Abstract
Class 1 integrons are central players in the worldwide problem of antibiotic resistance, because they can capture and express diverse resistance genes. In addition, they are often embedded in promiscuous plasmids and transposons, facilitating their lateral transfer into a wide range of pathogens. Understanding the origin of these elements is important for the practical control of antibiotic resistance and for exploring how lateral gene transfer can seriously impact on, and be impacted by, human activities. We now show that class 1 integrons can be found on the chromosomes of nonpathogenic soil and freshwater Betaproteobacteria. Here they exhibit structural and sequence diversity, an absence of antibiotic resistance genes, and a phylogenetic signature of lateral transfer. Some examples are almost identical to the core of the class 1 integrons now found in pathogens, leading us to conclude that environmental Betaproteobacteria were the original source of these genetic elements. Because these elements appear to be readily mobilized, their lateral transfer into human commensals and pathogens was inevitable, especially given that Betaproteobacteria carrying class 1 integrons are common in natural environments that intersect with the human food chain. The strong selection pressure imposed by the human use of antimicrobial compounds then ensured their fixation and global spread into new species.
The rapid appearance and rise of the class 1 integron is one of the most stunning examples of evolution in action, driven by the power of natural selection. Class 1 integrons appeared in a number of different locations, and on different plasmids and transposons, coincident with the widespread use of antibiotics (5, 29). They are now found in 40 to 70% of gram-negative pathogens isolated from clinical contexts (7, 18) and at similar frequencies in pathogens and commensals isolated from livestock (6, 10). The rapid spread of class 1 integrons through gram-negative and, more recently, into gram-positive species has been facilitated by their location on mobile DNA elements, such as plasmids and transposons, coupled with the selective advantage conferred by their associated antibiotic resistance genes (5, 26, 35). They have created a worldwide crisis in the management of bacterial infections.
There are at least 90 distinct integron classes, mostly located on chromosomes, and about 10% of sequenced bacterial genomes carry these elements (3, 19). All integrons capture mobile gene cassettes using site-specific recombination mediated by an integron-integrase (intI) (12). This integrase type catalyzes recombination between a primary recombination site (attI) and a corresponding 59-base element site (59-be or attC) carried on mobile gene cassettes. Through this activity, chromosomal integrons can accumulate up to hundreds of cassettes in a tandem array extending from attI, thus contributing extensively to the diversity of bacterial genomes (9, 14, 27). Chromosomal integrons do exhibit intragenomic, intercellular, and interspecies movement over evolutionary time frames but are generally stable in particular phylogenetic lineages (3, 9, 14, 19, 20, 27).
The first integrons to be described, classes 1, 2, and 3, exhibit a number of features not typical of the more numerically dominant chromosomal integron classes. They are carried on transposons and/or plasmids and most commonly contain from 0 to 6 cassettes drawn from a pool of about 100 cassettes in total, almost all of which encode antibiotic resistance determinants (24, 30). There is experimental evidence (26) that the antibiotic resistance genes found in class 1, 2, and 3 integrons were acquired by capturing gene cassettes from the vast pool of diverse cassettes that are prevalent in microbial communities (3, 19, 21, 28).
While potential sources of resistance genes may have been identified, the origin of the class 1 recombination platform, now so abundant in clinical pathogens, is not known. Since the class 1 integron has had a major role in the spread of antibiotic resistance and led to worldwide difficulties in controlling bacterial infection, understanding the origin of these elements is important both for practical control of antibiotic resistance and for exploring the means by which bacterial lateral gene transfer can seriously impact on, and be impacted by, human activities.
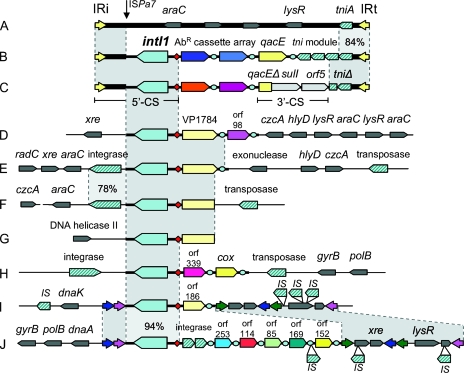
Clues to this origin lie in features that are common to class 1 integrons from clinical contexts. The nucleotide sequences for their integrase genes (intI1) are identical, or nearly so, strongly suggesting that these elements were derived from a very recent common ancestor. Here we will use the term “clinical” class 1 integrons to refer to the group of integrons which share this nucleotide sequence and are commonly found in pathogens and commensals. Clinical class 1 integrons are also associated with transposition functions of a type exemplified by Tn402 (23). The left-hand end of clinical class 1 integrons (Fig. 1B and C) includes a conserved noncoding sequence beyond the end of intI1 that terminates in a 25-bp sequence (IRi) which is an inverted repeat of another sequence (IRt) located at the right-hand end of most extant clinical class 1 integrons (Fig. 1B and C). These inverted repeats define the boundary of the clinical class 1 integron, since they are the recognition sites required for Tn402-mediated transposition. The immediate common ancestor of clinical class 1 integrons was probably similar to a Tn402-like transposon, consisting of (in order) IRi, intI1, attI1, gene cassette(s), qacE (a determinant of quaternary ammonium compound resistance), a complete tni transposition module, and IRt (Fig. 1B) (17, 31). The majority of extant class 1 integrons from clinical contexts carry various modifications to the right hand end of this ancestral element, including deletions in qacE, incorporation of a sulfonamide resistance gene (sulI), and partial deletions of the tni module (Fig. 1C). Nevertheless, all clinical class 1 integrons carry IRi through to attI1 (the 5′ conserved segment [5′-CS]) and retain at least some evidence of Tn402-like transposon sequences (4, 13, 22).
FIG. 1.
Schematic maps of class 1 integrons and associated elements, including Tn6008 from Enterobacter cloacae JKB7 (A), an example of a Tn402 integron (B), and a typical clinical class 1 integron (C). Below these, class 1 integrons from Hydrogenophaga strain PL2G6 (D), Aquabacterium strain PL1F5 (E), Acidovorax strain MUL2G8 (F), Imtechium strain PL2H3 (G), Azoarcus strain MUL2G9 (H), Thauera strain B4 (I), and Thauera strain E7 (J) are shown. Symbols are as follows: red diamonds, attI1 sites; blue circles, 59-be; block arrows, genes (showing the direction of transcription, with diagonal blue stripes indicating transposases, recombinases, or IS elements and solid colors indicating cassette-encoded genes or the class 1 integron-integrase gene intI1); colored arrowheads, inverted repeat regions, including IRi and IRt associated with Tn402 (yellow) and those found in Thauera chromosomal integrons (pink, blue and green); shaded regions, nucleotide homology between elements. Where this homology falls below 99%, the percent identity is given. The vertical arrow indicates the breakpoint for sequence homology between clinical and chromosomal class 1 integrons and is also the insertion point for ISPa7. Selected genes outside the integron are named. Generally, these genes have phylogenetic relationships that are consistent with vertical inheritance from a common betaproteobacterial ancestor. Other symbols are defined in the text.
Further clues to the origin of clinical class 1 integrons emerged recently, when two class 1 integrons were discovered in environmental bacteria isolated from sediment samples. These class 1 integrons appeared to predate the association with Tn402-like transposition functions and were chromosomally located (31), suggesting that the ancestor of the clinical class 1 integron was more like a typical chromosomal integron. We predict this ancestor had the following characteristics: (i) it was from an environment that intersects the human food chain; (ii) it was present in a bacterial group whose members carry diverse intI1 relatives; (iii) these intI1 genes should exhibit signs of mobilization; (iv) the ancestral integrons should contain diverse gene cassettes, not exclusively antibiotic resistance determinants; (v) they should lack the Tn402-like transposition features typical of clinical class 1 integrons. Here we show that diverse class 1 integrase genes can be routinely recovered from groundwater or lake sediment and that the chromosomes of various Betaproteobacteria contain class 1 integrons possessing the features described above.
MATERIALS AND METHODS
Sampling sites.
Samples used in this study were obtained from a variety of sources, including urban lakes, a groundwater treatment plant, and human fecal samples. Collection of sediment from Lake Yerbury has been previously described (31). Sediment was also collected from Lake Parramatta, situated in North Parramatta, NSW, Australia. This lake was constructed in 1856 by damming Hunt's Creek. It collects runoff from a low-density urban area. No hospitals or animal production facilities are in the catchment. Sample biofilms from granulated activated charcoal filters were obtained from a groundwater treatment plant in Botany, NSW, Australia. This plant was designed to remove and destroy volatile organic compounds from polluted groundwater and to treat the water for reuse on the Botany Industrial Park. The history and conformation of the plant have been described elsewhere (8). Fecal samples were obtained from healthy human volunteers under Macquarie University Ethics Review Committee approval reference HE01Apr2005-R03921.
Screening samples for intI1 diversity.
Samples were stored at 4°C before extraction of DNA, usually within 2 days of collection. DNA was extracted from sediment, biofilms, charcoal filters, and fecal samples using a bead beating method (36). The PCR competence of the resulting DNAs was confirmed by amplification of 16S rRNA genes using primers f27 and r1492 (16). The presence of intI1 in each sample was initially assessed using primers HS463a and HS464 (31).
CE-SSCP screening of environmental DNA.
Positive DNA samples were processed for capillary electrophoresis single strand conformation polymorphism (CE-SSCP) analysis by performing intI1 PCR using fluorochrome-labeled primer HS463a (5′-6-carboxyfluorescein-CTGGATTTCGATCACGGCACG) and unlabeled HS464. DNA (∼50 ng) from each sample was used as a template for amplification of the 473-bp intI1 fragment in 50-μl reaction mixtures with Pfu Turbo DNA polymerase (Stratagene) using the buffer supplied with the enzyme, 25 pmol of each primer, and a final Mg2+ concentration of 2 mM. The following thermal cycle was performed in a Hybaid Omne PCR machine: 94°C for 3 min (1 cycle); 94°C for 30 s, 60°C for 30 s, and 72°C 90 s (35 cycles); 72°C 5 for min (1 cycle). intI1 positive controls were used in every amplification and subsequent analysis. These included a typical Tn402-like class 1 integron-integrase gene located on plasmid R388 (13, 31) (accession no. BR000038), an isolate of Escherichia coli (KC2) also containing a clinical class 1 intI1, and two chromosomal class 1 integrase genes from Azoarcus communis MUL2G9 and Burkholderiales strain MUL2G11, respectively (31) (accession numbers DQ372711 and DQ372715). The efficiency of PCR was assessed by electrophoresis on 2% agarose. All positive samples generated a single band consistent with the expected size of 473 bp.
PCR products were diluted to between 1:10 and 1:100 with sterile water, depending on the strength of the PCR amplification. One microliter of the diluted sample was added to 10 μl of HiDi formamide containing Liz500 internal lane standards (Applied Biosystems). Samples were denatured at 95°C for 10 min and snap-chilled on ice for 10 min before loading into an Applied Biosystems 3130xl capillary sequencer using a 1.6-kV injection voltage. Samples were separated using 7% conformation analysis polymer (Applied Biosystems) in 50-cm capillaries at 15 kV for 50 min. Run temperatures were set at 25°C after determining the optimal temperature for resolution of diversity in the samples. The results were analyzed using GeneMapper software (Applied Biosystems).
Cloning of PCR products and screening by CE-SSCP.
Seven environmental samples, assessed by CE-SSCP as containing divergent class 1 integrase genes, were used as a source of intI1 clones. The 473-bp intI1 fragment was amplified from each environmental DNA sample using Red Hot DNA polymerase as above, but using unlabeled HS463a primer. The resulting PCR products were purified using Promega PCR purification columns, ligated into T-tailed plasmid vectors (pCR2.1-TOPO; Invitrogen), and used to transform competent E. coli TOP10 cells. Recombinant cells were identified using blue/white selection on ampicillin-5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside LB agar plates. All procedures were as specified by the manufacturer (TOPO TA cloning kit; Invitrogen). Twenty random clones were chosen from each environment, and their inserts were amplified with Pfu polymerase from boiled, single-colony lysates using PCR with primers HS464 and famHS463a. PCR products were prepared for CE-SSCP as described above.
DNA sequencing of selected clones.
Clones containing intI1 sequence variants were identified both within and between environments, based on relative mobility after CE-SSCP. Two examples of each mobility class, where available, were sequenced from each environment. Plasmids were purified from overnight liquid cultures using Wizard miniprep plasmid purification columns (Promega) according to the manufacturer's instructions. DNA sequencing reactions were performed at the Macquarie University sequencing facility using dye terminator technology and the flanking primer PCRNf (9). Sequences were determined on an Applied Biosystems 3130xl capillary sequencer, edited, and confirmed as intI1 sequences by interrogation of the NCBI DNA sequence database using BLAST algorithms (http://www.ncbi.nlm.nih.gov/BLAST/).
Recovery of intI-positive bacteria from environmental and fecal samples.
Lake sediment and granulated charcoal samples were serially diluted in phosphate buffer, spread onto Plate count agar (PCA) plates, and incubated at 25°C for 5 to 7 days. A total of 192 colonies per sample were picked into microtiter trays containing 100 μl PCA broth and incubated at 25°C for 48 h. Bacterial boil lysates were used as templates for intI1 PCR (31). Isolates from fecal samples were obtained using a method described previously (2) and screened for intI1 using colony PCR. Positive isolates were stored on PCA or L-agar plates and as glycerol stocks. These isolates were used as templates for intI1 and 16S rRNA PCR and were identified by sequencing of their 16S rRNA genes. Sequencing of intI1 and 16S rRNA genes was performed using BigDye v3.1 chemistry run on an ABI Prism 377 (PE Biosystems).
Construction and screening of fosmid libraries.
The intI1-positive isolates from Lake Parramatta were identified as members of the genera Imtechium, Aquabacterium, and Hydrogenophaga. The intI1-positive isolates from the groundwater treatment plant were identified as members of the genus Thauera, and the human fecal isolate of interest was identified as Enterobacter cloacae. Fosmid DNA libraries were constructed from the genomic DNA of each of these isolates. Construction of libraries and their screening for intI1 were conducted as outlined by Stokes et al. (31). Fosmids were purified from intI1-containing clones and used as templates for DNA sequencing across the integron region and flanking DNA. Fosmids from Thauera isolates E7 and B4 were sequenced at the Macquarie University sequencing facility by primer walking from intI1 and the insert termini. Fosmids from Lake Parramatta isolates and the E. cloacae isolate were commercially sequenced by Macrogen, Korea.
Sequence assembly.
Sequences of the Lake Parramatta and E. cloacae fosmids were assembled by Macrogen, Korea. Sequences of fosmids from the two Thauera isolates were assembled using AssemblyLIGN v1.0.9c (Oxford Molecular). Sequences were analyzed using bioinformatics software available through the Biomanager facility of ANGIS (http://www.angis.org.au/) and were annotated by hand after performing Blastn and Blastx searches through the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST/BLAST.cgi). Sequences were prepared as GenBank flat files for database submission using Sequin v5.00.
Sequence alignments and phylogenetic analysis.
Sequences were aligned using ClustalX (33) and the alignments edited manually to remove ambiguous regions. The final length of the alignment was 686 bp for 16S rRNA genes and 391 bp for intI1. Maximum likelihood phylogenetic analysis for both of these genes was performed with PHYML (11) using a discrete gamma model with eight rate categories plus invariable sites to estimate variation of rate among sites and the GTR model to estimate relative nucleotide substitution rates. Statistical support for the tree nodes represents the consensus of 1,000 trees reconstructed from bootstrap pseudo-replicates of the original data set.
Nucleotide sequence accession numbers.
DNA sequences for the intI1 sequence variants were deposited with GenBank as accession numbers EF470983 to EF471020. DNA sequences of the Thauera Lake Parramatta and E. cloacae fosmids were deposited with GenBank as accession numbers EU316185 and EU327987 to 327991.
RESULTS AND DISCUSSION
We recovered class 1 integrase genes (intI1) from environmental samples using PCR on DNA extracted from soils, sediments, and biofilms. These PCR products were analyzed with SSCP to identify sequence variants of intI1. PCR amplifications containing intI1 sequence variants were cloned and DNA sequenced to establish the extent of diversity in class 1 integrase genes.
On the basis of this analysis, samples from a groundwater treatment plant (8) and lake sediment were chosen for recovery of organisms carrying class 1 integrons. Pure cultures were screened for the presence of intI1 (31). In lake sediment samples, 1 to 3% of colonies were intI1 positive, while in biofilms from the groundwater treatment plant, the number of intI1-positive colonies reached 30%, despite our not using antibiotic selection during culturing. intI1-positive isolates were identified by 16S rRNA gene sequencing. All were nonpathogenic members of the Betaproteobacteria. Genomic DNA was purified from positive isolates belonging to the genera Imtechium, Hydrogenophaga, Aquabacterium, and Thauera and used to construct large insert fosmid libraries, which were screened to recover fosmids containing class 1 integrons (31). Fosmids were completely or partially DNA sequenced, and maps of the integrons and flanking DNA were constructed. More extensive maps were constructed for the two Thauera isolates by combining data from several overlapping fosmids. Maps were compared to typical clinical class 1 integrons and to previously described examples of chromosomal class 1 integrons from Azoarcus and Acidovorax (Fig. 1) (31).
DNA sequencing confirmed the presence of class 1 integrase genes and associated attI1 sites in all isolates. Homology with clinical class 1 integrons extended from attI1 through intI1 and into the flanking sequence downstream of intI1. This homology ended at exactly the same point for all environmental isolates, 107 bp beyond the intI1 stop codon (Fig. 1D to J; see also Fig. S1 in the supplemental data). This location is the target site for insertion of the ISPa7 element into plasmid-borne, clinical class 1 integrons carried by some Pseudomonas aeruginosa isolates (25).
Environmental class 1 integrons did not contain known antibiotic resistance gene cassettes. The integrons in Hydrogenophaga, Imtechium, and Aquabacterium carried a gene cassette whose open reading frame was homologous to a hypothetical protein (VP1784) from Vibrio parahaemolyticus. This cassette was also present in the previously described class 1 integron from Acidovorax strain MUL2G8 (31). The Hydrogenophaga integron carried a second gene cassette containing an open reading frame (orf 98) with no homology to any known gene. The two Thauera isolates each carried different cassette arrays. Isolate B4 had one cassette, encoding a hypothetical protein (orf 186). Isolate E7 carried six cassettes, the first of which encoded putative IS91-like transposition functions. The remaining cassettes contained ORFs encoding hypothetical or unknown proteins (Fig. 1I and J).
We concluded that these integrons were chromosomal, for the following reasons. The preponderance of gene cassettes encoding hypothetical functions is typically a characteristic of chromosomal integrons (3, 14, 28). Some integrons were closely linked to chromosomal genes, such as gyrB and the DNA polymerase III gene. In general, sequences outside the integron region showed extensive homology to betaproteobacterial genome sequences. Genes which are often chromosomal, such as the heavy metal efflux gene czcA, were closely linked to several of the integrons, as were various transcriptional regulators (lysR, xre, and araC) (Fig. 1). Finally, there was no evidence for any sequence motifs typical of plasmids.
The fosmid maps (Fig. 1D to J) showed that each putative chromosomal integron was in a different location, as evidenced by the different left-hand flanking regions. This was the case even for the two Thauera strains B4 and E7 (Fig. 1I and J), which were isolated from the same small biofilm sample and had nearly identical 16S rRNA gene sequences (see Fig. S2A in the supplemental material). Furthermore, the same arrangement of intI1 and a cassette homologous to VP1784 was found in four different genera (Fig. 1D to G). We concluded that class 1 integrons can move between chromosomal locations and between species, even over short evolutionary time scales, although the mechanism(s) mediating this lateral transfer is not clear. The conservation of the left-hand breakpoint (see Fig. S1 in the supplemental material) implies that site-specific recombination is involved. Interestingly, the Thauera integrons were bounded by two different types of inverted repeats (120 bp and 117 bp, respectively) (Fig. 1I and J), a feature typical of the termini of transposable elements.
Confirmation that these class 1 integrons have been mobilized comes from comparison of the phylogenetic tree of intI1 and the corresponding 16S rRNA gene phylogeny of the Betaproteobacteria. We constructed an intI1 tree by incorporating sequences obtained during the PCR-SSCP survey of intI1 diversity, sequences from environmental organisms, and sequences typical of clinical class 1 integrons. These data demonstrate that there is considerable diversity within the intI1 gene family, which comprises at least three distinct clades, each with >94% bootstrap support (see Fig. S2B in the supplemental material). intI1 from Hydrogenophaga, Aquabacterium, and Imtechium clustered with the near-identical intI1 genes from clinical class 1 integrons, represented by intI1 from plasmid R388. The intI1 genes from the Thauera isolates fell into two different clades. The betaproteobacterial tree generated using 16S rRNA gene sequences from database sources and from strains examined here was not congruent with the intI1 tree (see Fig. S2 in the supplemental material). Noncongruence with a reference phylogeny is typical of a gene that has undergone lateral gene transfer.
The mechanism(s) by which chromosomal class 1 integrons might move is unknown but appears to be independent of Tn402-like transposition, since Tn402 features were lacking in every case. However, the majority of integrons studied here were linked to other integrase-like genes or genes involved in DNA transposition, which might account for their mobility. In Thauera strain E7, transposase and helper protein genes were carried by a cassette within the array, and an IS element was inserted into a 59-be within the array. Beyond the right hand boundary of both Thauera integrons was a conserved region containing inverted repeats and transcriptional regulators. This region was a target for IS elements, with five independent insertions being observed (Fig. 1I and J).
The integrons we describe here have all the characteristics predicted for ancestors of clinical class 1 integrons. They occur in organisms that are not known pathogens or human commensals. These organisms are from water bodies, or are associated with plants, and thus intersect the human food chain. The Betaproteobacteria carry intI1 genes whose diversity is much greater than that described for clinical class 1 integrons and includes sequences identical to the latter. These betaproteobacterial integrons have been repeatedly mobilized and carry diverse gene cassettes that are not known antibiotic resistance determinants. Finally, they lack the inverted repeats associated with Tn402-like transposons that are characteristic of all clinical class 1 integrons (13, 15, 17).
We conclude that a chromosomal class 1 integron similar to those described here became incorporated into a plasmid-borne Tn402 transposon during a mobilization event (Fig. 2). This integron may have carried a gene cassette that conferred resistance to an antimicrobial agent, consequently conferring an enormous selective advantage if the newly formed plasmid were conjugated into a human commensal or pathogen. This first cassette may have encoded qacE, since it is a common feature of clinical class 1 integrons, and quaternary ammonium compounds have a longer history of use than antibiotics. Once resident in the human population, the chances of evolutionary success for such an element were vastly improved. The ability of class 1 integrons to access antibiotic resistance cassettes, coupled with the enhanced mobility conferred by the plasmid/transposon combination, allowed the rapid spread and structural diversification of clinical class 1 integrons, while still retaining the class 1 core that participated in the original mobilization event (Fig. 2).
FIG. 2.
Model for the origin and subsequent divergence of the class 1 integrons that are now widely disseminated in pathogens and human commensals. Stages in the hypothetical evolution are as follows. (A and B) The common ancestor of clinical class 1 integrons was a member of an integron pool that was repeatedly acquired by diverse Betaproteobacteria but not other lineages (Fig. 1) (21). (C and D) One betaproteobacterial chromosomal integron inserted/recombined into a Tn402-like element. This event occurred prior to, or concomitant with, the antibiotic era. Capture of qacE probably occurred around the same time. (E) sul1 and orf5 were captured. (F) Deletions, insertions, and other rearrangements involving qacE, sul1, and adjacent sequence generated the 3′-conserved segment (3′-CS). (G) Deletions and insertions involving tni generated Tn402 transposition-incompetent integrons, which conferred various antibiotic resistance phenotypes due to their diverse cassette arrays. (H) trans-mediated Tn402-like transposition events moved integrons into diverse plasmids and other transposons, such as the Tn21 family (22). These events generated further diversity and accelerated the penetration of class 1 integrons into a wide variety of pathogens and commensals.
A key prediction of this model is that representatives of the recipient Tn402-like element should still be recoverable. Such an element should carry flanking sequences that are present in clinical class 1 integrons but lacking in their betaproteobacterial ancestors. We have found such an element in a survey of fecal bacteria from healthy volunteers. An isolate of Enterobacter cloacae was recovered which contained a putative transposon (Tn6008) carrying IRi and part of the 5′-CS, extending up to the ISPa7 insertion point (Fig. 1A; see also Fig. S1 in the supplemental material). It also carried IRt and tniA (although not of the Tn402 type) but did not carry intI1 or any gene cassettes. Consequently, Tn6008 is a representative of the kind of element into which a chromosomal class 1 integron was originally inserted. Recombination between a Tn6008-like element and a betaproteobacterial class 1 integron at the ISPa7 insertion point would generate the 5′-CS typical of clinical class1 integrons (Fig. 1; see also Fig. S1 in the supplemental material). Tn6008 is from a commensal bacterium that also causes nosocomial infections, reinforcing the ease with which mobile elements can move between environments and providing a plausible route for entry of betaproteobacterial class 1 integrons into human commensal flora. This E. cloacae isolate was recovered in the absence of antibiotic selection, underscoring the fact that the current antibiotic resistance epidemic cannot be fully understood by only examining hospital isolates recovered using antibiotic resistance media.
While Tn6008 represents the kind of element into which a class 1 integron might have been inserted, it is not the immediate ancestor of clinical class 1 integrons, since it lacks the required set of transposition genes. However, further clues to the most immediate past event that linked the class 1 integron to a Tn402-like transposon can be found in the Thauera, Hydrogenophaga, and Aquabacterium integrons. Their left-hand boundaries agree with the model described above. In addition, these integrons also have a common right-hand boundary, 44 bases beyond the recombination point of the last gene cassette in each of their arrays. The 44 nucleotides leading up to this boundary are identical, or very nearly so, to a region of Tn402 in the same relative position (see Fig. S3 in the supplemental material). Thus, we predict that the Tn402-like transposon that captured the class 1 integron lacked these 44 bases, since they were brought into the transposon when the class 1 integron was captured.
Why was the class 1 integron the most successful in penetrating and propagating in the clinical environment? We postulate that the predisposition of class 1 integrons for mobilization and their presence in a bacterial group that is widespread and relatively abundant in water supplies were significant factors in their preeminent role in the integron-borne antibiotic resistance epidemic. However, given the apparent ubiquity of integrons in the environment, the selection of an integron with the ability to sequester antibiotic resistance genes was probably inevitable once antibiotics began to be widely used. In support of this idea, the other two integrons of clinical importance, classes 2 and 3, may also have recent environmental ancestors. A functional class 2 integron containing four gene cassettes, none of which encodes antibiotic resistance, has been isolated from Providencia stuartii (1). Similarly, a chromosomal class 3 integron has recently been recovered from Delftia (34). This genus is also in the Betaproteobacteria, and the class 3 integrases are the sister group of class 1 integrases. Finally, the Vibrio salmonicida resistance plasmid pRVS1 carries a tetracycline resistance gene cassette associated with an integron-integrase gene that is 99% identical to the chromosomal integrase in Pseudoalteromonas haloplanktis (32). Consequently, even if we could eliminate all integron-borne antibiotic resistance determinants from the human environment, new antibiotic resistance integrons would inevitably be selected from the vast pool of integrons and gene cassettes that are always endemic in natural environments.
Supplementary Material
Acknowledgments
This work was funded by the Australian Research Council and the National Health and Medical Research Council.
We thank A. Beattie, R. Frankham, and J. Koenig for reviewing early drafts and Michael Selleck for providing biofilms from the groundwater treatment plant.
Footnotes
Published ahead of print on 16 May 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Barlow, R., and K. Gobius. 2006. Diverse class 2 integrons from beef cattle sources. J. Antimicrob. Chemother. 581133-1138. [DOI] [PubMed] [Google Scholar]
- 2.Barlow, R., J. Pemberton, P. Desmarchelier, and K. Gobius. 2004. Isolation and characterization of integron-containing bacteria without antibiotic selection. Antimicrob. Agents Chemother. 48838-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boucher, Y., M. Labbate, J. Koenig, and H. Stokes. 2007. Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol. 15301-309. [DOI] [PubMed] [Google Scholar]
- 4.Brown, H., H. Stokes, and R. Hall. 1996. The integrons In0, In2 and In5 are defective transposon derivatives. J. Bacteriol. 1784429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies, J. 2007. Microbes have the last word. EMBO Rep. 8616-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebner, P., K. Garner, and K. Mathew. 2004. Class 1 integrons in various Salmonella enterica serovars isolated from animals and identification of genomic island SGI1 in Salmonella enterica var. Melagridis. J. Antimicrob. Chemother. 531004-1009. [DOI] [PubMed] [Google Scholar]
- 7.Essen-Zandbergen, A., H. Smith, K. Veldman, and D. Mevius. 2007. Occurrence and characteristics of class 1, 2 and 3 integrons in Escherichia coli, Salmonella and Campylobacter spp. in the Netherlands. J. Antimicrob. Chemother. 59746-750. [DOI] [PubMed] [Google Scholar]
- 8.Gillings, M., M. Holley, and M. Selleck. 2006. Molecular identification of species comprising an unusual biofilm from a groundwater treatment plant. Biofilms 319-24. [Google Scholar]
- 9.Gillings, M., M. Holley, H. Stokes, and A. Holmes. 2005. Integrons in Xanthomonas: a source of species genome diversity. Proc. Natl. Acad. Sci. USA 1024419-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein, C., M. Lee, S. Sanchez, C. Hudson, B. Phillips, B. Register, M. Grady, C. Liebert, A. Summers, D. White, and J. Maurer. 2001. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 45723-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52696-704. [DOI] [PubMed] [Google Scholar]
- 12.Hall, R., and C. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15593-600. [DOI] [PubMed] [Google Scholar]
- 13.Hall, R., H. Brown, D. Brookes, and H. Stokes. 1994. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J. Bacteriol. 1766286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes, A., M. Holley, A. Mahon, B. Nield, M. Gillings, and H. Stokes. 2003. Recombination activity of a distinctive integron-gene cassette system associated with Pseudomonas stutzeri populations in soil. J. Bacteriol. 185918-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kholodii, G., S. Mindlin, I. Bass, O. Yurieva, S. Minakhina, and V. Nikiforov. 1995. Four genes, two ends and a res region are involved in transposition of Tn5053: a paradigm for a novel family of transposons carrying either a mer operon or an integron. Mol. Microbiol. 171189-1200. [DOI] [PubMed] [Google Scholar]
- 16.Lane, D. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, London, England.
- 17.Liebert, C., R. Hall, and A. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Freijo, P., A. Fluit, F. Schmitz, V. Grek, J. Verhoef, and M. Jones. 1998. Class I integrons in gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J. Antimicrob. Chemother. 42689-696. [DOI] [PubMed] [Google Scholar]
- 19.Mazel, D. 2006. Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4608-620. [DOI] [PubMed] [Google Scholar]
- 20.Mazel, D., B. Dychinco, V. Webb, and J. Davies. 1998. A distinctive class of integron in the Vibrio cholerae genome. Science 280605-608. [DOI] [PubMed] [Google Scholar]
- 21.Michael, C., M. Gillings, A. Holmes, L. Hughes, N. Andrew, M. Holley, and H. Stokes. 2004. Mobile gene cassettes: A fundamental resource for bacterial evolution. Am. Nat. 1641-12. [DOI] [PubMed] [Google Scholar]
- 22.Partridge, S., G. Recchia, H. Stokes, and R. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 453014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radstrom, P., O. Skold, G. Swedberg, J. Flensburg, P. Roy, and L. Sundstrom. 1994. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7 Mu and the retro elements. J. Bacteriol. 1763257-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Recchia, G., and R. Hall. 1995. Gene cassettes, a new mobile element. Microbiology 1413015-3027. [DOI] [PubMed] [Google Scholar]
- 25.Riccio, M., L. Pallechi, J.-D. Docquier, S. Cresti, M. Catania, L. Pagani, C. Lagatolla, G. Cornaglia, R. Fontana, and G. Rossolini. 2005. Clonal relatedness and conserved integron structures in epidemiologically unrelated Pseudomonas aeruginosa strains producing the VIM-1 metallo-β-lactamase from different Italian hospitals. Antimicrob. Agents Chemother. 49104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowe-Magnus, D., A.-M. Guerot, and D. Mazel. 2002. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol. Microbiol. 431657-1669. [DOI] [PubMed] [Google Scholar]
- 27.Rowe-Magnus, D., A.-M. Guerot, L. Biskri, P. Bouige, and D. Mazel. 2003. Comparative analysis of superintergons: engineering extensive genetic diversity in the Vibrionaceae. Genome Res. 13428-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stokes, H., A. Holmes, B. Nield, M. Holley, K. Nevalainen, B. Mabbutt, B., and M. Gillings. 2001. Gene cassette PCR: sequence inedependent recovery of entire genes from environmental DNA. Appl. Environ. Microbiol. 675240-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stokes, H., and R. Hall. 1989. A novel family of potentially mobile elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 31669-1683. [DOI] [PubMed] [Google Scholar]
- 30.Stokes, H., D. O'Gorman, G. Recchia, M. Parsekhian, and R. Hall. 1997. Structure and function of 59-base element sites associated with mobile gene cassettes. Mol. Microbiol. 26731-745. [DOI] [PubMed] [Google Scholar]
- 31.Stokes, H., C. Nesbø, M. Holley, M. Bahl, M. Gillings, and Y. Boucher. 2006. Class 1 integrons predating the association with Tn402-like transposition genes are present in a sediment microbial community. J. Bacteriol. 1885722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szerkeres, S., M. Dauti, C. Wilde, D. Mazel, and D. Rowe-Magnus. 2007. Chromosomal toxin-antitoxin loci can diminish large scale genome reductions in the absence of selection. Mol. Microbiol. 631588-1605. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, J., T. Gibson, F. Plewniak, F. Jeanmougin, and D. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 244876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu, H., J. Davies, and V. Miao. 2007. Molecular characterization of class 3 integrons from Delftia spp. J. Bacteriol. 1896276-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, Z., L. Shi, C. Zhang, X. Li, Y. Cao, L. Li, and S. Yamasaki. 2007. Nosocomial infection caused by class1 integron-carrying Staphylococcus aureus in a hospital in South China. Clin. Microbiol. Infect. 13980-984. [DOI] [PubMed] [Google Scholar]
- 36.Yeates, C., and M. Gillings. 1998. Rapid purification of microbial DNA from soil for molecular biodiversity analysis. Lett. Appl. Microbiol. 2749-53. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.