Abstract
Enterohemorrhagic and enteropathogenic Escherichia coli (EHEC and EPEC, respectively) strains represent a major global health problem. Their virulence is mediated by the concerted activity of an array of virulence factors including toxins, a type III protein secretion system (TTSS), pili, and others. We previously showed that EPEC O127 forms a group 4 capsule (G4C), and in this report we show that EHEC O157 also produces a G4C, whose assembly is dependent on the etp, etk, and wzy genes. We further show that at early time points postinfection, these G4Cs appear to mask surface structures including intimin and the TTSS. This masking inhibited the attachment of EPEC and EHEC to tissue-cultured epithelial cells, diminished their capacity to induce the formation of actin pedestals, and attenuated TTSS-mediated protein translocation into host cells. Importantly, we found that Ler, a positive regulator of intimin and TTSS genes, represses the expression of the capsule-related genes, including etp and etk. Thus, the expression of TTSS and G4C is conversely regulated and capsule production is diminished upon TTSS expression. Indeed, at later time points postinfection, the diminishing capsule no longer interferes with the activities of intimin and the TTSS. Notably, by using the rabbit infant model, we found that the EHEC G4C is required for efficient colonization of the rabbit large intestine. Taken together, our results suggest that temporal expression of the capsule, which is coordinated with that of the TTSS, is required for optimal EHEC colonization of the host intestine.
Enterohemorrhagic Escherichia coli (EHEC) is an emerging pathogen causing outbreaks of food-borne gastroenteritis manifested by bloody diarrhea, which may progress to the potentially fatal hemolytic-uremic syndrome. The latter involves severe complications, such as renal impairment, hypertension, and central nervous system manifestations mainly caused by SLT toxins (3, 22). EHEC belongs to the family of the attaching and effacing (AE)-inducing pathogens, which includes the closely related species enteropathogenic E. coli (EPEC), Citrobacter rodentium, and rabbit EPEC. When colonizing the gut, these pathogens form AE lesions on the intestinal epithelial cell surface. AE lesions are characterized by localized destruction of the brush border microvilli, intimate bacterial attachment to host cells, and the formation of actin structures, termed pedestals, beneath the attached bacteria (24). This histopathology is dependent upon a type III protein secretion system (TTSS), which functions as a molecular syringe to translocate effector proteins from the bacterial cytoplasm directly into the cytoplasm of host epithelial cells (15). These effectors subvert normal host cell functions and are required for efficient host colonization (15, 34, 35). One of these effectors, Tir, is inserted into the host cell membrane to form a binding site for an outer membrane adhesin, intimin. Interaction of intimin with translocated Tir promotes tight bacterial attachment to the host cell and is essential for the formation of actin pedestals (15).
The TTSS and related proteins are encoded by 41 genes organized in several operons (operons LEE1 to LEE5) clustered in the locus of enterocyte effacement (LEE) (15). Alongside the TTSS and its effectors, EHEC and EPEC encode an array of additional confirmed and putative virulence factors including the Shiga toxins, EspP, Efa1, and others (11, 40). Ler is a transcriptional regulator encoded within the LEE1 operon, and it positively regulates most of the LEE operons (29). In addition, it positively and negatively regulates numerous non-LEE virulence genes, as well as housekeeping genes (1, 13, 26). Thus, Ler coordinates the expression of a large portion of the EHEC/EPEC virulon.
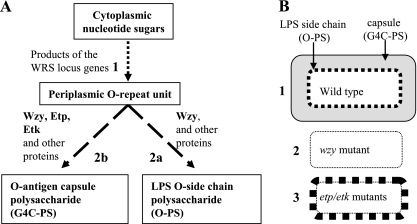
E. coli can use distinct pathways for polysaccharide export and capsule assembly, classified as group 1 to 4 capsules (50). The genes required for group 4 capsule (G4C) formation are organized into two loci; one of these loci is indistinguishable from those responsible for the expression of many different Wzy-dependent O antigens (2, 47, 48). Moreover, this locus frequently determines both the K and O serotypes (examples include O26, O55, O100, O111, O113, and O127) (17, 32, 33), and therefore, these G4Cs have also been termed “O-antigen capsules” (17). In these cases, both the lipopolysaccharide (LPS) O antigen (also known as KLPS) and the capsule (K antigen) are composed of the same repeat units, which consist of a sequence of three to five sugar residues. The genes in this locus, the Wzy-dependent repeat unit synthesis (WRS) locus, are required for the synthesis of the repeat unit, its delivery to the periplasm, and its polymerization by Wzy into both O antigen and G4C polysaccharide (G4C-PS) (Fig. 1A). Accordingly, mutants deficient in the WRS locus genes, including wzy, are expected to be both rough and uncapsulated (Fig. 1A and B).
FIG. 1.
Model of G4C, O-antigen, and capsule formation and expected phenotypes of etk, etp, and wzy mutants. (A) Steps in Wzy-dependent LPS O side chain and O-antigen capsule biogenesis. In step 1, the repeat units, composed of three to five sugar residues, are synthesized in the cytoplasm and then translocated to the periplasm. This step is mediated by a large number of proteins encoded mostly in the WRS locus. Steps 2a and 2b are alternative steps. In the first, the periplasmic repeat units are polymerized by Wzy into short polymers (15 to 20 repeat units), which are then ligated to the LPS core to form O side chains. In step 2b, the repeat units are polymerized, again by Wzy, into long polymers (>100 repeat units), which are then translocated to the bacterial surface to form capsules. Etp and Etk are specifically required for step 2b, whereas Wzy is required for both 2a and 2b. The 2a and 2b pathways compete for the same O repeat units. (B) Expected phenotypes upon inactivation of etp, etk, and wzy. (Phenotype 1) Wild-type EHEC can form both LPS O side chains (thick black broken line) and O-antigen capsules (gray zone around the bacteria), both indicated by arrows. (Phenotype 2) Mutants deficient in repeat unit synthesis or polymerization (wzy mutant), are expected to lack both LPS O side chains and O capsules (the thin dotted line represents the smooth outer membranes). (Phenotype 3) etk or etp mutants are expected to be deficient in O-capsule formation but to exhibit enhanced LPS O side chain formation (represented by the thick black broken line) since pathway 2b is no longer competing with pathway 2a (panel A) (32).
The second locus that is involved in G4C formation is the G4C (gfc) operon, also termed the “22-minute locus” because it is mapped to min 22 in the E. coli K-12 genetic map (50). This operon encodes seven proteins, including Etp and Etk, that are required for repeat unit polymerization into G4C-PS and for capsule assembly (Fig. 1A) (32). Interestingly, mutants deficient in G4C formation exhibit enhanced production of O antigen (17, 32), possibly because in these mutants all of the O repeat units produced are used for O-antigen production (Fig. 1B).
In this report, we demonstrate that, like EPEC O127, EHEC O157 also forms an O antigen, G4C, and that inactivation of etk or etp renders the bacteria noncapsulated. We also show that the capsules hinder EPEC and EHEC attachment to cultured epithelial cells and inhibit actin pedestal formation, probably via masking of the TTSS and intimin and thus attenuating Tir translocation and the intimin-Tir interaction. Interestingly, the transcription of the gfc operon is repressed by Ler, indicating that the expression of the TTSS by the G4C is inversely controlled by this virulence regulator. Furthermore, by using the infant rabbit model, we found that the EHEC capsule is required for efficient intestinal colonization. Taken together, our findings suggest that both EPEC and EHEC form a G4C and that temporal expression of this capsule improves the capacity of EHEC, and perhaps also EPEC, to colonize the host intestine.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The bacterial strains and plasmids used in this study are described in Table 1. For descriptions of the primers used, see Table S2 in the supplemental material.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmida | Description | Source or reference |
|---|---|---|
| Strains | ||
| E2348/69 | Wild-type EPEC O127:H6 | |
| OI899 | E2348/69 etk::kan | 32 |
| EDL933 Nalr (2280) | Nalr derivative of EDL933 | 11 |
| SK2472 | etk::kan mutant derivative of EDL933 Nalr | This study |
| TUV93-0 (1280) | Δstx1 Δstx2 mutant derivative of EDL933 | John Leong |
| DF1291 | TUV93-0 Δler::kan | 26 |
| SK2235 | TUV93-0 Δetp::cm | This study |
| SK1416 | TUV93-0 etk::kan | This study |
| B12-D5 (1853) | EHEC O157 Saki wzy::mini-Tn5kan2 | 41 |
| SK2526 | TUV93-0 wzy::mini-Tn5kan2, taken from B12-D5 | This study |
| CVD452 (63) | EPEC ΔescN::kan | 21 |
| SK2131 | TUV93-0 ΔescN::kan, taken from CVD425 | This study |
| Plasmids | ||
| pKD46 | Helper plasmid for gene deletion | 10 |
| pKD3 | Template for kan cassette amplification | 10 |
| pCX341 | Vector for blaM fusion formation | 30 |
| pCX392 | pCX341 expressing tir(EPEC)-blaM | This study |
| pKB3105 | pCX341 expressing tir(EHEC)-blaM | This study |
| pOI277 | pACYC184 expressing etk | 20 |
| pAP2064 | pSA10 expressing etp | 32 |
| pTU12 (881) | Plasmid expressing ler | 6 |
| pIR1 (501) | Vector for formation of gfp transcriptional fusions | 14 |
| pGY3159 | PLEE1(EHEC)-gfp-mut3 | This study |
In parentheses are the serial numbers in our strain collection.
Growth and infection conditions.
Bacteria were grown in Luria-Bertani (LB) broth without shaking at 37°C unless stated otherwise. Where appropriate, media were supplemented with ampicillin (100 μg ml−1), chloramphenicol (25 μg ml−1), or kanamycin (50 μg ml−1). When indicated, 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to bacterial cultures containing expression vectors carrying lacIq. The cell lines HeLa, Hep2, and Caco2 were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum, 100 U ml−1 penicillin, and 0.1 mg ml−1 streptomycin at 37°C in 5% CO2. Before infection, the cells were washed with phosphate-buffered saline (PBS) to remove antibiotics.
Strain construction.
To generate isogenic strains, we amplified DNA fragments containing tagged mutations (etp::Cm, etk::kan, and escN::kan from EPEC O127 and wzy::mini-Tn5kan2 from EHEC O157 Sakai) (Table 1) and electroporated them into EDL933-containing pKD46 (10). Mutants were selected, pKD46 was eliminated, and mutations were then verified by PCR analysis and sequenced as previously described (10). For the primers used, see Table S2 in the supplemental material.
Plasmid construction.
To construct pKB3105, a DNA fragment containing tir was amplified with primers #486 and #488 and an EDL933 DNA template. The amplified fragment was digested with EcoRI and KpnI and cloned into pCX341 digested with the same enzymes to generate a gene encoding a Tir-BlaM fusion protein. To construct pGY3159, a DNA fragment containing PLEE1 and ler was amplified with primers LEE1reg-F and Pler1-R and an EDL933 DNA template. The amplified fragment was digested with XbaI and BamHI and cloned into pIR1 digested with the same enzymes. For the primers used, see Table S2 in the supplemental material.
Electron microscopy.
Bacteria grown overnight in LB at 37°C were washed with 0.1 M cacodylate buffer (pH 7.0), labeled with cationized ferritin (2 mg/ml; Sigma) for 30 min, and finally fixed with 5% glutaraldehyde for 1 h at room temperature (24). Fixed samples were washed, postfixed with 1% osmium tetroxide for 2 h at room temperature, dehydrated in a graded series of ethanol solutions, and then embedded in Epon resin. Thin sections were stained with 2% uranyl acetate and lead citrate and examined under a JEOL 1200EX transmission electron microscope operating at 80 kV.
Preparation of total cellular polysaccharide and separation of capsule polysaccharide from LPS.
Total bacterial polysaccharides were extracted as previously described (32). Briefly, bacteria were grown overnight in 50 ml LB, the optical density (OD) of the cultures was adjusted to 1.0, and finally the cultures were centrifuged and resuspended in 500 μl of PBS. An equal volume of saturated phenol (pH 8.0) was added, and the mixture was incubated for 30 min at 70°C with occasional mixing, followed by centrifugation (1 h, 10,000 × g). The top, aqueous phase was recovered, 2 volumes of 100% ethanol was added to each sample, and the polysaccharides were allowed to precipitate for 1 h at −70°C. Next, the samples were centrifuged at 12,000 × g for 30 min, after which the pellets were washed with 500 μl of 70% ethanol, recentrifuged, and lyophilized. To separate capsule polysaccharide from LPS, the lyophilized total polysaccharide preparations were resuspended in 500 μl of water. LPS, which formed micelles, was precipitated by ultracentrifugation (1 h, 86,000 × g) (33). The supernatant containing the capsule polysaccharide was recovered, and residual LPS contamination was removed by phase partition with Triton X-114 as previously described (27). Briefly, Triton X-114 was added to the supernatant to a final concentration of 1%. The mixture was incubated at 4°C for 1 h with constant stirring to ensure a homogeneous solution. The mixture was then transferred to a 37°C water bath, incubated for 10 min to ensure phase partition, and centrifuged (1 h, 1,000 × g) at 25°C. The resulting aqueous phase, containing purified capsule polysaccharide, was carefully aspirated and lyophilized. If necessary, phase partition with Triton X-114 was repeated until the preparation was LPS free, as determined by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis with anti-O157.
Analysis of LPS and capsule polysaccharide.
Total cell polysaccharide was separated on 10% polyacrylamide gels containing 0.5% SDS (SDS-PAGE), and the gels were used for immunoblot analysis. SDS-PAGE analysis allowed visualization of the LPS without the interference of capsular polysaccharide, which cannot enter the gel because of its low net charge and high molecular weight (16, 33). For G4C-PS analysis by dot blotting, equivalent amounts of samples containing purified, LPS-free G4C-PS were applied directly to nitrocellulose membranes, allowed to dry, and then developed by immunoblotting with anti-O157 rabbit antisera (Statens Serum Institut, Copenhagen, Denmark) and secondary anti-rabbit immunoglobulin G conjugated to alkaline phosphatase (Sigma).
Evaluation of bacterial adhesion to epithelial cells and actin pedestal formation.
Adhesion was quantified as previously described (41). Briefly, epithelial cells grown on coverslips to 80% confluence were infected with 10 μl of overnight EHEC cultures (∼10−6 bacteria). At different time points postinfection, the monolayers were washed, fixed, treated for 2 min with 0.1% Triton X-100, and then stained with phalloidin-rhodamine. Coverslips were then washed and mounted on slides for immunofluorescence microscopy. Bacterial clusters on tissue culture cells consisting of eight or more bacteria were considered microcolonies, and the percentage of cells associated with microcolonies was determined. The numbers of microcolonies were scored as the sum of 10 microscopic fields unless otherwise stated. The efficiency of actin pedestal formation was measured as the fraction of attached bacteria forming phalloidin-labeled pedestals out of the total number of attached bacteria.
Attachment of EHEC to red blood cells (RBC).
Immobilized monolayer RBC were infected, fixed, and stained as previously described (39).
Determination of the efficiency of Tir translocation by EPEC.
Translocation assay by EPEC was carried out as previously described (30), with some modification. Briefly, HeLa cells seeded in 96-well plates were loaded with CCF2 (Invitrogen) for 1 h and then washed with CDMEM (CDMEM is a low-autofluorescing tissue culture medium [6, 30]) containing 2.5 mM probenecid (Sigma). The cells were then infected with overnight cultures diluted 1:50 in CCF2 loading buffer (each well was infected with 196 μl of a solution containing 186.6 μl CDMEM, 1 μM CCF2, 1.8 μl solution B, 2.7 μl solution C, 2.5 mM probenecid, 0.1 μM β-lactamase-inhibitory protein, and 4 μl overnight bacterial culture (solutions B and C are from the CCF2 loading kit [Invitrogen]). Infection was carried out in a prewarmed plate reader (set at 37°C), and the rate of CCF2 hydrolysis was measured at 5-min intervals as previously described (30). For each of the strains tested, we infected six wells with cultures generated from six different colonies and calculated the average rate of CCF2 production as previously described (30). The strains used include EPEC, an etk mutant containing the plasmids expressing tir-blaM from the tac promoter, and a complemented strain (etk mutant OI899) containing plasmids pOI277 and pCX392, expressing etk and tir-blaM, respectively. The basal expression of tir-blaM was used in these experiments (IPTG was not added, and the infection medium included glucose). As a negative control, we infected cells with EPEC etk::kan, containing pCX341, expressing unfused BlaM (30).
Determination of the efficiency of Tir translocation by EHEC.
For EHEC, we could not infect preloaded cells and detect translocation in real time as was done for EPEC. This is because the infection with EHEC is much slower and levels of CCF2 leakage from the cells become significant, distorting the results (our unpublished results). We therefore used endpoint measurements. EHEC bacteria containing a plasmid expressing tir-blaM (pKB3105) were grown overnight in LB, diluted 1:50 in CDMEM, and used to infect HeLa cells seeded in 96-well plates. At different time points postinfection, the wells were washed, overlaid with 20 μl CCF2 loading solution, and placed in a prewarmed plate reader (30°C). Per well, the loading solution contained 2 μM CCF2, 1.62 μl solution B, 1.6 μl solution C (solutions B and C were from the Invitrogen CCF2 loading kit), 2.5 mM probenecid, and 25 μg/ml chloramphenicol in CDMEC, for a final volume of 20 μl. Chloramphenicol was added to stop effector synthesis and translocation. The rate of CCF2 hydrolysis by BlaM (β-lactamase activity) was monitored for 30 min at 2.5-min intervals as previously described (30). We found that CCF2 reached a maximal intracellular concentration within 10 min (data not shown), and thus, for each time point, the rate of BlaM activity from 10 to 20 min postaddition of CCF2 was determined. This rate directly correlates with the Tir-BlaM concentrations in the HeLa cells. As a negative control, we infected cells with the EHEC etk::kan mutant containing pX341, which expresses unfused blaM.
Activity of the LEE1 promoter.
To measure LEE1 promoter activity, strains (wild-type EPEC and etp and etk mutants) containing pGY3159 were grown under repressive conditions [overnight at 30°C in LB supplemented with 20 mM (NH4)2SO4]. To activate PLEE1, cultures were washed, diluted 1:50 in CDMEM, and grown at 37°C in 96-well plates inside a microplate reader preset at 37°C (SPECTRAFluor Plus; TECAN). The fluorescence intensity (filter set at a 485-nm excitation wavelength and a 535-nm emission wavelength) and OD at 600 nm (OD600) were read at 5-min intervals during growth. Data were collected with the Magellan version 5.0 software (TECAN).
Analysis of expressed proteins.
Strains were grown as indicated, pelleted by centrifugation (5 min, 5,000 × g), lysed in 40 μl 2.5× SDS loading buffer (10 min, 100°C), and centrifuged for 2 min at 12,000 × g. Supernatant was loaded onto 12% SDS-PAGE gels and then subjected to immunoblotting with specific antibodies as indicated. Blots were developed with a secondary antibody conjugated to alkaline phosphatase or horseradish peroxidase (Sigma).
RNA analysis. Primer extension assays were carried out as described previously (12). Briefly, bacterial cultures grown to an OD600 of 1.0 were pelleted and resuspended in 10 mM Tris (pH 7.5)-1 mM EDTA. Lysozyme was added to 0.9 mg/ml, and the samples were subjected to three freeze-thaw cycles. Total RNA was isolated with an Ultraspec RNA kit according to the manufacturer's (BIOTECX Laboratories) instructions, except that 1 ml of reagent was used for cells with an OD600 of 12 to 16 U. The RNA samples (3 μg) were subjected to primer extension at 42°C for 45 min with avian myeloblastosis virus reverse transcriptase (CHIMWEx) and end-labeled primer 828 (GATTGCAGAAAGCTTGTG). The extension products and the sequencing reaction mixtures primed with end-labeled primer 828 were separated on a 6% sequencing gel.
Protein-DNA binding assay.
A modified enzyme-linked DNA-protein interaction assay (5) was used to measure protein-DNA binding. Briefly, three DNA fragments labeled at one end with biotin were generated by PCR. These included (i) the gfc regulatory region (with primers BioYmcDreg-F and YmcDreg-R), (ii) the LEE2 regulatory region (with primers BioLEE2reg2-F and 31R) as a positive control, and (iii) the etk coding region (with primers BioYccC-F and YccC-R) as a negative control. One hundred microliters of each of these fragments (500 pmol/ml) in PT buffer (PBS supplemented with 0.05% Tween 20 and 1 mM EDTA) was bound separately to streptavidin-coated 96-well plates (Sigma) by incubation for 1 h. Unbound DNA was removed by washing with PT. One hundred microliters of purified Ler-6His, at different concentrations in binding buffer (50 mM Tris [pH 7.4], 70 mM KCl, 5 mM EDTA, 1 mM dithiothreitol, 6% glycerol), was added to the fragment-bound wells for 1 h of incubation, and then unbound proteins were removed by washing with PT buffer. One hundred microliters of polyclonal anti-Ler antibodies was added, the wells were incubated for 1 h and washed with PT buffer, and then 100 μl of alkaline phosphatase-conjugated secondary anti-rabbit immunoglobulin G antibodies (Sigma) was added and the mixture was incubated for 1 h. The wells were washed with PT buffer and later with AP buffer (1 M Tris [pH 9.5], 0.5 mM MgCl2). Finally, 100 μl of AP buffer containing 10 mg/ml p-nitrophenylphosphate was added to each well and the plate was immediately inserted into a microplate reader (SPECTRAFluor Plus; TECAN). The OD405 was read every minute, and data were collected by the Magellan version 5.0 software (TECAN). The OD405 values were plotted against time, and the slopes of the linear parts of these plots were calculated, representing the level of binding. One hundred percent binding was set as the binding of the highest concentration of Ler (5 μM); higher concentrations gave similar results as the system was saturated.
Infection model.
Coinfection experiments with infant rabbits were performed as described previously (19). Briefly, 3-day-old infant rabbits were orogastrically inoculated with approximately equal numbers of wild-type and etk mutant cells at a dose equivalent to 5 × 108 CFU/90 g rabbit weight. At days 2 and 7 postinfection, infected rabbits were euthanized and their intestines were removed for bacterial enumeration. Tissue homogenates were plated onto sorbitol MacConkey plates to detect both EHEC strains and then replica plated onto sorbitol MacConkey plates containing kanamycin (50 μg ml−1) to enumerate the etk mutant cells. The data are expressed as a competitive index (CI) that represents the ratio of the number of etk to wild-type CFU postinfection to the number of etk to wild-type CFU present in the original inoculum. CI values were compared to a theoretical value of 1 (representing equal colonization of wild-type and mutant strains) by using the Wilcoxon signed rank test. Coinfection assays were performed with rabbits from two litters.
RESULTS
EPEC etk::kan mutant exhibits accelerated formation of actin pedestals.
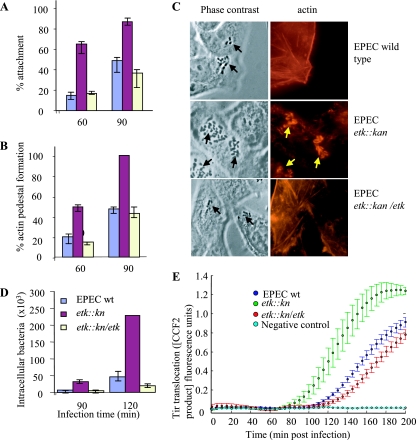
We have previously shown that EPEC forms a G4C and that Etk expression is essential for its formation (32). To investigate whether the capsule plays a role in EPEC-host cell interaction, we infected HeLa cells with different EPEC strains and compared the capacities of the bacteria to attach to the cells and to form actin pedestals. Bacteria were grown as static cultures in LB for 16 h, diluted 1:50 in DMEM, and then used to infect HeLa cells. At early time points (60 and 90 min) postinfection, we found that the etk::kan mutant exhibits better attachment efficiency and increased formation of actin pedestals (Fig. 2A, B, and C). Moreover, the etk mutant exhibits increased invasiveness at early time points postinfection (Fig. 2D). However, at 3 h postinfection, we could not discern any differences in attachment, pedestal formation, or invasiveness between the etk::kan mutant and wild-type strains (data not shown). Complementing the etk mutant with a plasmid expressing Etk restored the wild-type-like slower invasiveness, attachment, and pedestal formation kinetics (Fig. 2A, B, C, and D). Similar results were obtained when we used, instead of the etk mutant, etp mutants (data not shown). These results indicate that, at early time points postinfection, the G4C might inhibit EPEC attachment and actin pedestal formation.
FIG. 2.
The G4C interferes with EPEC interaction with host cells at early time points postinfection. HeLa cells were infected with overnight cultures (multiplicity of infection, ∼2) of wild-type (wt) or etk::kan mutant EPEC or with the mutant complemented with an Etk-expressing plasmid (etk::kan/etk). At different time points postinfection, cells were washed, fixed, and stained with phalloidin-rhodamine, and the attachment (A) and pedestal formation (B) levels were determined. Bacterial adhesion was evaluated as the percentage of cells with eight or more bacteria attached out of the total number of cells in the microscopic field. The mean percentages and standard deviations of 10 random microscopic fields (∼200 cells) are shown. The efficiency of actin pedestal formation was evaluated as the fraction of attached bacteria forming pedestals out of the total number of attached bacteria, and the mean percentages and standard deviations of five random microscopic fields (∼120 HeLa cells) are shown. Typical phase-contrast and corresponding actin-stained (red) images taken at 60 min postinfection are shown in panel C. The black arrows indicate bacteria, and the yellow arrows indicate actin pedestals. In a different experiment, cells were infected with similar overnight cultures and the invasiveness of the different strains at early time points postinfection was compared by using the gentamicin protection assay (D). The assay was carried out in triplicate as previously described (4), and standard deviations are shown by bars. In panel E, the same mutants, supplemented with the reporter tirEPEC-blaM gene, were used to infect HeLa cells preloaded with CCF2. The kinetics of translocation by the three mutants were determined in parallel and in real time as previously described (30). Each time point represents the average translocation by six different colonies, and the bars indicate standard errors.
EPEC etk::kan mutant exhibits increased Tir translocation.
The formation of actin pedestals by EPEC is dependent on the delivery of Tir into the host cell membrane, followed by Tir-intimin interaction. The enhanced actin pedestal formation by the etk mutant might indicate that the expression of tir, eae, or the TTSS is derepressed in the etk mutant. However, by Western blot analysis, we found that this is not the case; wild-type EPEC and the etk::kan mutant express similar levels of Tir, intimin, and the TTSS components EspB, EspA, and EscJ (data not shown), indicating that the expression of etk or G4C formation does not affect tir or eae at the transcriptional or translational level.
An alternative explanation for the inhibitory effect of the capsule is that it masks the TTSS and/or intimin. The former should result in attenuation not only of pedestal formation but also of Tir translocation into the host cells. To test this possibility, we introduced into the three EPEC strains (the wild type, the etk::kan mutant, and the mutant complemented with an etk-expressing plasmid) a plasmid expressing tir fused to the blaM translocation reporter gene (pCX392). We then compared the efficiency of Tir translocation by these strains by using a real-time, high-resolution translocation assay (30). The results showed that initial Tir translocation by the etk mutant was detected as early as ∼80 min postinfection, whereas Tir translocation by wild-type EPEC was detected only 40 min later, at ∼120 min postinfection (Fig. 2E). Also, the efficiency of translocation by the etk mutant was better, as indicated by the steeper slope of the curve of product accumulation (Fig. 2E). The complemented strain translocated Tir like the wild-type strain, with lower efficiency and delayed kinetics (Fig. 2E). As a control, we confirmed that the three strains express similar levels of Tir-BlaM (data not shown). These results indicate that the G4C attenuates the EPEC TTSS activity, most likely by partial masking of the needle complex. Taken together, our results suggest that in wild-type EPEC the G4C might mask the TTSS and intimin, which is smaller than the TTSS in size. The TTSS masking is presumably the cause of the delayed and attenuated Tir translocation, and the intimin masking is likely to further contribute to reduced pedestal formation by hindering Tir-intimin interaction.
EHEC O157:H7 forms a G4C.
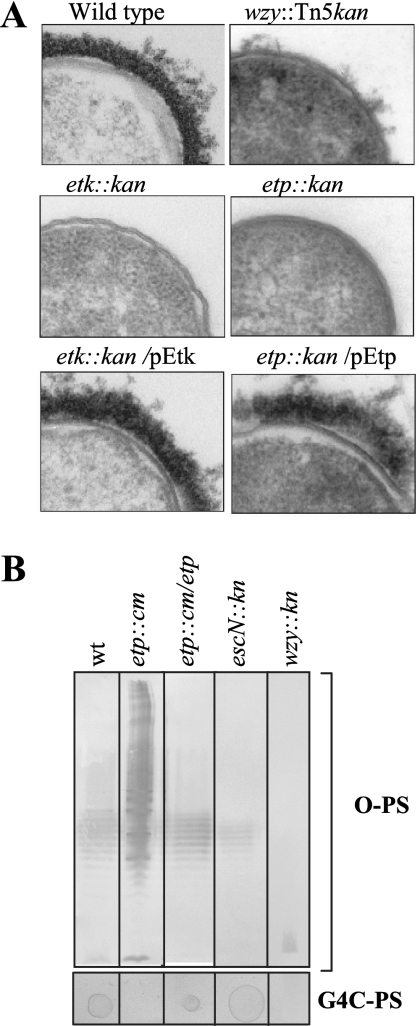
We next wished to examine whether the G4C also inhibits attachment and pedestal formation in the case of EHEC O157. We previously reported that EHEC expresses Etk (20), but whether it forms a G4C was not tested. Therefore, we first examined whether EHEC produces visible capsule structures. To this end, we constructed EHEC mutated in etk, etp, and wzy and subjected the different strains, as well as wild-type EHEC, to electron microscopy analysis as previously described (32). The results show that whereas wild-type EHEC is coated by capsule-like material, the etp, etk, and wzy mutants were not capsulated (Fig. 3A). The capsule reappeared upon complementation of the etk and etp mutants with plasmids expressing the corresponding wild-type genes (Fig. 3A). We did not attempt to complement the wzy mutant, as Wzy was already reported to be required for G4C formation in other E. coli serotypes (2, 47, 48). These results indicate that EHEC produces a capsule that is dependent on Etk, Etp, and Wzy. To corroborate these results, we extracted the total polysaccharides from the bacteria and separated the G4C-PS from the LPS by differential ultracentrifugation and phase partition as described in Materials and Methods. We then evaluated the amounts of LPS-free G4C-PS and the LPS O side chain (O-PS) by dot blotting and Western blotting, respectively (Fig. 3B). As predicted, we found that wild-type EHEC and a TTSS mutant (escN::kan) produced both G4C-PS and O-PS (Fig. 3B). The wzy mutant produced neither G4C-PS nor O-PS (Fig. 3B), confirming previous reports that Wzy is required for G4C formation (2, 47, 48). Notably, the etp mutant did not produce G4C-PS but exhibited increased O-PS production (Fig. 3B). Similar results were obtained with the etk mutants instead of the etp mutant (data not shown). Introduction of a complementing plasmid into the etp mutant restored G4C-PS production, which was associated with reduced O-PS synthesis (Fig. 3B). We did not attempt to complement the wzy mutant, as Wzy was already reported to be required for G4C formation in other E. coli serotypes (2, 47, 48). These results indicate that EHEC O157 strain EDL933 produces an O antigen, G4C, and confirmed that (i) Etp and Etk are required for the formation of this capsule and (ii) Wzy is required for the production of both G4C-PS and O-PS. These conclusions were further supported by results from an agglutination assay and a Percoll buoyancy assay, which were carried out as previously described (32) (data not shown).
FIG. 3.
EHEC O157 produces an O-antigen G4C. Different EHEC strains were grown overnight in LB at 30°C and processed for electron microscopy as previously described (32). Representative ferritin-stained thin sections are shown in panel A. The strains tested include wild-type (wt) EPEC; wzy, etk, and etp mutants; and complemented mutants, as indicated above each frame. In panel B, total polysaccharides of EHEC were extracted and the G4C-PS was separated from the LPS as described in Materials and Methods. LPS O-antigen levels were analyzed by SDS-PAGE and Western blotting with anti-O157 antibody (B, upper panel, O-PS). Aliquots of the LPS-free G4C-PS preparation (10 μl) were spotted onto a membrane and dried, and the amount of G4C-PS was estimated by dot blotting with anti-O157antiserum (B, lower panel). Care was taken to apply equivalent amounts of material derived from the same numbers of bacteria to all of the lanes and spots. The genotypes of the strains analyzed are indicated above the lanes.
G4C-deficient EHEC exhibits increased TTSS-dependent adherence and actin pedestal formation.
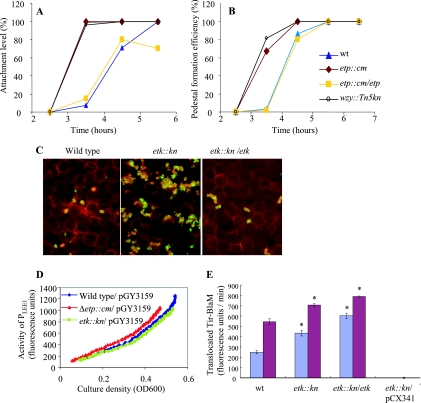
Previous reports indicated that EHEC rough mutants, including a wzy mutant, display increased binding to tissue culture cells (7, 9, 41, 42). According to these researchers’ interpretations of their findings, smooth LPS interferes with EHEC attachment. Our results, however, provide an alternative interpretation of these reported results, since the rough mutants should also be G4C deficient (Fig. 1 and 3). Therefore, it appears that the smooth LPS, the G4C, or both inhibit EHEC attachment to cultured monolayers. To differentiate among these possibilities, we infected Hep2 cells with wild-type EHEC or etp and wzy mutants and analyzed the kinetics of attachment and the formation of actin pedestals. We confirmed that, as previously reported by Tatsuno et al. (41), the wzy mutant attached to the cells and formed actin pedestals at an increased rate (Fig. 4A and B). Importantly, the etp mutants exhibited similarly increased rates of attachment and actin pedestal formation (Fig. 4A and B). The differences in attachment and pedestal formation between wild-type EHEC and the mutants were most prominent at 3 to 4 h postinfection but diminished at 5 to 6 h postinfection (Fig. 4A and B). Similar results were obtained when the etk mutant was used instead of the etp mutant (data not shown). Complementing the etp mutant with a plasmid expressing Etp restored the slower wild-type attachment kinetics (Fig. 4A and B). Similarly, increased attachment to host cells by the wzy, etk, and etp mutants was observed with other cell lines, including HeLa and Caco2 (data not shown). Given that the etp mutant is deficient in G4C formation but efficiently forms smooth LPS (Fig. 3B), we concluded that EHEC attachment to host cells is attenuated by the G4C and not by the smooth LPS.
FIG. 4.
The G4C interferes with EHEC interaction with host cells at early time points postinfection. (A and B) The efficiency of adherence (A) and of pedestal formation (B) by wild-type (wt) EHEC was compared to that of G4C-deficient etp and wzy mutants. Hep2 cells were infected with overnight cultures (OD, ∼1.0; multiplicity of infection, ∼2) of wild-type EHEC, the wzy::Tn5 and etp::Cm mutants, and the complemented etp::Cm/etp mutant. At different time points postinfection, the cells were fixed and the actin cytoskeleton was stained with rhodamine-phalloidin. The slides were analyzed by microscopy and scored for the percentage of Hep2 cells associated with more than eight attached bacteria (A, percent attachment) and for the percentage of Hep2 cells associated with more than five pedestals (B, percent actin pedestal formation). In all cases, the standard errors obtained in the experiments were below 14% (error bars are not shown to simplify the figure). In panel A, the wzy graph overlaps that of etp::Cm and thus it is hard to see it. (C) Similarly, we also infected monolayers of immobilized RBC, as described in the text, with wild-type EHEC, the etk::kan mutant, and a complemented mutant (etk::kan/etk). At 4 h postinfection, the infected RBC were fixed and stained (bacteria and RBC are in red, and EspA filaments are in green). The images shown were taken from a representative experiment out of two, and in both cases, the attachment of the etk mutant was increased by ∼6-fold in comparison to wild-type EHEC and by ∼14-fold in comparison to the complemented etk mutant (data not shown). (D) Activity of the LEE1 promoter in etp and etk mutants. A plasmid containing a gfp gene transcriptionally fused to the LEE1 regulatory region (pGY3159) was introduced into wild-type EHEC and the etp and etk mutants. Overnight bacterial cultures were diluted 1:50 in CDMEM in 96-well plates, which were immediately placed in a plate reader preset at 37°C. Bacteria were grown within the plate reader under infection conditions without shaking, and growth (OD600) and the activity of the LEE1 promoter (fluorescence levels) were monitored at 120-s intervals. Shown is a representative experiment out of three. (E) Tir translocation by the EHEC etk::kan mutant. EHEC strains containing a plasmid encoding TirEHEC-BlaM (pKB3105) were used to infect HeLa cells. At 3 h (blue columns) or 3:45 h (red columns) postinfection, the cells were loaded with CCF2 and the rate of CCF2 hydrolysis in the cells, reflecting the amount of translocated Tir-BlaM, was determined as described in Materials and Methods. The genotypes of the strains used are indicated below the columns. As a negative control, we used an etk::kan mutant containing a plasmid expressing unfused blaM (pCX341). Each column represents the average translocation by six different colonies, and the bars indicate the standard errors. A representative experiment out of three is shown. Mutants were compared to the wild type by both parametric and nonparametric unpaired tests, and significant differences (P < 0.05) are indicated by asterisks.
The major EHEC attachment mechanism involves the binding of intimin to translocated Tir, but TTSS-independent attachment mechanisms have also been reported (28, 42). However, EHEC attachment to RBC is exclusively TTSS dependent and mediated either by the EspA filament or by Tir-intimin interaction (39). To clarify whether the G4C interferes with TTSS-dependent attachment, we tested the adherence of different EHEC strains to immobilized RBC monolayers as previously described (39). We found that the absence of the G4C was associated with increased bacterial attachment to RBC (Fig. 4C), supporting the notion that the G4C interferes with TTSS-dependent cell attachment.
G4C-deficient mutants express normal levels of TTSS components but display increased translocation efficiency.
We tested the possibility that increased attachment and pedestal formation by the EHEC G4C mutants is due to enhanced expression of the TTSS genes. Ler, encoded in the LEE1 operon, is a positive regulator of tir, eae, and most of the TTSS genes. We therefore used the gfp reporter gene to compare the activities of the EHEC LEE1 promoter in the different mutants. We found that wild-type EHEC and the etp and etk mutants exhibited very similar growth rates and LEE1 expression levels (Fig. 4D and data not shown), suggesting that the inactivation of etp or etk is not associated with increased expression of ler and other LEE genes. This conclusion was corroborated by Western blot analysis with antibodies raised against the LEE-encoded proteins EscJ, EspA, EspB, and intimin (data not shown). Thus, as in the case of EPEC, the increased attachment and pedestal formation by the G4C EHEC mutants is not likely to be due to the increased expression of TTSS genes.
We next tested whether the G4C also inhibits Tir translocation in the case of EHEC by comparing the efficiency of Tir translocation by wild-type EHEC to that of the etk mutants. As a negative control, we used a TTSS-deficient strain (escN::kan). Strains containing a plasmid encoding tir-blaM (pKB3105) were used to infect Hep2 cells, and at different time points postinfection, the translocation level was determined (the assay was different from that used for EPEC, as described in Materials and Methods). The results show a significant, but not dramatic, increase in the rate of Tir-BlaM translocation by etk mutants (Fig. 4E). In contrast to the results obtained with EPEC, complementation of the mutant with the etk-expressing plasmid did not restore the reduced translocation (Fig. 4E). Interestingly, a similar increase in Tir translocation was seen when we used the etp mutant instead of the etk mutant (data not shown), but also in this case, for unknown reasons, we could not reverse the phenotype by complementation (data not shown). Possible reasons for failing to complement etp and etk mutants in this assay will be discussed later. Nevertheless, taken together, our results indicate that in both EPEC and EHEC, the G4Cs (O127 and O157 capsules, respectively) inhibit bacterial attachment to host cells, as well as actin pedestal formation, by TTSS masking and intimin shielding, resulting in reduced Tir translocation and Tir-intimin interaction.
The TTSS and G4C are conversely regulated.
We next investigated why the G4C inhibitory effect fades after prolonged infection (∼3 h for EPEC and ∼6 h for EHEC). A partial explanation for this apparent inconsistency might be the elongation of the EspA filament (15) to allow Tir delivery through the capsule. However, the capsule should still inhibit attachment via intimin masking and interfering with its interaction with the translocated Tir. Another scenario that can resolve this paradox is that upregulation of TTSS assembly is associated with the downregulation of G4C formation. To test this possibility, we examined whether the EHEC ler::kan mutant, deficient in the expression of most of the TTSS genes, can form G4C similar to that of wild-type EHEC.
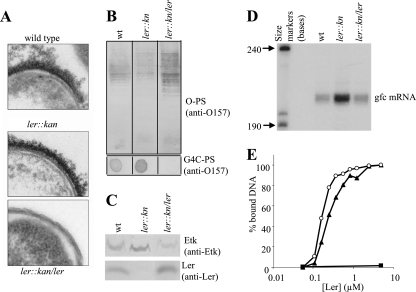
We first compared wild-type EHEC and the ler::kan mutant by electron microscopy and found that they appeared to be similar (Fig. 5A). However, since electron microscopy cannot be used to quantify capsular size, we could not exclude possible differences in the quantity of capsular polysaccharide between the two. Notably, a dramatic capsule disappearance was visualized upon the complementation of the ler mutant with a ler-expressing plasmid (Fig. 5A), resulting in modest ler overexpression (Fig. 5C). These results suggest that Ler might negatively regulate capsule production.
FIG. 5.
Ler represses the gfc promoter and G4C production. Wild-type (wt) EHEC, a ler::kan mutant (DF1291), and the mutant containing a plasmid containing ler (DF1291 containing pTU12) were grown under conditions that promote Ler expression. These strains were compared for capsule formation by electron microscopy, and representative electron micrographs of ferritin-stained thin sections are shown (A). These strains were also compared for LPS O-PS and G4C-PS production (B) and production of Ler and Etk (C). Amounts of O-PS and G4C-PS were assayed as described in the legend to Fig. 3, and the levels of Ler and Etk were assayed by Western blotting with anti-Ler or anti-Etk antibodies. To determine the gfc mRNA levels in these strains, equal amounts of extracted mRNA samples were subjected to a primer extension reaction and the reaction products were resolved in a 6% sequencing gel (D). Binding of Ler to the gfc regulatory region was tested by a modified enzyme-linked DNA-protein interaction assay as described in Materials and Methods (E). Different concentrations of purified Ler were incubated with a DNA fragment containing the gfc promoter region (black triangles), with negative control DNA (the etk coding region, black rectangles), and with positive control DNA (the LEE2 regulatory region, open circles), and binding was quantified. One hundred percent binding was defined as the binding of the highest concentration of Ler (5 μM; higher concentrations gave similar results as the system was saturated), and the relative binding of Ler to different fragments is shown. Purified Ler tends to gradually lose activity (unpublished observation). Thus, although we used freshly purified Ler, the concentration of active Ler might be lower than that of total Ler.
To further test this possibility, we extracted the total polysaccharides from wild-type EHEC and a ler mutant grown overnight in DMEM. The LPS and G4C-PS were then separated as previously described (see Materials and Methods). The amount of O antigen (LPS-associated O-PS) was then tested by Western blotting with anti-O157 antibody (Fig. 5B, upper panel), and the amount of capsular polysaccharide (LPS free, G4C-PS) was evaluated by dot immunoblot analysis with anti-O157 antibody (Fig. 5B, lower panel). Care was taken to apply equivalent amounts of material derived from the same numbers of bacteria in all of the lanes and spots. We found that the ler mutant produced more capsular polysaccharide (and less LPS-associated O antigen; Fig. 5B). Moreover, complementation of the ler mutant with a ler-expressing plasmid was associated with a dramatic reduction in capsule production (Fig. 5B, lower panel). Taken together, our findings suggest that Ler, or a product of a gene positively regulated by Ler (possibly a TTSS component), represses capsule formation.
Ler inhibits G4C formation by repressing gfc operon transcription.
Ler positively controls the expression of TTSS and other virulence-associated genes but also represses a large number of virulence and housekeeping genes (1). We therefore tested whether Ler represses the expression of the gfc operon. First, we extracted proteins from EHEC grown under Ler-expressing conditions (overnight static culture in DMEM) and used Western blot analyses with anti-Etk and anti-Ler antibodies to assess the Ler and Etk levels in the bacteria. Importantly, we found that Ler expression was directly correlated with Etk downregulation (Fig. 5C). In the ler::kan mutant, Ler was absent and the Etk levels were elevated. In contrast, in wild-type EHEC and the complemented ler mutant, Ler was produced and the Etk levels were reduced (Fig. 5C). Similar inhibition of Etk production by Ler was found when we used EPEC O127 instead of EHEC (data not shown). Given that the gfc operon is controlled by a single promoter (32), our results suggest that Ler might repress (directly or indirectly) the gfc promoter.
To further test the role of Ler in the repression of the gfc promoter, we extracted mRNA from similarly grown EHEC strains and used it to perform primer extension analyses with a primer complementing the ymcD gene mRNA (32). Primer extension was performed, and the products were resolved with a sequencing gel as previously described (see Materials and Methods and reference 32). We found markedly elevated gfc mRNA levels in the ler mutant, whereas in wild-type EHEC and the complemented ler mutant, the gfc mRNA levels were reduced (Fig. 5D). These results support the hypothesis that Ler directly or indirectly represses the gfc promoter.
To explore whether Ler directly represses gfc expression, we tested the ability of Ler to bind to DNA fragments containing the gfc promoter region (presumably the regulatory region of this promoter). The solid-phase DNA-binding assay (5) was used to compare Ler binding to three DNA fragments, (i) a fragment containing the gfc regulatory region, (ii) a fragment containing the LEE2 regulatory region (positive control), and (iii) a fragment consisting of part of the etk coding region (the negative control). We found that purified Ler efficiently binds to the gfc regulatory region and to the positive control DNA but not to the negative control DNA (Fig. 5E), supporting the hypothesis that Ler directly represses the gfc promoter.
Taken together, the above results indicate that the expression of TTSS operons and that of the gfc operon are conversely regulated at the transcription level and that Ler might directly repress the gfc promoter. Thus, these results explain why the inhibition of attachment by the G4C is temporary.
The G4C is required for efficient colonization of the infant rabbit intestine.
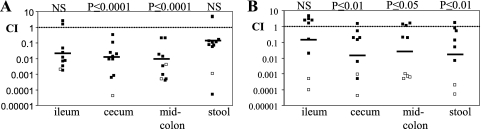
We next investigated how the G4C might contribute to bacterial fitness. One possibility is that the G4C contributes to E. coli fitness in the environment. Another possibility is that transient capsule expression contributes to host colonization. To test the latter possibility, we tested how a G4C deficiency would influence EHEC colonization of the host intestine in the infant rabbit model (35). Briefly, 3-day-old infant rabbits were coinoculated with the wild type and the etk mutant. The numbers of wild-type and etk mutant CFU in different intestinal sections were determined 2 and 7 days postinoculation, and CI values were calculated. At 2 days postinoculation, the CI values were 0.007 and 0.005 in the cecum and colon (Fig. 6A), indicating that the etk mutant has a markedly (>150-fold) reduced capacity to colonize these regions of the intestine. The CI in the ileum was ∼0.02 (Fig. 6A), but the difference did not reach statistical significance. Thus, the G4C appears to be most important for efficient colonization of regions distal to the ileum. At 7 days postinoculation, the results were very similar although the reduction in colonization by the etk mutant compared with the wild type was slightly less marked than that observed on day 2 (Fig. 6B). These results suggest that G4C is required for efficient colon colonization by EHEC.
FIG. 6.
Attenuated intestinal colonization by an EHEC etk mutant. Infant rabbits were coinfected with EDL933 Nalr and an isogenic etk::kan mutant (SK2472). At days 2 (A) and 7 (B) postinfection, CI values were determined. Each point represents an animal, and the bars represent the geometric mean. Open symbols represent animals in which no etk mutant CFU was recovered from the lowest dilution; the results were adjusted to represent the recovery of 1 CFU at that dilution. The data were compared to a theoretical CI of 1 with the Wilcoxon signed rank test, and the P values obtained are shown. Nonsignificant results are marked as NS.
DISCUSSION
Formation of G4C by EPEC and EHEC.
The gfc operon comprises seven genes, ymcD, ymcC, ymcB, ymcA, yccZ, etp, and etk (20, 32, 45). In this report, we show that EHEC O157, including strains EDL933 (shown here) and Sakai (data not shown), form an O-antigen capsule whose assembly is dependent on Etp and Etk. The existence of this O-antigen capsule in EHEC O157 has been overlooked, probably because it cannot be identified by standard serological tests. We did not test the involvement of the other gfc genes in G4C assembly by EHEC, but it is likely that, as in EPEC O127 (32), the products of these genes are also required for G4C assembly in EPEC O157. Given their role in G4C formation, we suggest that ymcD, ymcC, ymcB, ymcA, and yccZ be renamed gfcA, gfcB, gfcC, gfcD, and gfcE, respectively.
G4C inhibits the interaction of EPEC and EHEC with host cells.
EHEC O157 rough mutants display increased binding to tissue culture cells (7, 9, 41, 42). On the basis of these results, it was suggested that LPS plays an inhibitory role in attachment. Our new findings suggest, however, that this is not the case and that it is the G4C that inhibits attachment. Moreover, we showed that also in EPEC the G4C inhibits attachment to host cells and the formation of actin pedestals. This inhibition is probably mediated by the masking of the TTSS and intimin to reduce both Tir translocation and intimin-Tir interaction. The inhibition of Tir translocation is only partial, perhaps because the EHEC and EPEC TTSSs are fitted with the EspA extension filament (38), allowing protein injection through the capsule. This might indicate that intimin masking plays a more dominant role in inhibition of attachment and pedestal formation than that of TTSS masking.
Masking of intimin-like adhesins and TTSS by capsule and other bacterial surface structures.
The thickness of the G4C of EPEC and EHEC was not determined, but it probably resembles that of other capsules, which may typically extend 100 to 1,000 nm from the bacterial surface, depending on its type and composition (37). Masking by capsule of E. coli adhesins similar to intimin in size, protruding ∼10 nm beyond the outer membrane (23), was previously reported by Schembri et al. (37). In fact, on the basis of their results, these authors predict that intimin might also be subjected to masking by encapsulation. This phenomenon is not restricted to E. coli and was also reported in other bacteria, including Klebsiella pneumoniae (36) and Neisseria (46). Shielding of TTSSs by capsule was never reported before, but two reports describe TTSS shielding by other bacterial surface structures (49). The activity of the Yersinia TTSS can be inhibited via masking by the surface protein YadA (31), and the activity of the Shigella TTSS can be masked and inhibited by the LPS O antigen (49).
Solving the shielding dilemma.
The shielding concept leaves the bacteria with an obvious dilemma. They cannot translocate protein or adhere without the assistance of adhesin proteins and the TTSS, but at the same time, the shields contribute to bacterial fitness by providing protection against countermeasures at the disposal of a mammalian host. Yersinia solved the shielding dilemma by careful adjustment of the respective lengths of the TTSS needle and YadA, such that the length of the TTSS needle is the minimum that still allows efficient protein translocation through the YadA shield (31). In the case of Shigella, glucosylation of the LPS O antigen, via a bacteriophage-encoded enzyme, shortens the O antigens and thus enhances the TTSS function without compromising the protective properties of the LPS (49). Interestingly, we found that some Shigella strains contain an intact gfc operon and can express Etk (unpublished results). It remains to be seen whether Shigella also produces an O-antigen capsule and, if so, whether it also plays a role in TTSS masking and whether, like the LPS O antigen, it is subjected to glucosylation.
EPEC and EHEC display a dual solution to the shielding dilemma. First, the capsule only partially inhibits protein translocation, probably reflecting the capacity of these pathogens to fit the TTSS with the EspA filament, which can traverse, and inject proteins through, the capsule. Thus, it is expected that, upon the assembly of elongated EspA filaments, these bacteria can use the TTSS for protein translocation even in their capsulated form. Intimin, however, is expected to remain masked in capsulated bacteria. The second solution of EPEC/EHEC to this dilemma is to reduce the capsule size upon the expression of intimin and the TTSS. Taken together, these findings suggest that it is possible that, at some initial infection phase, the bacteria can efficiently translocate Tir and other effectors, whereas intimin is still shielded by the capsule. Thus, the rate of capsule thinning may modulate a time gap between Tir translocation and its clustering beneath the bacteria by intimin.
Inhibition of gfc expression by Ler.
Importantly, we found that the production of the G4C and the LEE proteins (including TTSS components, Tir, and intimin) is inversely coordinated by Ler at the transcriptional level. This ensures that activation of the TTSS-related genes is associated with repression of the gfc promoter. Ler exhibits a general structure that is similar to that of H-NS (14, 29), and like H-NS, it appears to polymerize on the target DNA to cover stretches of ∼200 bp, sometimes without clear boundaries (6, 18, 29). Its positive effect on LEE gene expression is mediated by its binding to specific regions in the LEE and thereby negating the H-NS-mediated repression (8, 18, 43, 44). This antirepressor activity is probably mediated by interfering with a specific H-NS-DNA complex (8, 18, 43). How Ler mediates the repression of the gfc promoter is not clear. Given that Ler binds the regulatory region of the gfc promoter, we speculate that Ler might interfere with the binding of the RNA polymerase to this promoter. Clearly, detailed investigation of the Ler-mediated repression of the gfc promoters is needed to solve the repression mechanism, but this study was outside the scope of the present work.
Role of the G4C in improving bacterial fitness.
Polysaccharide capsules were implicated in protecting bacteria from desiccation and were found to be involved in virulence by protecting the bacteria from phagocytosis, complement, and bactericidal-peptide-like defensins. It is not clear, however, how the G4C contributes to the fitness of EPEC or EHEC. Previous reports show some disadvantages associated with G4C formation, which leads to the formation of LPS with less O antigen. In E. coli O111 and O127, this reduction in LPS O antigen is associated with hypersensitivity to complement, whereas noncapsulated mutants become complement resistant (16, 32). An additional disadvantage associated with capsule formation was found in this study, namely, interfering with the function of adhesins and TTSS. However, the fact that the bacteria maintain the capsule indicates that this surface structure contributes to bacterial fitness under some conditions. In support of this notion, by using the infant rabbit model, we found that the G4C-deficient mutant has a markedly reduced capacity to colonize the host intestine, particularly in regions distal to the ileum. These findings, together with the results presented in this report, suggest that dynamic, temporal, and spatial regulation of the production of the TTSS and the G4C is important for optimal EHEC colonization of the host intestine. However, it is not clear how the G4C promotes EHEC colonization. Recently, Lacour and colleagues (25) showed that etk mutants are more sensitive to the antibacterial peptide polymyxin B, raising the possibility that the G4C protects EHEC from some antibacterial factors encountered in the host gastrointestinal tract. It is also possible that the capsule promotes initial transient attachment to the host intestinal tissue. The latter might explain why we could not restore reduced Tir translocation upon complementation (Fig. 4E) since, if this is the case, thick capsule might improve initial attachment but still inhibit translocation. In contrast, thin or patchy capsule will promote initial attachment without inhibition of translocation and thus enhance translocation. Nevertheless, we do not have any evidence to support the above hypothesis.
Supplementary Material
Acknowledgments
We thank J. Leong, M. Stevens, and J. Kaper for bacterial strains and G. Frankel and B. Kenny for providing antibodies.
This work was supported by grants from the United States-Israel Binational Science Foundation, the Center of Study of Emerging Disease, the EraNet-PathoGenomic program, and the Abisch-Frenkel Foundation. T.B. was supported by a Boehringer Ingelheim Fonds scholarship, and G.Y. was supported by the Einstein Scholarship sponsored by the Isaac Kaye Foundation. M.K.W. and J.M.R. were supported by HHMI and NIH.
Footnotes
Published ahead of print on 23 May 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abe, H., A. Miyahara, T. Oshima, K. Tashiro, Y. Ogura, S. Kuhara, N. Ogasawara, T. Hayashi, and T. Tobe. 2008. Global regulation by horizontally transferred regulators establishes the pathogenicity of Escherichia coli. DNA Res. 1525-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amor, P. A., and C. Whitfield. 1997. Molecular and functional analysis of genes required for expression of group IB K antigens in Escherichia coli: characterization of the his-region containing gene clusters for multiple cell-surface polysaccharides. Mol. Microbiol. 26145-161. [DOI] [PubMed] [Google Scholar]
- 3.Bell, B. P., M. Goldoft, P. M. Griffin, M. A. Davis, D. C. Gordon, P. I. Tarr, C. A. Bartleson, J. H. Lewis, T. J. Barrett, J. G. Wells, et al. 1994. A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers. The Washington experience. JAMA 2721349-1353. [PubMed] [Google Scholar]
- 4.Ben-Ami, G., V. Ozeri, E. Hanski, F. Hofmann, K. Aktories, K. M. Hahn, G. M. Bokoch, and I. Rosenshine. 1998. Agents that inhibit Rho, Rac, and Cdc42 do not block formation of actin pedestals in HeLa cells infected with enteropathogenic Escherichia coli. Infect. Immun. 661755-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benotmane, A. M., M. F. Hoylaerts, D. Collen, and A. Belayew. 1997. Nonisotopic quantitative analysis of protein-DNA interactions at equilibrium. Anal. Biochem. 250181-185. [DOI] [PubMed] [Google Scholar]
- 6.Berdichevsky, T., D. Friedberg, C. Nadler, A. Rokney, A. Oppenheim, and I. Rosenshine. 2005. Ler is a negative autoregulator of the LEE1 operon in enteropathogenic Escherichia coli. J. Bacteriol. 187349-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilge, S. S., J. C. Vary, Jr., S. F. Dowell, and P. I. Tarr. 1996. Role of the Escherichia coli O157:H7 O side chain in adherence and analysis of an rfb locus. Infect. Immun. 644795-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39664-678. [DOI] [PubMed] [Google Scholar]
- 9.Cockerill, F., III, G. Beebakhee, R. Soni, and P. Sherman. 1996. Polysaccharide side chains are not required for attaching and effacing adhesion of Escherichia coli O157:H7. Infect. Immun. 643196-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dziva, F., P. M. van Diemen, M. P. Stevens, A. J. Smith, and T. S. Wallis. 2004. Identification of Escherichia coli O157: H7 genes influencing colonization of the bovine gastrointestinal tract using signature-tagged mutagenesis. Microbiology 1503631-3645. [DOI] [PubMed] [Google Scholar]
- 12.Elgrably-Weiss, M., S. Park, E. Schlosser-Silverman, I. Rosenshine, J. Imlay, and S. Altuvia. 2002. A Salmonella enterica serovar Typhimurium hemA mutant is highly susceptible to oxidative DNA damage. J. Bacteriol. 1843774-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott, S. J., V. Sperandio, J. A. Giron, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 686115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34941-952. [DOI] [PubMed] [Google Scholar]
- 15.Garmendia, J., G. Frankel, and V. F. Crepin. 2005. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect. Immun. 732573-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldman, R. C., K. Joiner, and L. Leive. 1984. Serum-resistant mutants of Escherichia coli O111 contain increased lipopolysaccharide, lack an O antigen-containing capsule, and cover more of their lipid A core with O antigen. J. Bacteriol. 159877-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldman, R. C., D. White, F. Orskov, I. Orskov, P. D. Rick, M. S. Lewis, A. K. Bhattacharjee, and L. Leive. 1982. A surface polysaccharide of Escherichia coli O111 contains O-antigen and inhibits agglutination of cells by O-antiserum. J. Bacteriol. 1511210-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haack, K. R., C. L. Robinson, K. J. Miller, J. W. Fowlkes, and J. L. Mellies. 2003. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect. Immun. 71384-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho, T. D., and M. K. Waldor. 2007. Enterohemorrhagic Escherichia coli O157:H7 gal mutants are sensitive to bacteriophage P1 and defective in intestinal colonization. Infect. Immun. 751661-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ilan, O., Y. Bloch, G. Frankel, H. Ullrich, K. Geider, and I. Rosenshine. 1999. Protein tyrosine kinases in bacterial pathogens are associated with virulence and production of exopolysaccharide. EMBO J. 183241-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 927996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2123-140. [DOI] [PubMed] [Google Scholar]
- 23.Kelly, G., S. Prasannan, S. Daniell, K. Fleming, G. Frankel, G. Dougan, I. Connerton, and S. Matthews. 1999. Structure of the cell-adhesion fragment of intimin from enteropathogenic Escherichia coli. Nat. Struct. Biol. 6313-318. [DOI] [PubMed] [Google Scholar]
- 24.Knutton, S., P. H. Williams, D. R. Lloyd, D. C. Candy, and A. S. McNeish. 1984. Ultrastructural study of adherence to and penetration of cultured cells by two invasive Escherichia coli strains isolated from infants with enteritis. Infect. Immun. 44599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lacour, S., P. Doublet, B. Obadia, A. J. Cozzone, and C. Grangeasse. 2006. A novel role for protein-tyrosine kinase Etk from Escherichia coli K-12 related to polymyxin resistance. Res. Microbiol. 157637-641. [DOI] [PubMed] [Google Scholar]
- 26.Li, M., I. Rosenshine, S. L. Tung, X. H. Wang, D. Friedberg, C. L. Hew, and K. Y. Leung. 2004. Comparative proteomic analysis of extracellular proteins of enterohemorrhagic and enteropathogenic Escherichia coli strains and their ihf and ler mutants. Appl. Environ. Microbiol. 705274-5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, S., R. Tobias, S. McClure, G. Styba, Q. Shi, and G. Jackowski. 1997. Removal of endotoxin from recombinant protein preparations. Clin. Biochem. 30455-463. [DOI] [PubMed] [Google Scholar]
- 28.Low, A. S., F. Dziva, A. G. Torres, J. L. Martinez, T. Rosser, S. Naylor, K. Spears, N. Holden, A. Mahajan, J. Findlay, J. Sales, D. G. Smith, J. C. Low, M. P. Stevens, and D. L. Gally. 2006. Cloning, expression, and characterization of fimbrial operon F9 from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 742233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33296-306. [DOI] [PubMed] [Google Scholar]
- 30.Mills, E., K. Baruch, X. Charpentier, S. Kobi, and I. Rosenshine. 2008. Real-time analysis of effector translocation by the type III secretion system of enteropathogenic Escherichia coli. Cell Host Microbe 3104-113. [DOI] [PubMed] [Google Scholar]
- 31.Mota, L. J., L. Journet, I. Sorg, C. Agrain, and G. R. Cornelis. 2005. Bacterial injectisomes: needle length does matter. Science 3071278. [DOI] [PubMed] [Google Scholar]
- 32.Peleg, A., Y. Shifrin, O. Ilan, C. Nadler-Yona, S. Nov, S. Koby, K. Baruch, S. Altuvia, M. Elgrably-Weiss, C. M. Abe, S. Knutton, M. A. Saper, and I. Rosenshine. 2005. Identification of an Escherichia coli operon required for formation of the O-antigen capsule. J. Bacteriol. 1875259-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson, A. A., and E. J. McGroarty. 1985. High-molecular-weight components in lipopolysaccharides of Salmonella typhimurium, Salmonella minnesota, and Escherichia coli. J. Bacteriol. 162738-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ritchie, J. M., C. M. Thorpe, A. B. Rogers, and M. K. Waldor. 2003. Critical roles for stx2, eae, and tir in enterohemorrhagic Escherichia coli-induced diarrhea and intestinal inflammation in infant rabbits. Infect. Immun. 717129-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritchie, J. M., and M. K. Waldor. 2005. The locus of enterocyte effacement-encoded effector proteins all promote enterohemorrhagic Escherichia coli pathogenicity in infant rabbits. Infect. Immun. 731466-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schembri, M. A., J. Blom, K. A. Krogfelt, and P. Klemm. 2005. Capsule and fimbria interaction in Klebsiella pneumoniae. Infect. Immun. 734626-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schembri, M. A., D. Dalsgaard, and P. Klemm. 2004. Capsule shields the function of short bacterial adhesins. J. Bacteriol. 1861249-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekiya, K., M. Ohishi, T. Ogino, K. Tamano, C. Sasakawa, and A. Abe. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. USA 9811638-11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw, R. K., S. Daniell, G. Frankel, and S. Knutton. 2002. Enteropathogenic Escherichia coli translocate Tir and form an intimin-Tir intimate attachment to red blood cell membranes. Microbiology 1481355-1365. [DOI] [PubMed] [Google Scholar]
- 40.Spears, K. J., A. J. Roe, and D. L. Gally. 2006. A comparison of enteropathogenic and enterohaemorrhagic Escherichia coli pathogenesis. FEMS Microbiol. Lett. 255187-202. [DOI] [PubMed] [Google Scholar]
- 41.Tatsuno, I., K. Nagano, K. Taguchi, L. Rong, H. Mori, and C. Sasakawa. 2003. Increased adherence to Caco-2 cells caused by disruption of the yhiE and yhiF genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 712598-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres, A. G., and J. B. Kaper. 2003. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect. Immun. 714985-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torres, A. G., G. N. Lopez-Sanchez, L. Milflores-Flores, S. D. Patel, M. Rojas-Lopez, C. F. Martinez de la Pena, M. M. Arenas-Hernandez, and Y. Martinez-Laguna. 2007. Ler and H-NS, regulators controlling expression of the long polar fimbriae of Escherichia coli O157:H7. J. Bacteriol. 1895916-5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Umanski, T., I. Rosenshine, and D. Friedberg. 2002. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 1482735-2744. [DOI] [PubMed] [Google Scholar]
- 45.Vincent, C., B. Duclos, C. Grangeasse, E. Vaganay, M. Riberty, A. J. Cozzone, and P. Doublet. 2000. Relationship between exopolysaccharide production and protein-tyrosine phosphorylation in gram-negative bacteria. J. Mol. Biol. 304311-321. [DOI] [PubMed] [Google Scholar]
- 46.Virji, M., K. Makepeace, I. R. Peak, D. J. Ferguson, M. P. Jennings, and E. R. Moxon. 1995. Opc- and pilus-dependent interactions of meningococci with human endothelial cells: molecular mechanisms and modulation by surface polysaccharides. Mol. Microbiol. 18741-754. [DOI] [PubMed] [Google Scholar]
- 47.Wang, L., C. E. Briggs, D. Rothemund, P. Fratamico, J. B. Luchansky, and P. R. Reeves. 2001. Sequence of the E. coli O104 antigen gene cluster and identification of O104 specific genes. Gene 270231-236. [DOI] [PubMed] [Google Scholar]
- 48.Wang, L., H. Curd, W. Qu, and P. R. Reeves. 1998. Sequencing of Escherichia coli O111 O-antigen gene cluster and identification of O111-specific genes. J. Clin. Microbiol. 363182-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.West, N. P., P. Sansonetti, J. Mounier, R. M. Exley, C. Parsot, S. Guadagnini, M. C. Prevost, A. Prochnicka-Chalufour, M. Delepierre, M. Tanguy, and C. M. Tang. 2005. Optimization of virulence functions through glucosylation of Shigella LPS. Science 3071313-1317. [DOI] [PubMed] [Google Scholar]
- 50.Whitfield, C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 7539-68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.