled-coil linker region protrudes and could therefore confirm former biochemical structure predictions (26, 56). In the present study, we constructed hybrid proteins consisting of different Oca family TLDs (i.e., a linking region and the transmembrane beta-barrel region) and the YadA passenger domain to address two issues. First, we wanted to find proof for our assumption that the TLD is an autonomous functional translocator unit by exchanging the YadA TLD with related TLDs of Oca family members. Second, we wanted to reveal the contribution of the TLDs to serum resistance and virulence in the mouse.
All three constructed YadA-hybrid proteins were translocated across the OM and exposed their trimeric YadA passenger domain, as shown by detection with the YadA MAb 8D1. However, the YadA-UspA1 hybrid protein showed ca. 20% less MAb 8D1 reactivity, as demonstrated by ELISA and immunoblotting data, suggesting less surface exposure in comparison to wild-type YadA. In spite of this slight difference it can be concluded that the TLDs of YadA, EibA, Hia, and UspA1 are able to translocate the YadA passenger domain, which means that they can replace each other without a significant loss of autotransporter function. Upon comparing the amino acid sequence of the TLD regions of EibA, Hia, and UspA1 with that of YadA, we found homologies of only 44, 23, and 18%, respectively. In spite of this low degree of relatedness, YadA passenger domain translocation was efficient, indicating that the TLDs had similar structures. This finding is in accordance with crystal structure studies on the beta-barrels of monomeric autotransporters NalP and EspP, which showed almost superimposable structures by a homology of only 15% (5, 6, 36). Since the two independently analyzed crystallographic structures of YadA and Hia membrane anchors also showed a high degree of similarity (26, 56), we conclude from our results that processes of membrane insertion and passenger domain translocation should be very similar and conserved in the Oca protein family.
Recently, the involvement of Omp85/YaeT in autotransporter assembly in the OM has been described, suggesting that C-terminal sequences of OM proteins might possess species-specific recognition sites for Omp85/YaeT, an effect that was observed when trying to express meningococcal porin PorA in E. coli (41). Considering the involvement of Omp85/YaeT, our results with YadA-hybrid proteins are interesting, because we have not found strong evidence for species specificity in the function of trimeric autotransport. Possibly, this species specificity of Omp85/YaeT observed for porin proteins does not apply to Oca family proteins.
To study the promiscuous autotransporter capability of Oca family TLDs in more detail, we constructed two additional YadA-UspA1 hybrids, YadA-UspA1-3 and YadA-UspA1-2, by replacing the entire UspA1 linker region or only its proximal coiled-coil part by the homologous YadA region, respectively. In contrast to the functional chimeric YadA-UspA1 autotransporter, the YadA-UspA1-3 and YadA-UspA1-2 chimeras were not detectable on the bacterial surface, in the OM fraction, or in bacterial whole-cell lysates. Previous work has shown that a full-length YadA with specific deletions of either the proximal coiled-coil or distal hairpin-loop segment of the linker region is not functionally expressed and cannot be detected in the cytosol or the membrane fraction (42). Furthermore, the crystallographic structure of the YadA C terminus displayed the trimeric alpha-helical coiled-coil traversing the beta-barrel pore, indicating a structural and functional unit of the TLD (56). On the other hand, however, it could also be shown in previous work that the hairpin-loop region of the linker alone is sufficient to allow oligomerization and OM insertion of a FLAG-tagged truncated YadA membrane anchor (42). The results of the three YadA-UspA1 hybrid proteins demonstrate that the UspA1 beta-barrel absolutely requires its cognate UspA1 linker region for translocation of the YadA passenger domain. This again indicates that the linker region and the four transmembrane beta-barrel strands form a coherent autotransporter module, i.e., a functional TLD, with translocation competence for foreign passenger proteins. This demonstrates also that the function of TLDs is sensitive to changes in amino acid sequence, as has also been shown by Grosskinsky et al.; in that study, the exchange of the highly conserved glycine residue G389 in the YadA TLD led to a severe impairment of YadA translocation (16). Furthermore, Meng et al. could show that amino acid exchanges in the hairpin loop of the Hia TLD led to changes in structural stability (26).
For Oca family members a temperature-sensitive oligomerization stability is known, which can be demonstrated by SDS-PAGE: UspA1 depolymerizes completely after the OM sample is boiled for 5 min (12), whereas for Hia harsh formic acid pretreatment is required for disintegration of the trimers in SDS-PAGE (13). The EibA oligomer has also been shown to remain completely stable after boiling (46). Interestingly, this oligomer stability could also be observed for the corresponding YadA chimeras. Although, the YadA oligomer could be separated into trimer and monomer bands after boiling and SDS-PAGE, the YadA-UspA1 hybrid disintegrated completely into its monomeric form. Strikingly, YadA-EibA and YadA-Hia hybrids completely remained in their oligomeric form after boiling. Treatment of the samples with 8 M urea disintegrated YadA and YadA-UspA1 completely but not the YadA-EibA and YadA-Hia hybrids (results not shown). From this we conclude that the TLDs of these trimeric autotransporters determine the heat stability of the oligomeric form. The oligomers also remained stable after a mild tryptic digestion, indicating that oligomerization is controlled by the TLDs and not by the passenger domain. Recently, it was demonstrated for Oca family member Hia that a stably oligomerized passenger domain is required for cell adhesion (13). Therefore, yersiniae producing YadA chimeras were tested for adherence capability to collagen and HEp-2 cells. Interestingly, the YadA hybrids showed no significant differences in the two adherence tests. These results do not exclude that the YadA hybrid proteins might differ in their passenger domain structure (e.g., in their packing density). Meng et al. could show that amino acid exchanges in the hairpin loop of the Hia linker region lead to changes in structural stability of Hia but not in adhesion ability for cells or ECM (26). Furthermore, bacterial AA, a YadA-dependent phenotype, could not be observed for the YadA-UspA1 hybrid producing yersiniae. The ELISA data and the immunoblot analysis indicated slightly reduced surface concentrations of YadA-UspA1 hybrid protein in the OM. This might suggest an impaired formation or reduced stability of the trimeric YadA head structure in YadA-UspA1, which could lead to the loss of AA.
Another feature of the YadA molecule and several other Oca family members (e.g., UspA1, UspA2, EibA, and DsrA) is their ability to confer resistance to serum-mediated killing (38, 53). However, the exact mechanism of serum resistance mediated by Oca family members is still unclear. For example, it was possible to demonstrate that the passenger domains of UspA1 and UspA2 are involved in the binding of complement inhibitor factors C4BP, C3, and vitronectin, but the physiological relevance of these binding activities still needs to be fully elucidated (3, 32, 33). Also, the binding of the complement inhibitor factor H through YadA has been under debate, but until now these findings could not be corroborated (7, 10). In contrast to EibA, the Hia protein does not confer serum resistance. However, a detailed analysis of EibA-mediated serum resistance is lacking. Previously, we could demonstrate that the YadA head-neck region is not necessary for conferring serum resistance, but we were not able to exactly localize it to the stalk or TLD of YadA (42). Therefore, we presumed that the YadA TLD could be important for this process, since beta-barrel pores from other OM proteins have already been shown to be involved in serum resistance, such as, for example, Ail, another Yersinia OM protein, where probably complement inhibiting factors such as factor H or C4b-binding protein bind to these loops (27). Interestingly, all strains expressing YadA hybrid genes showed reduced resistance to 50% normal human serum. This supports our previous conclusion that the TLD of YadA could contribute to serum resistance. Possibly, the linker region of YadA or the surface-exposed loops linking the transmembrane beta-strands are involved in this phenomenon by binding a complement inhibitor factor, such as, for example, factor H. Although the surface- exposed loops of the TLD of the trimeric autotransporters are shorter than those of Ail or OmpX, the contribution of TLD to serum resistance cannot be excluded. The YadA stalk region could also be involved in the binding of factor H or C4BP. Thus, distortion of the coiled-coil structure of the stalk of YadA due to the fused non-YadA TLD may affect the binding of complement inhibitors and favor complement activation.
Finally, we were interested to find out whether the degree of YadA hybrid protein-mediated serum resistance correlated with the degree of mouse virulence. A comparison of bacterial loads in the Peyer's patches, spleen, and liver after peroral challenge and in the spleen and liver after intravenous or intraperitoneal challenge of BALB/c mice clearly showed a highly significant attenuation of all YadA-hybrid producing strains compared to the wild-type YadA producing WA(pYVO8-A1). Interestingly, the chimeric YadA-UspA1 producing strain seemed to be even more attenuated than the YadA mutant strain. Although we have no convincing explanation for this yet, it is conceivable that chimeric YadA-UspA1 transport and insertion into the OM could cause more envelope stress than the other YadA chimeras, which might affect the type 3 protein secretion apparatus in contrast to the YadA-negative mutant.
In summary, nonautologous TLDs fused to the N-terminal YadA might lead to structural changes or distortions of the YadA stalk and head region with concomitant attenuation of virulence function. Moreover, serum resistance seems to contribute essentially to the virulence function of YadA in the mouse model.
Acknowledgments
Eric J. Hansen of the Southwestern Medical Center (Dallas) is gratefully acknowledged for the gift of the M. catarrhalis strain.
This study was supported by the Deutsche Forschungsgemeinschaft (RO1239/4-2) and the Munich Center for Integrated Protein Science.
Footnotes
Published ahead of print on 16 May 2008.
REFERENCES
- 1.Abdallah, A. M., N. C. Gey van Pittius, P. A. Champion, J. Cox, J. Luirink, C. M. Vandenbroucke-Grauls, B. J. Appelmelk, and W. Bitter. 2007. Type VII secretion: mycobacteria show the way. Nat. Rev. Microbiol. 5883-891. [DOI] [PubMed] [Google Scholar]
- 2.Aebi, C., I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, G. H. McCracken, Jr., and E. J. Hansen. 1997. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect. Immun. 654367-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attia, A. S., S. Ram, P. A. Rice, and E. J. Hansen. 2006. Binding of vitronectin by the Moraxella catarrhalis UspA2 protein interferes with late stages of the complement cascade. Infect. Immun. 741597-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 5.Barnard, T. J., N. Dautin, P. Lukacik, H. D. Bernstein, and S. K. Buchanan. 2007. Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nat. Struct. Mol. Biol. 141214-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein, H. D. 2007. Are bacterial “autotransporters” really transporters? Trends Microbiol. 15441-447. [DOI] [PubMed] [Google Scholar]
- 7.Biedzka-Sarek, M., R. Venho, and M. Skurnik. 2005. Role of YadA, Ail, and lipopolysaccharide in serum resistance of Yersinia enterocolitica serotype O:3. Infect. Immun. 732232-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capecchi, B., J. Adu-Bobie, F. Di Marcello, L. Ciucchi, V. Masignani, A. Taddei, R. Rappuoli, M. Pizza, and B. Arico. 2005. Neisseria meningitidis NadA is a new invasin which promotes bacterial adhesion to and penetration into human epithelial cells. Mol. Microbiol. 55687-698. [DOI] [PubMed] [Google Scholar]
- 9.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.China, B., M. P. Sory, B. T. N′Guyen, M. de Bruyere, and G. R. Cornelis. 1993. Role of the YadA protein in prevention of opsonization of Yersinia enterocolitica by C3b molecules. Infect. Immun. 613129-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cianciotto, N. P. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13581-588. [DOI] [PubMed] [Google Scholar]
- 12.Cope, L. D., E. R. Lafontaine, C. A. Slaughter, C. A. Hasemann, Jr., C. Aebi, F. W. Henderson, G. H. McCracken, Jr., and E. J. Hansen. 1999. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol. 1814026-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotter, S. E., N. K. Surana, S. Grass, and J. W. St Geme. 2006. Trimeric autotransporters require trimerization of the passenger domain for stability and adhesive activity. J. Bacteriol. 1885400-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotter, S. E., N. K. Surana, and J. W. St. Geme. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13199-205. [DOI] [PubMed] [Google Scholar]
- 15.Cotter, S. E., H. J. Yeo, T. Juehne, and J. W. St. Geme. 2005. Architecture and adhesive activity of the Haemophilus influenzae Hsf adhesin. J. Bacteriol. 1874656-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosskinsky, U., M. Schutz, M. Fritz, Y. Schmid, M. C. Lamparter, P. Szczesny, A. N. Lupas, I. B. Autenrieth, and D. Linke. 2007. A conserved glycine residue of trimeric autotransporter domains plays a key role in Yersinia adhesin A autotransport. J. Bacteriol. 1899011-9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 18.Heesemann, J., C. Keller, R. Morawa, N. Schmidt, H. J. Siemens, and R. Laufs. 1983. Plasmids of human strains of Yersinia enterocolitica: molecular relatedness and possible importance for pathogenesis. J. Infect. Dis. 147107-115. [DOI] [PubMed] [Google Scholar]
- 19.Heise, T., and P. Dersch. 2006. Identification of a domain in Yersinia virulence factor YadA that is crucial for extracellular matrix-specific cell adhesion and uptake. Proc. Natl. Acad. Sci. USA 1033375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. a'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6370-378. [DOI] [PubMed] [Google Scholar]
- 22.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 7751-59. [DOI] [PubMed] [Google Scholar]
- 23.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 195989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koretke, K. K., P. Szczesny, M. Gruber, and A. N. Lupas. 2006. Model structure of the prototypical non-fimbrial adhesin YadA of Yersinia enterocolitica. J. Struct. Biol. 155154-161. [DOI] [PubMed] [Google Scholar]
- 25.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405914-919. [DOI] [PubMed] [Google Scholar]
- 26.Meng, G., N. K. Surana, J. W. St. Geme, and G. Waksman. 2006. Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter. EMBO J. 252297-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, V. L., K. B. Beer, G. Heusipp, B. M. Young, and M. R. Wachtel. 2001. Identification of regions of Ail required for the invasion and serum resistance phenotypes. Mol. Microbiol. 411053-1062. [DOI] [PubMed] [Google Scholar]
- 28.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1702575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mota, L. J., and G. R. Cornelis. 2005. The bacterial injection kit: type III secretion systems. Ann. Med. 37234-249. [DOI] [PubMed] [Google Scholar]
- 30.Mota, L. J., L. Journet, I. Sorg, C. Agrain, and G. R. Cornelis. 2005. Bacterial injectisomes: needle length does matter. Science 3071278. [DOI] [PubMed] [Google Scholar]
- 31.Mougous, J. D., M. E. Cuff, S. Raunser, A. Shen, M. Zhou, C. A. Gifford, A. L. Goodman, G. Joachimiak, C. L. Ordonez, S. Lory, T. Walz, A. Joachimiak, and J. J. Mekalanos. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 3121526-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nordstrom, T., A. M. Blom, A. Forsgren, and K. Riesbeck. 2004. The emerging pathogen Moraxella catarrhalis interacts with complement inhibitor C4b binding protein through ubiquitous surface proteins A1 and A2. J. Immunol. 1734598-4606. [DOI] [PubMed] [Google Scholar]
- 33.Nordstrom, T., A. M. Blom, T. T. Tan, A. Forsgren, and K. Riesbeck. 2005. Ionic binding of C3 to the human pathogen Moraxella catarrhalis is a unique mechanism for combating innate immunity. J. Immunol. 1753628-3636. [DOI] [PubMed] [Google Scholar]
- 34.Nummelin, H., M. C. Merckel, J. C. Leo, H. Lankinen, M. Skurnik, and A. Goldman. 2004. The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel beta-roll. EMBO J. 23701-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oomen, C. J., P. van Ulsen, P. Van Gelder, M. Feijen, J. Tommassen, and P. Gros. 2004. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 231257-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pallen, M. J., R. R. Chaudhuri, and I. R. Henderson. 2003. Genomic analysis of secretion systems. Curr. Opin. Microbiol. 6519-527. [DOI] [PubMed] [Google Scholar]
- 38.Pilz, D., T. Vocke, J. Heesemann, and V. Brade. 1992. Mechanism of YadA-mediated serum resistance of Yersinia enterocolitica serotype O3. Infect. Immun. 60189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pohlner, J., R. Halter, K. Beyreuther, and T. F. Meyer. 1987. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature 325458-462. [DOI] [PubMed] [Google Scholar]
- 40.Pukatzki, S., A. T. Ma, D. Sturtevant, B. Krastins, D. Sarracino, W. C. Nelson, J. F. Heidelberg, and J. J. Mekalanos. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 1031528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robert, V., E. B. Volokhina, F. Senf, M. P. Bos, P. Van Gelder, and J. Tommassen. 2006. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS. Biol. 4e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roggenkamp, A., N. Ackermann, C. A. Jacobi, K. Truelzsch, H. Hoffmann, and J. Heesemann. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 1853735-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roggenkamp, A., H. R. Neuberger, A. Flugel, T. Schmoll, and J. Heesemann. 1995. Substitution of two histidine residues in YadA protein of Yersinia enterocolitica abrogates collagen binding, cell adherence, and mouse virulence. Mol. Microbiol. 161207-1219. [DOI] [PubMed] [Google Scholar]
- 44.Roggenkamp, A., K. Ruckdeschel, L. Leitritz, R. Schmitt, and J. Heesemann. 1996. Deletion of amino acids 29 to 81 in adhesion protein YadA of Yersinia enterocolitica serotype O:8 results in selective abrogation of adherence to neutrophils. Infect. Immun. 642506-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandt, C. H., and C. W. Hill. 2000. Four different genes responsible for nonimmune immunoglobulin-binding activities within a single strain of Escherichia coli. Infect. Immun. 682205-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandt, C. H., and C. W. Hill. 2001. Nonimmune binding of human immunoglobulin A (IgA) and IgG Fc by distinct sequence segments of the EibF cell surface protein of Escherichia coli. Infect. Immun. 697293-7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skurnik, M., I. Bolin, H. Heikkinen, S. Piha, and H. Wolf-Watz. 1984. Virulence plasmid-associated autoagglutination in Yersinia spp. J. Bacteriol. 1581033-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Surana, N. K., D. Cutter, S. J. Barenkamp, and J. W. St Geme. 2004. The Haemophilus influenzae Hia autotransporter contains an unusually short trimeric translocator domain. J. Biol. Chem. 27914679-14685. [DOI] [PubMed] [Google Scholar]
- 49.Tahir, Y. E., P. Kuusela, and M. Skurnik. 2000. Functional mapping of the Yersinia enterocolitica adhesin YadA: identification of eight NSVAIG-S motifs in the amino-terminal half of the protein involved in collagen binding. Mol. Microbiol. 37192-206. [DOI] [PubMed] [Google Scholar]
- 50.Tamm, A., A. M. Tarkkanen, T. K. Korhonen, P. Kuusela, P. Toivanen, and M. Skurnik. 1993. Hydrophobic domains affect the collagen-binding specificity and surface polymerization as well as the virulence potential of the YadA protein of Yersinia enterocolitica. Mol. Microbiol. 10995-1011. [DOI] [PubMed] [Google Scholar]
- 51.Thanabalu, T., E. Koronakis, C. Hughes, and V. Koronakis. 1998. Substrate-induced assembly of a contiguous channel for protein export from Escherichia coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 176487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veiga, E., E. Sugawara, H. Nikaido, V. de Lorenzo, and L. A. Fernandez. 2002. Export of autotransported proteins proceeds through an oligomeric ring shaped by C-terminal domains. EMBO J. 212122-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Visser, L. G., P. S. Hiemstra, M. T. van den Barselaar, P. A. Ballieux, and R. van Furth. 1996. Role of YadA in resistance to killing of Yersinia enterocolitica by antimicrobial polypeptides of human granulocytes. Infect. Immun. 641653-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voulhoux, R., M. P. Bos, J. Geurtsen, M. Mols, and J. Tommassen. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299262-265. [DOI] [PubMed] [Google Scholar]
- 55.Voulhoux, R., and J. Tommassen. 2004. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res. Microbiol. 155129-135. [DOI] [PubMed] [Google Scholar]
- 56.Wollmann, P., K. Zeth, A. N. Lupas, and D. Linke. 2006. Purification of the YadA membrane anchor for secondary structure analysis and crystallization. Int. J. Biol. Macromol. 393-9. [DOI] [PubMed] [Google Scholar]