Abstract
Colonization of the Hawaiian squid Euprymna scolopes by the marine bacterium Vibrio fischeri requires the symbiosis polysaccharide (syp) gene cluster, which contributes to symbiotic initiation by promoting biofilm formation on the surface of the symbiotic organ. We previously described roles for the syp-encoded response regulator SypG and an unlinked gene encoding the sensor kinase RscS in controlling syp transcription and inducing syp-dependent cell-cell aggregation phenotypes. Here, we report the involvement of an additional syp-encoded regulator, the putative sensor kinase SypF, in promoting biofilm formation. Through the isolation of an increased activity allele, sypF1, we determined that SypF can function to induce syp transcription as well as a variety of biofilm phenotypes, including wrinkled colony formation, adherence to glass, and pellicle formation. SypF1-mediated transcription of the syp cluster was entirely dependent on SypG. However, the biofilm phenotypes were reduced, not eliminated, in the sypG mutant. These phenotypes were also reduced in a mutant deleted for sypE, another syp-encoded response regulator. However, SypF1 still induced phenotypes in a sypG sypE double mutant, suggesting that SypF1 might activate another regulator(s). Our subsequent work revealed that the residual SypF1-induced biofilm formation depended on VpsR, a putative response regulator, and cellulose biosynthesis. These data support a model in which a network of regulators and at least two polysaccharide loci contribute to biofilm formation in V. fischeri.
In nature, the preferred lifestyle of many bacterial cells is growth within a biofilm, a community of microbes attached to a surface and/or to each other and embedded in a matrix consisting primarily of secreted polysaccharides. Biofilm cells exhibit different properties than individual, planktonic cells, such as increased resistance to antimicrobial agents, altered gene expression, and reduced metabolic rates (reviewed in references 11, 13, 14a, and 40). Because of these properties, and the fact that many human infections occur in the context of a biofilm, a critical area of research is in understanding how biofilms form, persist, and disperse.
To date, a number of organisms have been intensively studied for their biofilm-forming properties, including Pseudomonas aeruginosa (17, 25, 36, 46), Staphylococcus spp. (1, 3, 30, 48), and Vibrio cholerae (23, 42, 45, 50, 51). In these model systems, it has become apparent that numerous traits, such as motility and polysaccharide production, contribute to optimal biofilm formation. It is further evident that the regulatory control over these processes can be quite complex. For example, V. cholerae, which forms biofilms in the natural environment and during intestinal colonization of individuals with cholera (14, 41), uses multiple regulators to control biofilm formation. One of these is VpsR, a putative two-component response regulator protein that alters the transcriptome to produce a biofilm-competent state under specific environmental conditions (5, 22, 49). Two important targets of VpsR are the vpsI and vpsII (vibrio polysaccharide) loci, which produce VPS, the secreted polysaccharide that makes up the bulk of the V. cholerae biofilm matrix (49). Additional regulators, such as VpsT, a putative response regulator, HapR, a quorum-sensing regulator, and proteins that modulate the levels of the second messenger cyclic diguanylate (c-di-GMP), also control this locus (5, 9, 18, 54); thus, a complex network of regulation controlling biofilm formation exists.
While not traditionally considered an important model for biofilm formation, Vibrio fischeri contains a novel locus recently shown to be involved in polysaccharide production and biofilm formation. V. fischeri requires this 18-gene cluster, termed syp for symbiosis polysaccharide, to colonize its symbiotic host, the squid Euprymna scolopes (Fig. 1A) (53). In this highly specific model of symbiosis, E. scolopes acquires its bacterial symbiont soon after hatching (reviewed in references 29, 37, and 43). V. fischeri cells from seawater form a biofilm-like aggregate on the surface of the symbiotic organ before entering pores and migrating to the sites of colonization. The syp cluster and an unlinked regulator, RscS, appear to be critical for the initial aggregation outside the pores; disruption of rscS or a representative syp gene resulted in a loss of aggregate formation and a defect in colonization (44, 52, 53). Furthermore, induction of the syp cluster by multicopy expression of RscS resulted in substantially increased aggregation on the surface of the symbiotic organ; it also caused, in laboratory culture, a variety of cell-cell aggregation phenotypes, including wrinkled colony and pellicle formation (52). These data revealed a link between symbiotic aggregation and biofilm formation in culture and suggested that symbiotic aggregation represents a type of natural biofilm formation that is crucial for V. fischeri to colonize E. scolopes.
FIG. 1.
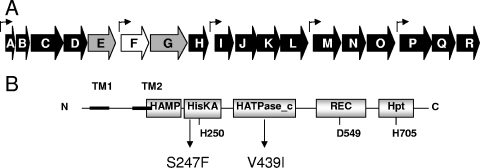
The syp cluster and the SypF protein. (A) The syp cluster contains 18 genes (sypA to -R) that encode putative polysaccharide synthesis and transport proteins as well as four putative regulatory proteins, three of which are two-component regulators. The genes that are the subject of the manuscript are shown with gray (sypE and sypG) and white (sypF) arrows, while the remaining genes are shown with black arrows. The small bent arrows above the genes indicate known and putative promoters. (B) Domain structure of the hybrid sensor kinase SypF. SypF contains conserved histidine (H250, H705) and aspartate (D549) residues typical of hybrid sensor kinases. The putative SypF1 protein contains two substitutions, S247F in the HisKA domain and V439I in the H_ATPase domain, relative to that encoded within the annotated genome.
Control of syp expression appears complex. The unlinked regulator rscS encodes the sensor kinase portion of a two-component regulatory system. Two-component systems are widely used by bacteria to respond to their environment (reviewed in references 4, 7, and 20). A signal is sensed by the sensor kinase, resulting in autophosphorylation; the phosphoryl group is then passed to a response regulator. The activated response regulator then performs some function, such as inducing transcription, to elicit a cellular response. Indeed, we have found that RscS works upstream of SypG, a syp-encoded σ54-dependent response regulator, to induce syp transcription and biofilm formation (21).
Located immediately upstream of sypG are genes for two other putative two-component regulators, sypE and sypF (Fig. 1A). The sypE gene encodes a putative response regulator that lacks a DNA-binding domain, while sypF encodes a putative sensor kinase. Our data to date suggest that SypE affects phenotypes induced by RscS and SypG; however, it appears to affect these phenotypes either positively or negatively, depending on which regulator is overexpressed, through an as-yet-unknown mechanism (21). A role for SypF has not yet been investigated. SypF appears to be a hybrid sensor kinase, with two putative transmembrane regions with a 120-amino-acid chain between them, a HAMP sensory domain, and putative domains typically involved in a phosphorelay (HisKA, HATPase_c, REC, and Hpt) (2, 35) (Fig. 1B).
Due to the complexity of the putative SypF protein and the intriguing location of its gene between two regulators required for biofilm formation, we sought a deeper understanding of its function. As part of this work we identified an increased activity allele, termed sypF1, that strongly induced biofilm-associated phenotypes. We found that these phenotypes depended in part on the syp locus and in part on another putative polysaccharide regulator, vpsR. These experiments thus reveal another layer in the complex control exerted by V. fischeri over polysaccharide production and biofilm formation.
MATERIALS AND METHODS
Media.
V. fischeri strains were grown in SWT (53), LBS (15, 38), or HEPES minimal medium (HMM) (34) containing 0.3% Casamino Acids and 0.2% glucose (53). Escherichia coli strains were grown in LB (10) or brain heart infusion medium (Difco, Detroit, MI). The following antibiotics were added as needed, at the final concentrations indicated: ampicillin (Ap), 100 μg ml−1; chloramphenicol (Cm), 25 μg ml−1 for E. coli and 2.5 μg ml−1 for V. fischeri; erythromycin (Em), 150 μg ml−1 for E. coli and 5 μg ml−1 for V. fischeri; kanamycin (Kn), 50 μg ml−1 for E. coli and 100 μg ml−1 for V. fischeri; tetracycline (Tc), 15 μg ml−1 for E. coli and 5 μg ml−1 [in LBS] or 30 μg ml−1 [in HMM] for V. fischeri. For solid media, agar was added to a final concentration of 1.5%.
Strain and plasmid construction.
Wild-type V. fischeri ES114 (6) was used as the parent strain for these studies. Strains derived from ES114 and plasmids used in this study are described in Tables 1 and 2, respectively. E. coli strains TAM1 λ pir (Active Motif, Carlsbad, CA), TOP10 F′ (Invitrogen, Carlsbad, CA), and DH5α (47) were used for cloning, and E. coli CC118 (19) containing pEVS104 (39) was used to carry out triparental matings as described previously (44).
TABLE 1.
V. fischeri strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| ES114 | Wild-type isolate from E. scolopes | 6 |
| KV1715 | vpsR::pEAH11 | 22 |
| KV1787 | ΔsypG | 22 |
| KV1838 | sypN::pTMB54 | 53 |
| KV2283 | sypI::pTMO89 | This study |
| KV2481 | sypB::pESY33 | This study |
| KV3001 | Wild-type attTn7::PsypAlacZ | 21 |
| KV3030 | sypB::pESY33 attTn7::PsypAlacZ | This study |
| KV3031 | sypN::pTMB54 attTn7::PsypAlacZ | This study |
| KV3232 | ΔsypG attTn7::PsypAlacZ | 21 |
| KV3291 | ΔsypG vpsR::pEAH11 | This study |
| KV3299 | ΔsypE | 21 |
| KV3378 | ΔrscS | Gesvai and Visicka |
| KV3513 | vpsR::pEAH11 attTn7::PsypAlacZ | This study |
| KV3514 | sypI::pTMO89 attTn7::PsypAlacZ | This study |
| KV3586 | ΔsypE vpsR::pEAH11 | This study |
| KV3619 | ΔsypG vpsR::pEAH11 attTn7::PsypAlacZ | This study |
| KV3620 | ΔsypE attTn7::PsypAlacZ | 21 |
| KV3621 | ΔrscS attTn7::PsypAlacZ | This study |
| KV3769 | VFA0884::pCLD28 | This study |
| KV3965 | ΔsypG ΔsypE | This study |
| KV3968 | ΔsypG ΔsypE attTn7::PsypAlacZ | This study |
| KV3970 | VF0352::pCLD49 | This study |
| KV4146 | VF0349::pCLD57 | This study |
K. Gesvain and K. L. Visick, submitted for publication.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristics | Reference |
|---|---|---|
| pCLD6 | Mini-Tn7 delivery plasmid; mob; Knr Cmr attTn7::PsypA-lacZ | 21 |
| pCLD19 | pESY20 + 2 kb upstream and downstream of sypE | 21 |
| pCLD28 | pESY20 + 5′,3′-truncated VFA0884 | This study |
| pCLD29 | sypF1 subcloned into pKV69; Cmr Tcr | This study |
| pCLD42 | vpsR subcloned into pKV69; Cmr Tcr | This study |
| pCLD49 | pESY20 + 5′,3′-truncated VF0352 | This study |
| pCLD54 | Wild-type sypF subcloned into pKV69; Cmr Tcr | This study |
| pCLD57 | pESY20 + 5′,3′-truncated VF0349 | This study |
| pCR2.1-TOPO | Commercial cloning vector; Apr Knr | Invitrogen |
| pEAH11 | pEVS122 + 5′,3′-truncated vpsR (VF0454) | 22 |
| pESY20 | pEVS122 cut with EcoRI (partial), filled in, and self-ligated | 31 |
| pEVS107 | Mini-Tn7 delivery plasmid; mob; Knr Emr | 27 |
| pEVS122 | R6Kγ oriV oriTRP4, Emr, lacZα | 12 |
| pJET1 | Commerical cloning vector; Apr | Fermentas |
| pKV69 | Mobilizable vector; Cmr Tcr | 44 |
| pMAC3 | pKV69 expressing SypF-V439I; Cmr Tcr | This study |
| pMAC4 | pKV69 expressing SypF1; Cmr Tcr | This study |
To overexpress sypF, we amplified the sypF gene using primers sypFexpF (AAGCGGGGACACCACCC) and 1026RTR (GAGGATAGCCAGAGAAGTGG), cloned the fragment into PCR cloning vector pCR2.1-TOPO (Invitrogen, Carlsbad, CA), and then subcloned it into the mobilizable pKV69 (44) to obtain pMAC3. A similar approach was used to generate a vpsR overexpression construct pCLD42, using primers VpsRF (AAGGATCCCATTTGTGAGACGAGATAAGG) and VpsRR (AAGGATCCCATTTGTGAGACGAGATAAGG) and PCR cloning vector pJET1 (Fermentas, Glen Burnie, MD) as an intermediate cloning vector. The sypF overexpression plasmid pMAC3 was passaged through E. coli mutator strain CC130 (26) and introduced into ES114 by conjugation. The resulting colonies were examined for wrinkled colony formation. We isolated the plasmid (pMAC4) from one wrinkled colony and subcloned the sypF portion into pKV69 to obtain the sypF1 overexpression plasmid pCLD29. The sequences of the sypF and vpsR genes in these constructs were obtained using forward, reverse, and/or gene-specific primers using the Genomics Core sequencing facility at Northwestern University. Sequence analysis revealed a single point mutation in pMAC3 (resulting in a V439I substitution), so a wild-type copy of sypF was cloned using sypFexpF and VFA10265R (CCTGCCCTGACATATCTG), yielding pCLD54.
We constructed several V. fischeri mutants using a vector integration approach (8) as follows: we cloned an internal fragment of the target gene into the suicide vector pEVS122 (12) or its derivative pESY20 (31), generally by first cloning the appropriate PCR product into pCR2.1-TOPO. The resulting plasmid was introduced into V. fischeri. The resulting Emr colonies were presumptive mutants, generated through homologous recombination to produce two truncated, nonfunctional copies of the gene. The mutants were subsequently verified by Southern analysis using the vector sequences as probe as described previously (53). The internal fragments for disrupting VFA0884, VF0349, and VF0352 were generated with primer sets VFA0884-c-F (CCGGTTGCAATTGAACTTTTAT)/VFA0884-c-R (GAGTTAAAGGAATGTCTTGCG), VF0349-c-F (CGTCTGCGCGTGCTACTC)/VF0349-c-R (AGCTGCTGCTTCAATAACGG), and VF0352-c-F (GTACCCTAACAAAGGCATTTG)/VF0352-c-R (CTGATACCGTAAGGAGTAAAG), respectively.
The ΔsypG ΔsypE double mutant was constructed by allelic replacement, using the ΔsypG parent strain and the ΔsypE deletion construct pCLD19, as previously described (21).
β-Galactosidase assay.
Strains were grown (in triplicate) with shaking in HMM with Tc at 28°C overnight and then subcultured into fresh medium and grown for 20 h. Aliquots (1 ml) of cells were removed, concentrated, resuspended in Z-buffer, and lysed. The β-galactosidase activity (28) and the protein concentration (24) of each sample were assayed. β-Galactosidase units are reported as units of activity per mg of protein.
Crystal violet assay of biofilm formation.
Strains were grown with shaking in HMM with Tc at 28°C overnight and then subcultured into fresh medium to an optical density at 600 nm (OD600) of 0.1. Cultures were then grown for 48 h under static or shaking conditions and stained with 1% crystal violet. Tubes were rinsed with deionized H2O and photographed. We quantified staining by adding 2 ml 100% ethanol and 1 g glass beads (1 mm), vortexing, and then measuring the OD600.
Pellicle assay.
Strains were grown with shaking in HMM with Tc at 28°C overnight and then subcultured to an OD6oo of 0.1 in 3 ml of fresh medium in 12-well microtiter dishes. Cultures were then grown at room temperature for 48 h. The strength of the pellicle was evaluated by disrupting the air-liquid interface with a sterile pipette tip after 48 h of incubation. Cultures with no pellicle were scored as −; cultures with an easily broken pellicle were scored as +; cultures with an intermediate strength were scored as ++; cultures with a strong pellicle that was able to be lifted from the culture intact were scored as +++. To facilitate photography of the pellicles after they had formed, 25 μl of 100 mg/ml Coomassie blue was added to provide contrast, and the pellicles were disturbed so that they could be visualized.
RESULTS
An increased activity allele of sypF induces wrinkled colony formation.
Biofilm formation by V. fischeri is controlled by at least three regulators, a sensor kinase (RscS) and two response regulators (SypG and SypE) encoded within the syp locus (Fig. 1A) (53). Between the two syp genes lies sypF, which encodes a putative sensor kinase. The juxtaposition of these latter genes suggested a common function. Thus, we hypothesized that SypF might act upstream of SypG and/or SypE to control biofilm formation. To probe the role of SypF, we assayed the biofilm phenotypes of a sypF mutant, as well as a wild-type strain of V. fischeri overexpressing sypF (from the multicopy plasmid pCLD54). Like the wild type and vector control, these strains did not induce wrinkled colony formation, pellicle production, adherence to glass, or syp transcription, although overexpression of sypF caused a slight aggregation of cells during growth with shaking in a minimal medium (HMM) (data not shown).
We thus hypothesized that our culture conditions might not include a strong activating signal recognized by this regulator. Therefore, we sought a constitutively active allele of sypF through random mutagenesis experiments (see Materials and Methods). Of over 10,000 colonies screened, we found a single colony that exhibited a wrinkled colony phenotype (Fig. 2A). We verified through subcloning experiments that the sypF portion of the plasmid was responsible for the wrinkled colony phenotype. The sequence of the sypF1 allele carried by pCLD29 differed from that of sypF in the annotated genome at two positions, resulting in protein changes S247F and V439I. S247F is located within the HisKA domain and is three residues N-terminal to the conserved histidine predicted to be the site of phosphorylation; a change at this position could impact the predicted kinase activity of SypF (Fig. 1). The latter substitution, V439I, appears unimportant, as a strain carrying plasmid pMAC3, which carries sypF with only this change, behaved like a strain carrying wild-type sypF on pCLD54 (i.e., no impact on biofilm formation) (Fig. 2A and data not shown; see also Fig. 3A, 4A, and 5A, below). For the purposes of these studies, we used pMAC3 to control for the impact of SypF1 (expressed from pCLD29) on biofilm formation by V. fischeri.
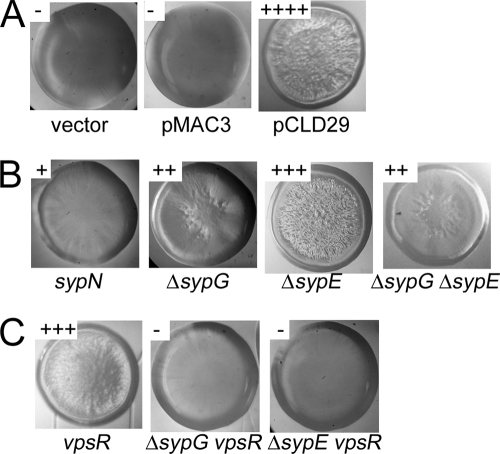
FIG. 2.
Wrinkled colony morphology. Wrinkled colony formation was assessed by streaking strains on LBS with Tc and 0.3% glycerol. (A) Colonies formed by strains carrying the vector control, pMAC3 (sypF overexpression) or pCLD29 (sypF1 overexpression). (B) Colonies formed by pCLD29-containing syp mutant strains, as indicated. (C) Colonies formed by pCLD29-containing vpsR mutant strains and vpsR syp double mutant strains.
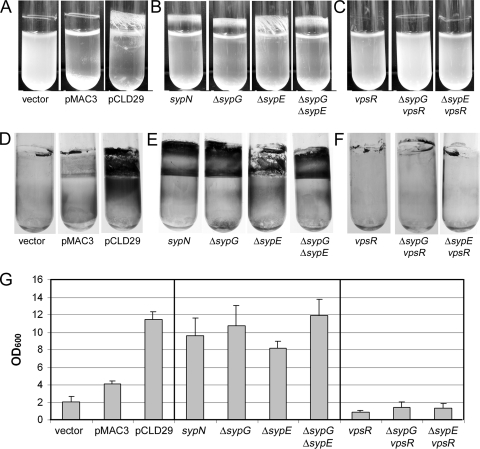
FIG. 3.
Pellicle formation. Pellicles were assayed by culturing strains in HMM with Tc in 12-well microtiter dishes for 72 h. A pipette tip was dragged over the air-liquid interface to visualize the pellicle, and Coomassie blue was added to enhance the contrast. (A) Pellicle formation (or lack thereof) by strains carrying the vector, pMAC3, or pCLD29. (B) Pellicle formation by pCLD29-containing syp mutant strains. (C) Pellicle formation by pCLD29-containing vpsR mutant and vpsR syp double mutant strains. Scores were assigned as described in Materials and Methods.
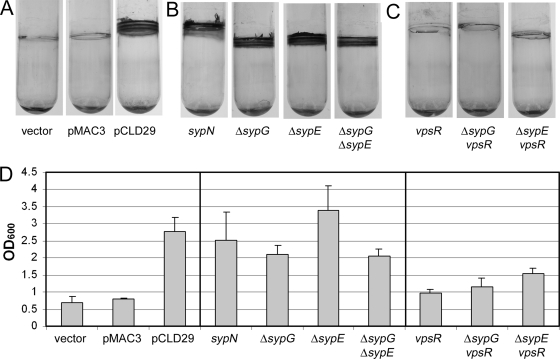
FIG. 4.
Crystal violet staining of glass-attached material following static growth. Glass-attached materials following growth of cells under static conditions were assayed by subculturing into 2 ml HMM with Tc and incubating at room temperature for 48 h and then staining with 1% crystal violet. (A) Crystal violet staining of glass-attached material from cultures of vector-, pMAC3-, and pCLD29-containing cells. (B) Crystal violet staining of glass-attached material from cultures pCLD29-containing syp mutants. (C) Crystal violet staining of glass-attached material from cultures of pCLD29-containing vpsR mutant and vpsR syp double mutant strains. (D) Quantification of the crystal violet-stained material from tubes shown in panels A to C (and additional tubes not shown), solubilized as described in Materials and Methods. Error bars represent standard deviations.
FIG. 5.
Crystal violet staining of glass-attached material following growth with shaking. Glass-attached materials following growth of cells under shaking conditions were assayed by subculturing into 2 ml HMM with Tc and incubating at 28°C for 48 h (A to C) and then staining with 1% crystal violet (D to F). (A and D) Cultures of vector-, pMAC3-, and pCLD29-containing cells. (B and E) Cultures of pCLD29-containing syp mutants, as indicated. (C and F) Cultures of pCLD29-containing vpsR mutant and vpsR syp double mutant strains, as indicated. (G) Quantification of the crystal violet-stained materials from tubes shown in panels D to F (and additional tubes not shown), solubilized as described in Materials and Methods. Error bars represent standard deviations.
Wrinkled colony formation by SypF1 depends on multiple regulators.
Because the sypF1 allele mediated wrinkled colony formation, a phenotype previously associated with syp function, we asked whether SypF1-induced wrinkled colony formation required an intact syp cluster. We introduced pCLD29 or the vector control into mutants defective for representative syp structural genes: sypB, which encodes a putative OmpA-like outer membrane protein, and sypI and sypN, which encode putative glycosyltransferases. In these mutants, the ability of SypF1 to induce wrinkling was substantially reduced (Fig. 2B and data not shown). These data suggest that the syp cluster contributes to SypF1-induced wrinkling.
Our previous work revealed that wrinkled colonies induced by overexpressing the sensor kinase RscS were dependent upon the syp-encoded response regulator SypG, and to a lesser extent, SypE, a non-DNA-binding response regulator (21). To determine if these response regulators were required for SypF1-mediated wrinkled colony formation, we assayed the impact of SypF1 in mutants deleted for sypG or sypE. Deletion of either regulator caused a substantial reduction in, but not elimination of, wrinkling induced by SypF1 (Fig. 2B). To determine whether sypE and sypG together accounted for the ability of SypF1 to induce wrinkled colony formation, we constructed a strain in which both were deleted and then evaluated the resulting colony morphology induced by sypF1. Wrinkled colony formation in this strain was also substantially reduced, but again not eliminated (Fig. 2B). From these data, we conclude that SypE and SypG contribute to wrinkling induced by SypF1, but that another factor is also involved.
SypF1 induces pellicle formation.
Another phenotype associated with the syp cluster is the formation of strong pellicles at the air-liquid interface of static cultures (52). We therefore assessed the ability of SypF1 to induce pellicle formation. We grew the sypF1-overexpressing strains statically in a minimal medium, HMM, for 48 h in a 12-well polystyrene microtiter dish. We found that overexpression of sypF1 in the wild-type strain caused the formation of a strong pellicle that could be lifted from the culture as an intact aggregate. To aid comparisons between this pellicle and those formed in other mutant backgrounds, we applied a sterile pipette tip to the surface of the pellicle and assigned a score (see Materials and Methods) to the resistance encountered; we scored the pellicles produced by pCLD29-containing wild-type cultures as +++ (Fig. 3A).
To determine whether these pellicles depended upon syp structural genes, we assayed pellicle formation by the SypF1-overexpressing syp mutants (sypB, sypI, and sypN). The syp structural mutations substantially disrupted pellicle formation induced by SypF1: the pellicles that formed were very thin and easily broken (scored as +) (Fig. 3B and data not shown). These data support the conclusion that the product of the syp locus makes an important contribution to cell-cell interactions under these conditions.
We next assayed the dependence of SypF1-induced pellicle formation on sypG and sypE. Deletion of sypG substantially reduced, but did not eliminate, pellicle formation induced by SypF1 (scored as +) (Fig. 3B); this pellicle was easily broken into segments, supporting the hypothesis that Syp is important for cell-cell interactions. Deletion of sypE also reduced pellicle formation, though not as severely as the sypG mutant (scored as ++). Finally, a strain in which both sypG and sypE were deleted still retained some SypF1-induced pellicle formation. These data further support the hypothesis that SypF1 may function through an additional regulator.
SypF1 increases adherence to glass.
A third phenotype associated with the syp locus is adherence to a glass surface (21, 52, 53), as assayed by crystal violet staining of cultures grown in HMM. Therefore, we asked whether SypF1 overexpression caused an increase in crystal violet-stainable material of statically grown V. fischeri cultures. We found that wild-type cultures carrying pCLD29 exhibited enhanced crystal violet staining at the air-liquid interface (Fig. 4A and D).
To determine if the SypF1-mediated enhancement of adherence to glass was dependent on syp, we evaluated attachment by pCLD29-containing syp mutants. We found that the sypB, sypI, and sypN genes were dispensable for this phenotype (Fig. 4B and data not shown). This result was somewhat surprising, since SypF1-mediated pellicle formation, a phenotype that occurs under similar static conditions, was disrupted by the loss of these genes. Like the structural genes, sypG and sypE did not appear to be important for the SypF1-induced adherence to glass under static conditions (Fig. 4B). Together with the pellicle data, our results suggested that the role of the syp cluster under these conditions involves promoting cell-cell, rather than cell-surface, interactions. They also supported the existence of a separate pathway, downstream from SypF, involved in mediating adherence to glass.
We also evaluated the ability of SypF1 to induce attachment to glass when cells were grown with shaking, a phenotype first observed in V. fischeri with the overexpression of the response regulator SypG (53). Under these conditions, SypF1 induced substantial cell clumping: the cells appeared aggregated, both at the air-liquid interface and the bottom of the tube, with the rest of the broth remaining relatively clear (Fig. 5A). Like SypG overexpression, SypF1 also induced a robust staining pattern with crystal violet (Fig. 5D and G).
Disruptions of sypB, sypI, and sypN each altered the appearance of the crystal violet-stained material and reduced aggregation (Fig. 5B and E and data not shown). Similarly, the loss of the regulators sypG, sypE, or both altered staining and decreased clumping (Fig. 5B, E, and G), though the sypE mutant had a less severe defect. Despite the different staining pattern, quantification revealed no significant differences in the amounts of crystal violet staining in these mutants relative to the control (Fig. 5G). We speculate that the qualitative difference in staining may result from apparent differences in the aggregation properties of the mutant cultures relative to the wild type (Fig. 5A). Taken together, these results further indicate that, under SypF1-overexpressing conditions, the syp regulators play more important roles in cell-cell adherence (as measured by aggregation) than in cell-surface attachment (as measured by crystal violet staining).
sypF1 induces syp transcription.
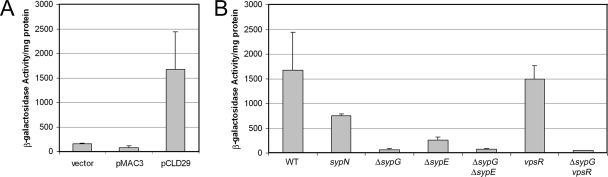
To begin to understand the mechanism by which sypF1 overexpression promotes the observed biofilm phenotypes, we asked whether pCLD29 induced transcription of the syp locus and, if so, whether induction depended upon SypG or SypE. We introduced pCLD29 and the vector control into strains that carried a PsypA-lacZ transcriptional reporter (21) and measured the resulting β-galactosidase activity. Relative to the vector control, pCLD29 induced a 10-fold increase in β-galactosidase activity of the wild-type reporter strain (Fig. 6A). This induction was completely dependent on SypG, as its loss eliminated the observed induction (Fig. 6B). The induction was also somewhat dependent on SypE, as a deletion of this gene reduced SypF1-mediated induction by sixfold (Fig. 6B). However, disruptions of other syp genes (sypB, sypI, and sypN) also caused reductions in sypA transcription (Fig. 6B and data not shown), making it somewhat difficult to interpret the sypE results. In any event, because SypE has no predicted DNA-binding domain, the effect of SypE is likely to be indirect. Not surprisingly, induction of the syp cluster was also eliminated in the sypG sypE double mutant (Fig. 6B). These data suggest that SypF1 primarily acts upstream of SypG in controlling syp transcription.
FIG. 6.
β-Galactosidase activity. Transcription of the syp locus was monitored using a fusion of the sypA promoter region upstream of a promoterless lacZ gene, inserted in the chromosome at the Tn7 site of wild-type and mutant cells. Cells were grown in HMM with Tc for 24 h. (A) Transcription from PsypA-lacZ in cells carrying the vector control, pMAC3, or pCLD29. (B) Transcription from PsypA-lacZ in pCLD29-containing wild-type (WT), syp, or vpsR mutant cells, as indicated. Error bars represent standard deviations.
Some SypF1-induced phenotypes also depend upon vpsR.
Together, our data support a role for syp and syp regulators in the phenotypes induced by SypF1, but it is clear that there are syp-independent effects. Because SypF is a sensor kinase, we hypothesized that it might signal through an additional response regulator(s) to promote the observed phenotypes. V. fischeri encodes 40 putative response regulators, including SypG and SypE. Of these, a prime candidate was VF0454. This gene encodes a protein with high sequence similarity to the V. cholerae gene vpsR (68% identity, 84% similarity over 441 amino acids [with one gap] of the V. fischeri ES114 protein with V. cholerae O1 biovar El tor [e-value, 6e-174]), which controls the vps polysaccharide locus (22, 35, 49).
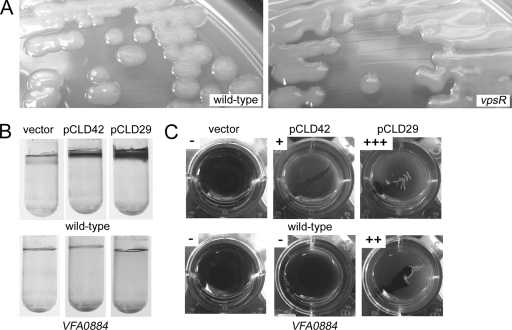
Careful observation of a VF0454 vector integration mutant revealed that it formed colonies that were opaque and mucoid compared to the wild type (Fig. 7A and data not shown), suggesting that VF0454 may serve as a repressor of exopolysaccharide production. Therefore, we designated this gene vpsR, for vibrio polysaccharide regulator. Loss of vpsR also impacted the biofilm phenotypes induced by sypF1 overexpression. Most significantly, the increase in crystal violet-stainable material produced under both static and shaking conditions appeared to be completely abrogated in the vpsR mutant (Fig. 4C and D and 5F and G). Loss of vpsR also altered wrinkled colony formation and reduced the strength of pellicle formed (Fig. 2C and 3C). The resulting pellicle was substantially defective in attaching to the surface of the well, suggesting a role for VpsR in cell-surface attachment. Finally, despite the loss of vpsR, the pCLD29-containing cultures, when grown in HMM with shaking, retained their characteristic clumping, although the amount appeared somewhat reduced (Fig. 5C). Together these data support an important role for VpsR in some SypF-induced phenotypes, particularly the cell-surface phenotypes revealed by crystal violet staining, and a lesser role in others. These effects appeared to be independent of SypF1-mediated syp transcription, as loss of vpsR did not alter this activity (Fig. 6C).
FIG. 7.
Biofilm phenotypes of vpsR mutant and vpsR overexpression strains. Biofilms were produced upon disruption or overexpression of vpsR. (A) Colony morphology of vector (pKV69)-containing wild-type and vpsR mutant cells. (B) Crystal violet staining of glass-attached material from static cultures of wild-type or VFA0884 mutant cells carrying the vector control, pCLD42 (vpsR overexpressing), or pCLD29. (C) Pellicle formation by wild-type or VFA0884 mutant cells with vector, pCLD42, or pCLD29. Scores were assigned as described in Materials and Methods. Cells were grown as described in the legends for Fig. 2, 3, and 4.
Because most SypF-induced phenotypes were not abolished by loss of sypG and sypE, or of vpsR, we asked whether SypF worked through both pathways. To determine if these regulators together could account for all of the SypF-induced phenotypes, we constructed sypG vpsR and sypE vpsR double mutants. Like the single vpsR mutant, these double mutants exhibited a mucoid phenotype, though this phenotype was lost when SypF1 was overexpressed (data not shown). The formation of wrinkled colonies and pellicles was completely abrogated in both pCLD29-containing double mutants (Fig. 2C and 3C). Crystal violet staining of double mutant cultures grown under either static or shaking conditions looked similar to that of the vpsR single mutant, indicating VpsR is responsible for these phenotype (Fig. 4C and 5C). Furthermore, the shaken cultures themselves were substantially less aggregated, appearing more similar to the vector control cultures (Fig. 5C). We obtained similar results when we evaluated SypF1-induced biofilm formation in a mutant defective for both vpsR and a syp structural gene, sypP (data not shown). These data support the hypothesis that syp and a vpsR-dependent pathway together account for the SypF1 overexpression phenotypes.
VpsR overexpression impacts biofilm formation.
To further evaluate the role of VpsR in biofilm formation, we constructed a plasmid that overexpresses the vpsR gene. After introducing this plasmid, pCLD42, into wild-type cells, we evaluated biofilm-associated phenotypes. Overexpression of VpsR was not sufficient to induce wrinkled colony morphology or alter crystal violet staining resulting from cultures grown under shaking conditions (data not shown). However, VpsR-overexpressing cultures were able to form weak pellicles (Fig. 7). Additionally, pCLD42 caused an increase in crystal violet staining from cultures grown under static conditions (Fig. 7). Together, these data support VpsR as an activator of biofilm formation.
Cellulose plays a role in V. fischeri biofilm formation.
In V. cholerae, VpsR controls a large polysaccharide gene locus, vps (49, 51). Only a portion of this locus is conserved in V. fischeri (16). To determine whether this vps-like locus was responsible for the phenotypes induced by either SypF1 or VpsR overexpression, we disrupted two representative genes, VF0349 and VF0352. However, neither of these mutations disrupted the observed biofilm phenotypes (data not shown).
Subsequent exploration of the biofilms induced by these regulators revealed an increase in Congo red binding, a phenotype indicative of, among other things, cellulose biosynthesis. V. fischeri contains a cellulose biosynthetic gene cluster (VFA0881-885) similar to that found in Salmonella (32, 33, 35). To evaluate the role of this locus in the observed phenotypes, we constructed a mutant defective for VFA0884, which encodes the catalytic subunit of a putative cellulose synthase, and introduced either pCLD29 (sypF1) or pCLD42 (vpsR). Loss of VFA0884 disrupted the Congo red binding induced by these regulators (data not shown). Furthermore, this mutation eliminated glass attachment induced by SypF1 or VpsR (Fig. 7B). Finally, the pellicles formed by SypF1 in the VFA0884 mutant were reduced to levels similar to the vpsR mutant; VpsR-induced pellicles were completely dependent on VFA0884 (Fig. 7C). Together, these data support a role for cellulose in biofilm formation by V. fischeri under these conditions.
DISCUSSION
The significance of microbial biofilms as major survival strategies under both environmental and host conditions is well-accepted. Control of biofilm formation in numerous systems is complex (e.g., V. cholerae [5, 9, 50, 51, 54]). Here, we have shown that biofilm formation in V. fischeri also depends on multiple regulators. Our discovery of an increased activity allele, sypF1, revealed a role for this putative hybrid sensor kinase in the transcriptional control of syp, a polysaccharide locus necessary for symbiotic colonization. In addition, multicopy expression of sypF1 induced biofilm phenotypes, including the formation of wrinkled colonies on solid medium and cell-cell aggregating, surface attachment, and pellicle formation in liquid medium. Surprisingly, we found that SypF1 appears to work through at least two response regulators, SypG and VpsR: disruption of both regulators was required for the complete loss of both wrinkled colony and pellicle formation induced by SypF1 overexpression. Multicopy expression of VpsR could also induce biofilm phenotypes (Fig. 7B and C), and these were found to be dependent on cellulose biosynthesis. Together, these data extend our knowledge of the complex control of biofilm formation by V. fischeri and suggest that SypF may coordinate syp activity through SypG with cellulose through VpsR.
Perhaps the simplest phenotype induced by overexpression of sypF1 was transcription of the syp locus, as monitored with a lacZ reporter fused downstream from the sypA promoter. Reporter activity was increased 10-fold by SypF1, and this induction depended completely on SypG, a putative DNA-binding response regulator thought to directly activate syp transcription (53) (E. A. Hussa and K. Visick, unpublished data). Transcription did not depend fully upon any other regulator, although loss of SypE impacted the overall levels of induction, as did, to a lesser extent, loss of representative Syp structural proteins (encoded by sypB, sypI, and sypN). Perhaps these results indicate a form of feedback control on syp transcription. Importantly, syp transcription did not depend on VpsR, a response regulator predicted to control polysaccharide production: loss of the vpsR gene did not impact sypA transcription induced by SypF1 overexpression. Finally, SypF1-mediated sypA transcription also did not depend upon RscS, a sensor kinase known to act upstream of SypG (21) (C. Darnell and K. Visick, unpublished data). Together, these data support the hypothesis that SypF1 activates syp transcription by working upstream of SypG.
Despite this apparent connection between SypF and SypG with respect to syp transcription, a number of biofilm phenotypes induced by overexpression of sypF1 did not depend exclusively on SypG, or in the case of attachment to glass, on SypG at all. Biofilm phenotypes are complex, requiring many factors to produce a complete structure (including, for example, motility, pili, exopolysaccharide, secreted proteins, and in some cases DNA) (reviewed in references 11 and 13). Furthermore, different phenotypes require different subsets of these factors. Therefore, to evaluate the role of SypF in biofilm formation, we assessed three major phenotypes (wrinkled colony morphology, surface attachment, and pellicle formation). Through this comprehensive approach, combined with mutant analyses, we were able to uncover a role for VpsR. Whereas loss of either SypG or VpsR severely impacted only a subset of the SypF1-induced phenotypes, loss of both regulators abolished all of them. Surprisingly, although VpsR (but not SypG) was required for attachment to glass induced by SypF1 under shaking conditions (Fig. 5), VpsR overexpression did not induce a similar phenotype (data not shown). These results suggest the involvement of other factors controlled by SypF1.
SypF also appeared to work upstream of SypE, though the impact of sypE disruption was less severe than that of sypG disruption and similar to disruption of syp structural genes, suggesting a less important regulatory role for SypE. At this time, our data support the hypothesis that SypE contributes to SypF-mediated biofilm formation, but whether SypE can be considered a direct downstream regulator remains uncertain, as its loss eliminates none of the observed phenotypes. Indeed, our preliminary data suggest that an additional RR may also be involved in the complex phenotypes induced by SypF; this possibility is currently under investigation (Darnell and Visick, unpublished). However, from the data presented here, we conclude that SypG and VpsR appear to represent major regulators involved in relaying the SypF-initiated signal.
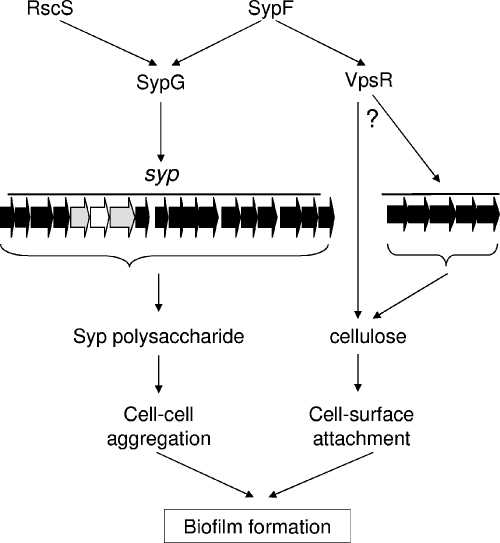
Using these data and previous work (21, 52, 53) as a guide, we have developed a working model for the roles of the various syp regulators described to date (Fig. 8). SypF and RscS each have the potential to serve as phospho-donors to SypG, which we propose directly activates syp transcription. We previously reported that SypE may serve, under some circumstances, as an inhibitor of SypG activity, although SypE is required for full RscS and SypF activity (reference 21 and this work). As a result of these activities, a syp-dependent polysaccharide is produced, contributing to the formation of wrinkled colonies and pellicles and other phenotypes. In addition, SypF may directly or indirectly activate VpsR. Although the role of this protein as a response regulator is unclear, given the relative lack of conservation of key residues involved in phosphorylation (22, 49), genetic evidence supports the role of the V. cholerae homolog as a response regulator (23). Regardless of its specific activity, the V. fischeri protein apparently functions upstream of cellulose biosynthesis and thus biofilm formation. In other bacteria such as Salmonella, cellulose biosynthesis is controlled through posttranscriptional mechanisms, including through the availability of c-di-GMP (32, 33). Therefore, our simple model (Fig. 8) shows VpsR as a transcriptional regulator of the cellulose biosynthetic locus or as an indirect regulator of cellulose biosynthesis.
FIG. 8.
Model for the roles of syp regulators in biofilm formation in V. fischeri. Transcription of the syp locus can be induced by overexpression of RscS or SypF (SypF1), acting through SypG. The Syp structural proteins contribute to the production of a Syp polysaccharide, involved in cell-cell aggregation phenotypes, such as wrinkled colonies and pellicles. SypE contributes by an unknown mechanism to these phenotypes; it also appears to inhibit SypG activity (not depicted). SypF overexpression may activate VpsR, ultimately promoting cellulose operon (VFA0881-885) expression or cellulose synthesis. The result of these interactions is enhanced cell-surface interactions. Block arrows indicate genes (depicted in gray for sypE and sypG and in white for sypF), and line arrows indicate activation. The question mark indicates that the level at which VpsR exerts its effect is not yet known.
We propose that cellulose is responsible for initial attachment during static growth and that subsequent cell-cell interactions depend upon the Syp polysaccharide (Fig. 8). This hypothesis is supported by the following data: loss of VpsR or VFA0884 (but not SypG) disrupted surface attachment induced by SypF1 during static growth. The subsequent development of pellicles depended heavily on SypG. Disruption of vpsR did not prevent pellicle formation, but the pellicles that formed failed to attach properly to the well. These data suggest different roles for the two polysaccharides.
Similar to VpsR in V. cholerae (VpsRVC), in which this protein has been most thoroughly investigated (5, 49), VpsR in V. fischeri (VpsRVF) appears to be an orphan response regulator encoded within an analogous region of the large chromosome. The two proteins exhibit high sequence similarity yet appear to play distinct roles. VpsRVF controls cellulose biosynthesis; in contrast, V. cholerae doesn't contain a cellulose locus. While VpsRVC activates transcription of the vps polysaccharide locus, VpsRVF, appears to serve both as a positive regulator of cellulose biosynthesis and as a negative regulator of an unknown polysaccharide, as evidenced by the increased mucoidy of a vpsR mutant. A further understanding of VpsRVF function awaits a more thorough investigation of its regulon through array studies.
It is possible that by overexpressing an increased activity allele of sypF, we are creating artifactual phenotypes: an overabundance of active SypF may phosphorylate response regulators that are not biologically relevant. However, SypG is encoded directly downstream of sypF, suggesting a likely connection between these regulators. VpsR also seems to be a relevant target, given that it is unlinked to a known sensor kinase and it clearly plays a role in polysaccharide production in its own right. Finally, the vpsR mutant exhibited a small defect in colonization of the squid when competed with the wild-type strain, suggesting a biologically relevant role (22). Thus, while SypF1 may improperly donate phosphoryl groups to other response regulators that contribute to phenotypes not identified here, this work nonetheless reveals regulators and phenotypes involved in biofilm formation by V. fischeri.
Taken together, this work elucidates a role for SypF in controlling the syp locus and thus contributing to the production of a syp-generated polysaccharide. It also provides evidence for a second distinct arm for polysaccharide production coordinated by SypF through VpsR, resulting in cellulose production. These two polysaccharides make differential contributions to the observed SypF-induced biofilms. Thus, this work further demonstrates the complexity of biofilm formation and regulation in V. fischeri, a nontraditional model of biofilm formation.
Acknowledgments
We thank Mark Clementz, Deborah Muganda, and Emily Yip for their experimental contributions to this work and Alan Wolfe and Christine Anderson for their review of the manuscript.
This work was supported by NIH grant GM59690 awarded to K.L.V.
Footnotes
Published ahead of print on 9 May 2008.
REFERENCES
- 1.Al Laham, N., H. Rohde, G. Sander, A. Fischer, M. Hussain, C. Heilmann, D. Mack, R. Proctor, G. Peters, K. Becker, and C. von Eiff. 2007. Augmented expression of polysaccharide intercellular adhesin in a defined Staphylococcus epidermidis mutant with the small-colony-variant phenotype. J. Bacteriol. 1894494-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begun, J., J. M. Gaiani, H. Rohde, D. Mack, S. B. Calderwood, F. M. Ausubel, and C. D. Sifri. 2007. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 3e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beier, D., and R. Gross. 2006. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 9143-152. [DOI] [PubMed] [Google Scholar]
- 5.Beyhan, S., K. Bilecen, S. R. Salama, C. Casper-Lindley, and F. H. Yildiz. 2007. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J. Bacteriol. 189388-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 1723701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calva, E., and R. Oropeza. 2006. Two-component signal transduction systems, environmental signals, and virulence. Microb. Ecol. 51166-176. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, A. 1962. Episomes. Adv. Genet. 11101-145. [Google Scholar]
- 9.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 1861574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 11.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn, A. K., M. O. Martin, and E. Stabb. 2005. Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid 54114-134. [DOI] [PubMed] [Google Scholar]
- 13.Dunne, W. M., Jr. 2002. Bacterial adhesion: seen any good biofilms lately? Clin. Microbiol. Rev. 15155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faruque, S. M., K. Biswas, S. M. Udden, Q. S. Ahmad, D. A. Sack, G. B. Nair, and J. J. Mekalanos. 2006. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. USA 1036350-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Geszain, K., and K. L. Visick. 28 May 2008. Multiple factors contribute to keeping levels of the symbiosis regulator RscS low. FEMS Microbiol. Lett. doi: 10.1111/j.1574-6968.2008.01209. [DOI] [PMC free article] [PubMed]
- 15.Graf, J., P. V. Dunlap, and E. G. Ruby. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 1766986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grau, B. L., M. C. Henk, K. L. Garrison, B. J. Olivier, R. M. Schulz, K. L. O'Reilly, and G. S. Pettis. 2008. Further characterization of Vibrio vulnificus rugose variants and identification of a capsular and rugose exopolysaccharide gene cluster. Infect. Immun. 761485-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guvener, Z. T., and C. S. Harwood. 2007. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 661459-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50101-104. [DOI] [PubMed] [Google Scholar]
- 19.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1726557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoch, J. A., and K. I. Varughese. 2001. Keeping signals straight in phosphorelay signal transduction. J. Bacteriol. 1834941-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hussa, E. A., C. Darnell, and K. L. Visick. 25 April 2008. RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J. Bacteriol. 1904576-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hussa, E. A., T. M. O'Shea, C. L. Darnell, E. G. Ruby, and K. L. Visick. 2007. Two-component response regulators of Vibrio fischeri: their identification, mutagenesis and characterization. J. Bacteriol. 1895825-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lauriano, C. M., C. Ghosh, N. E. Correa, and K. E. Klose. 2004. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J. Bacteriol. 1864864-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193265-275. [PubMed] [Google Scholar]
- 25.Ma, L., H. Lu, A. Sprinkle, M. R. Parsek, and D. J. Wozniak. 2007. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J. Bacteriol. 1898353-8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manoil, C., and J. Beckwith. 1985. TnphoA: a transposon probe for protein export signals. Proc. Natl. Acad. Sci. USA 828129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCann, J., E. V. Stabb, D. S. Millikan, and E. G. Ruby. 2003. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl. Environ. Microbiol. 695928-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, New York, NY.
- 29.Nyholm, S. V., and M. J. McFall-Ngai. 2004. The winnowing: establishing the squid-Vibrio symbiosis. Nat. Rev. Microbiol. 2632-642. [DOI] [PubMed] [Google Scholar]
- 30.O'Gara, J. P., and H. Humphreys. 2001. Staphylococcus epidermidis biofilms: importance and implications. J. Med. Microbiol. 50582-587. [DOI] [PubMed] [Google Scholar]
- 31.O'Shea, T. M., A. H. Klein, K. Geszvain, A. J. Wolfe, and K. L. Visick. 2006. Diguanylate cyclases control magnesium-dependent motility of Vibrio fischeri. J. Bacteriol. 1888196-8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romling, U. 2002. Molecular biology of cellulose production in bacteria. Res. Microbiol. 153205-212. [DOI] [PubMed] [Google Scholar]
- 33.Ross, P., R. Mayer, and M. Benziman. 1991. Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 5535-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruby, E. G., and K. H. Nealson. 1977. Pyruvate production and excretion by the luminous marine bacteria. Appl. Environ. Microbiol. 34164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lostroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 1023004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakuragi, Y., and R. Kolter. 2007. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 1895383-5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stabb, E. V. 2006. The Vibrio fischeri-Euprymna scolopes light organ symbiosis, p. 204-218. In F. L. Thompson, B. Austin, and J. Swings (ed.), The biology of vibrios. ASM Press, Washington, DC.
- 38.Stabb, E. V., K. A. Reich, and E. G. Ruby. 2001. Vibrio fischeri genes hvnA and hvnB encode secreted NAD+-glycohydrolases. J. Bacteriol. 183309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stabb, E. V., and E. G. Ruby. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358413-426. [DOI] [PubMed] [Google Scholar]
- 40.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56187-209. [DOI] [PubMed] [Google Scholar]
- 41.Tamplin, M. L., A. L. Gauzens, A. Huq, D. A. Sack, and R. R. Colwell. 1990. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl. Environ. Microbiol. 561977-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visick, K. L., and E. G. Ruby. 2006. Vibrio fischeri and its host: it takes two to tango. Curr. Opin. Microbiol. 9632-638. [DOI] [PubMed] [Google Scholar]
- 44.Visick, K. L., and L. M. Skoufos. 2001. A two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J. Bacteriol. 183835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 643648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waite, R. D., A. Papakonstantinopoulou, E. Littler, and M. A. Curtis. 2005. Transcriptome analysis of Pseudomonas aeruginosa growth: comparison of gene expression in planktonic cultures and developing and mature biofilms. J. Bacteriol. 1876571-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smither, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cystosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 173469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yarwood, J. M., D. J. Bartels, E. M. Volper, and E. P. Greenberg. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 1861838-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 1831716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53497-515. [DOI] [PubMed] [Google Scholar]
- 51.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 964028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yip, E. S., K. Geszvain, C. R. DeLoney-Marino, and K. L. Visick. 2006. The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol. Microbiol. 621586-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yip, E. S., B. T. Grublesky, E. A. Hussa, and K. L. Visick. 2005. A novel, conserved cluster of genes promotes symbiotic colonization and σ54-dependent biofilm formation by Vibrio fischeri. Mol. Microbiol. 571485-1498. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5647-656. [DOI] [PubMed] [Google Scholar]