Abstract
We measured the relative fitness among a set of experimentally evolved Escherichia coli strains differing by a small number of adaptive mutations by directly measuring allelic frequencies in head-to-head competitions using a mass spectrometry-based method. We compared the relative effects of mutations to the same or similar genes acquired in multiple strains when expressed in allele replacement strains. We found that the strongest determinant of fitness among the evolved strains was the impact of beneficial mutations to the RNA polymerase β and β′ subunit genes. We also identified several examples of epistatic interactions between rpoB/C and glpK mutations and identified two other mutations that are beneficial only in the presence of previously acquired mutations but that have little or no adaptive benefit to the wild-type strain. Allele frequency estimation is shown to be a highly sensitive method for measuring selection rates during competitions between strains differing by as little as a single-nucleotide polymorphism and may be of great use for investigating epistatic interactions.
It is still difficult to understand the mechanisms used to adapt to a defined metabolic challenge, even in well-characterized organisms such as Escherichia coli. Genome-scale metabolic models have accurately predicted metabolic phenotypes such as growth and secretion rates after adaptation (6), but these models do not include the capacity to predict what genetic changes could induce the flux shifts that produce the end point phenotype. The mutations acquired during adaptation to a defined challenge have been identified following several adaptive evolution experiments (9, 25, 26), but in bacterial systems the mutations have been found to occur in multiple, distinct biological processes, including mutations in genes without an easily understood role in adaptation to the imposed pressure. Adding to the challenge of discovering the biochemical mechanism of a particular mutation's effect on phenotype, selection may involve pleiotropic consequences or epistatic interactions with other mutations (13, 21), which cannot generally be predicted. Epistatic interactions or the dependence of the fitness value of some mutations on the presence or absence of other mutations can be especially important to consider since they are thought to create the potential for divergent endpoint genotypes among replicate adaptive evolution experiments (12, 27).
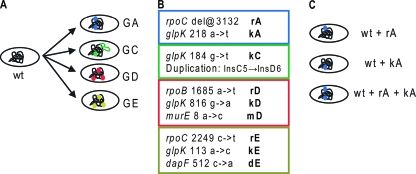
In the present study, we compared the fitness phenotype between similar sets of mutations acquired by four endpoint strains from parallel laboratory evolutions of E. coli K-12 on glycerol minimal medium for 44 days (5). The mutations fixed in these endpoint strains were previously identified by whole-genome sequencing and were found to define the full genetic basis for adaptation (9). All four strains have acquired a mutation of the glycerol kinase gene (glpK), three strains have a mutation of one of two RNA polymerase genes (rpoB/C), and two strains have a mutation of the peptidoglycan synthesis genes (dapF/murE) (Fig. 1A). The growth rates of the strains were relatively similar (0.46 to 0.58 h−1), indicating that they had achieved similar fitness gains over the wild-type strain. However, none of these mutations are to the same loci, suggesting the endpoint strains had reached different peaks on the fitness landscape. We performed competition experiments, as described below, which measure the relative fitness among the endpoint strains to establish whether they have meaningfully distinct phenotypes.
FIG. 1.
Origin of the strains utilized in the present study. (A) Parallel adaptive evolutions produced the four endpoint strains analyzed. (B) The mutations acquired by the endpoint strains were identified by whole-genome resequencing and are listed in the colored boxes. The short-hand notation for each mutation indicates the effected gene (r = rpoB/C, k = glpK, m = murE, d = dapF) and the endpoint strain it was found in (i.e., kA = glpK mutation identified in strain GA) (9). (C) Allele replacement strains were then constructed using gene gorging (8, 9) to introduce the mutations to the wild-type genetic background. An allele replacement strain was created that carries each mutation identified in isolation, as well as strains that carry multiple or all mutations identified in an endpoint strain.
We also compared the fitness phenotype induced by mutations at similar loci. This allows us to compare how relative fitness among each type of mutation correlates to the relative fitness among the endpoints. The outcomes also revealed several instances of synergistic epistasis between pairs of coacquired adaptive mutations. This study represents the first time that relative fitness between parallel-evolved strains has been correlated to the fitness effects among the individual mutations that constitute the full genetic basis of adaptation. Although these results alone are not sufficient to explain the physiological mechanism mediating the genotype-phenotype relationship, they contribute significantly to the design of future studies that may since the relative fitness among similar mutations can be used to analyze which enzyme activity changes caused by the mutations correlate to increased fitness, and knowledge of epistatic interactions justifies study of the interaction networks of the effected proteins.
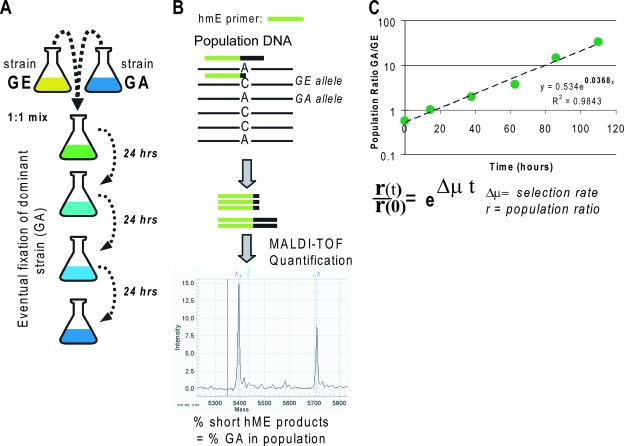
We measured relative fitness using head-to-head competition experiments between pairs of strains and detected the change in population frequency by measuring the allele frequency of each strain's known mutations. Competition experiments are the standard method for assessing relative fitness between closely related strains of bacteria in a defined environment (10, 14, 17, 21) and entail culturing two strains together in the same vessel to compete directly for resources, while monitoring the rate of population frequency change as one strain selectively outcompetes the other (Fig. 2A). The population composition is often monitored by using a detectable or selectable genetic marker in one of the competed strains to allow discriminatory colony counting. Because of the labor involved in generating strains with selectable markers, often all measured selection rates are generated relative to a marked wild-type strain rather than by direct competition (14, 17). Other recently developed methods that can be used to monitor competitions include fluorescent protein expression (7) and a plasmid sequence-based code (10) to trace lineages. In the present study, competitions were monitored by directly measuring the frequencies of the mutant alleles, which allowed us to measure direct head-to-head competitions between strains that differed by as little as a single mutation and to use multiple allelic markers in instances where strains differed by multiple mutations (see Table S1 in the supplemental material). This approach provides significantly increased sensitivity to detect fitness differences compared to optical-density-based growth rate measurements of individual strains that were previously used to study glycerol-evolved strains (14).
FIG. 2.
Design of the competition experiments. (A) The two strains to be competed are inoculated into a single vessel to compete directly for media resources. The culture is propagated for 3 days by serial passage, with samples collected daily to determine the current population frequency. (B) The population frequency at each time point is based on the allele frequency of the known mutations in each strain. DNA isolated daily from the competing culture is subjected to heterogeneous mass-extension assays (Sequenom, Inc., San Diego, CA) to determine the frequency of the mutations that distinguish the competing strains. The 200-bp region around each mutation is PCR amplified (results not shown), followed by PCR extension of an hME primer that binds directly upstream of the mutation. Extension of the primer is performed with nucleoside triphosphate mix missing a species of nucleotide that complements one of the alleles, causing amplification of mutation-harboring alleles to terminate at different loci than the wild type, producing products with distinct masses. All of the primers are listed in Table S2 in the supplemental material. The relative amounts of the extension products are determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry from the ratio of the product peak areas. (C) We calculate the estimated selection rate as the Malthusian parameter of the change in the population ratio over time (14). Assay bias was negligible in most cases (see Table S3 in the supplemental material).
MATERIALS AND METHODS
Strains.
Strains GA, GC, GD, and GE were studied because of the similarities between their sets of mutations. Further descriptions of these strains have previously been published (5, 9).
Competition experiments.
The two strains to be competed were cultured separately overnight from frozen stocks in M9-glycerol medium. After sufficient culture density was reached by both inoculums (>0.2 OD), cultures were combined based on their relative optical densities to produce the desired population ratio. Competitions were initiated by inoculating flasks of 250 ml of prewarmed M9-glycerol media with the combined culture. The initial population ratio was typically prepared to be approximately 1:1 of each strain, though the initial ratio was occasionally varied to favor the less-fit strain to allow more time points to be sampled before fixation of the dominant strain. The initial ratio was not observed to affect the observed selection rate in several independent competitions begun with various initial ratios (1:1,000, 1:100, 1:10, and 1:1) (see Table S5 in the supplemental material).
Competition cultures were typically maintained for 3 days by serial passage once per day. Cultures were maintained at 30°C with aeration, identical to evolution conditions (9). The volume of spent culture passed each day was estimated based on the previous day's growth rate, and the number of hours until the subsequent passage, to produce an OD of ∼0.3. The population frequency of each strain was measured from the SNP allele frequency in samples of spent culture collected each day (5 to 20 ml). We recorded the exact time of collection (hour:minute). Competitions between each pair of strains were performed in duplicate or triplicate.
Competitions were limited to 3 days because the selection rates of competitions between allele replacement strains were often observed to change after longer periods of time. This was interpreted to be due to the emergence of new adaptive mutations within the competing cultures, since allele replacement strains with single mutations still have high adaptation potential. As described in the statistical methods section, data points collected after a clear selection rate shift were discarded. The selection rate shifts generally differed between biological replicates of the experiments, in terms of the time of emergence, the strain affected, and the magnitude of the shift.
Allelotyping.
The DNA from competing cultures was isolated by using Qiagen DNEasy kit, using ultrapure water to elute DNA from the column. All samples were diluted to 5 to 10 ng/μl DNA concentration.
Allelotyping reactions were performed as described elsewhere (9) (Fig. 2B) using Sequenom hME protocols. Assay primers are given in Table S2 in the supplemental material. All samples were assayed in triplicate or quadruplicate, and the measurements averaged. The accurate detection limit is reached at ca. 5% population frequency, putting a limit on the population frequency range that can be accurately sampled. Furthermore, some genome loci have proven difficult to develop working hME assays for, possibly due to local DNA conformation. Techniques for measuring frequencies of these loci, including glpK 113 a→c (acquired by evolved strain GE), have been developed by modifying the related hMC resequencing protocols, as described elsewhere (9).
The small bias in some assays due to primer bias and/or other unidentified sources was corrected using normalization curves generated by assaying mixtures of purified endpoint and wild-type DNA manually combined to the desired ratios (1:9, 4:6, 6:4, and 9:1). The best-fit curves between actual and detected allelic frequencies were exponential (power-trend line in MS Excel), with an average exponent value of 1.25 ± 0.28.
Calculation of the selection rate.
The Malthusian parameter is assumed to equal the difference in growth rate between the two competing strains, assuming maintained exponential growth in a constant environment with no quorum sensing or other cell-cell interactions (14). Correspondingly, strain fitness is based solely on growth rate, and the selection rate, r, is equal to the difference in the growth rate between the two strains: rAB = mA − mB = [ln(Af/Ai) − ln(Bf/Bi)]/ Δt = [ln(Af/Bf) − ln(Ai/Bi)]/Δt. The variables A and B refer to the population frequencies of the competed strains at times initial (i) and final (f), and Δt is the time passed. The selection rate of each competition is calculated as the slope of the best-fit line on a plot of the natural log of the population frequency ratios versus time (Fig. 2C). The data are presented in terms of the selection rate to allow values within a set of competitions to be directly compared to facilitate analysis of epistatic and nontransitive interactions.
Data analysis and statistical methods.
There were three different types of replication in the competition experiments: (i) biological replicates corresponding to separate competition experiments with the same two strains, (ii) multiple assays used to measure the population frequencies within the same competition experiment, and (iii) technical replicates corresponding to separate measurements of the individual allelic frequencies using the hME method. The raw population frequency data for each individual replicate competition experiment and allelotyping assay was first log transformed.
Biological replicate competitions that either did not have sufficient number of reliable data points or strongly deviated from linearity were eliminated by manual inspection of individual population frequency curves. Similarly, outlier data points for individual competitions that were indicative of assay saturation or sudden slope change due to selection to fix a mutation acquired during the experiment were eliminated. After filtering out outliers, the total number of individual data points used to estimate the selection rates ranged between 18 and 144, depending on the competition experiment. For each pair of strains competed there were two to five biological replicates, one to four separate assays depending on the number of mutations in the strains, and three technical replicates per biological replicate and assay (see Table S1 in the supplemental material).
For each pair of strains competed we fitted a linear mixed-effects model to all of the data obtained for that pair simultaneously (i.e., all biological and technical replicates and assays). This allowed us to use all of the data effectively to estimate the selection rates, as well as standard errors for these rates. The model we used for estimating selection rates for individual competitions assumed that all replicates/assays have the same slope but may have different intercepts corresponding to different initial ratios between the starting strains. We used a linear mixed-effect model with the assay effect treated as a fixed effect and the biological replicates treated as random effects (4). The mixed-effect model was fit using the restricted maximum-likelihood approach implemented in the nmle package in R (22). A summary of the results obtained using the mixed-effect model, including the estimated selection rates, is shown in Table 1.
TABLE 1.
Summary of selection rates for different competitionsa
| Competition | Strain characteristic(s) | S (102) | SE | df | t | P |
|---|---|---|---|---|---|---|
| GA vs GC | Endpoints | 9.13 | 0.32 | 61 | 28.93 | 2.5E-37 |
| GA vs GE | Endpoints | 4.99 | 0.11 | 137 | 46.30 | 1.5E-85 |
| GD vs GA | Endpoints | 2.63 | 0.11 | 136 | 23.48 | 1.1E-49 |
| GD vs GC | Endpoints | 8.24 | 0.23 | 106 | 35.11 | 3.4E-60 |
| GD vs GE | Endpoints | 7.13 | 0.28 | 115 | 25.54 | 3.3E-49 |
| GE vs GC | Endpoints | 0.73 | 0.04 | 59 | 19.22 | 4.4E-27 |
| kA vs kD | glpK | 0.74 | 0.10 | 80 | 7.82 | 1.8E-11 |
| kA vs kE | glpK | 4.47 | 0.38 | 13 | 11.73 | 2.7E-08 |
| kC vs kA | glpK | 0.49 | 0.06 | 43 | 7.62 | 1.7E-09 |
| kC vs kD | glpK | 4.58 | 0.64 | 17 | 7.18 | 1.6E-06 |
| kC vs kE | glpK | 4.83 | 0.63 | 29 | 7.70 | 1.7E-08 |
| kD vs kE | glpK | 0.66 | 0.04 | 45 | 14.94 | 4.6E-19 |
| rkA vs rkE | glpK+rpoB/C | 9.39 | 0.27 | 122 | 34.83 | 3.1E-65 |
| rkD vs rkA | glpK+rpoB/C | 2.07 | 0.05 | 99 | 37.89 | 9.9E-61 |
| rkD vs rkE | glpK+rpoB/C | 10.41 | 0.33 | 112 | 31.66 | 1.1E-57 |
| dE vs wt | murE/dapF | 0.22 | 0.09 | 14 | 2.58 | 2.2E-02 |
| GD vs rkD | murE/dapF | 4.98 | 0.21 | 31 | 23.33 | 3.2E-21 |
| GE vs rkE | murE/dapF | 2.60 | 0.06 | 31 | 41.59 | 9.2E-29 |
| mD vs wt | murE/dapF | −0.06 | 0.05 | 29 | −1.21 | 2.4E-01 |
| rA vs rE | rpoB/C | 4.56 | 0.18 | 63 | 25.84 | 3.1E-35 |
| rD vs rA | rpoB/C | 1.37 | 0.04 | 56 | 34.25 | 3.0E-39 |
| rD vs rE | rpoB/C | 5.14 | 0.29 | 57 | 17.89 | 4.7E-25 |
S, selection rate; SE, standard error (95% confidence interval); df, degrees of freedom for the mixed-effect model fit; t, Student test t value; P, Student t test P value (null hypothesis S = 0).
In order to detect possible assay-specific effects in individual competitions (i.e., cases where different assays give significantly different selection rates), we modified the mixed-effect model so that the slope (or selection rate) was allowed to depend on the specific assay utilized. Statistically significant assay effects (P < 0.01) were detected in 9 of the 29 competitions. However, most of the assay effects were relatively small in magnitude (see Table S3 in the supplemental material).
RESULTS
Competition experiments were performed between all pairwise combinations of glycerol-evolved endpoint strains and between allele replacement strains with similar mutations (8, 9) (Fig. 1). The outcomes are reported as the selection rate (in units of 1/h), which corresponds to the estimated absolute growth rate difference between the two strains (Fig. 3).
FIG. 3.
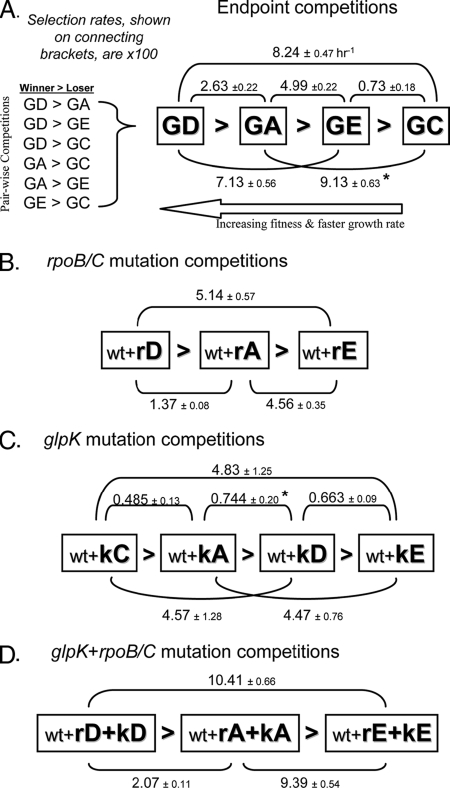
Head-to-head competition results. The selection rate constant between each pairwise competition, ±2 standard errors, is listed on the bracket connecting two strains. Displayed values have been increased ×100. Each box represents a strain within the set. The outcomes within the four sets of competitions are internally consistent. (A) Endpoint strain competition outcomes. The results of pairwise competitions are on the left and combined in the diagram on the right with the measured selection rates displayed on the connecting brackets. The increased strain fitness correlates roughly to a higher measured growth rate. The mutations acquired by each endpoint strain are listed in Fig. 1B. (B) Outcome of competitions between allele replacement strains harboring one of the identified rpoB/C mutations. Mutations are described by the abbreviations shown in Fig. 1B, which reference the source endpoint strain and the gene affected. (C) Outcome of glpK allele replacement strain competitions. (D) Competitions between double allele replacement strains that carry both the glpK and the rpoB/C mutations identified in the same endpoint strain. The asterisk indicates that the value does not agree well with estimates extrapolated from the rest of the set and may represent systematic measurement error (see Discussion).
Competitions between the four endpoint strains yielded an internally consistent fitness hierarchy, with reproducible selection rates that were significantly different from zero in all cases (mixed-effect linear model t test, P < 10−26; see Materials and Methods for details). The relative fitness of each endpoint strain was clearly distinct from the other strains. For example, the GE strain was found to be significantly less fit than the GA strain, although the growth rates of these strains were essentially indistinguishable when measured by optical density (14).
We next compared the fitness effects of similar mutations acquired by different strains by competing allele replacement strains that harbor each mutation individually. Like endpoint strains, the set of three rpoB/C mutation allele replacement strains (Fig. 3B) and the set of four glpK mutation allele replacement strains (Fig. 3C) proved to have an internally consistent fitness hierarchy, with selection rates that were reproducible and significantly different from zero (P < 10−6). This demonstrates that the individual mutations each have a distinct impact on the fitness of the organism.
The rpoB/C+glpK strains have a fitness hierarchy that matches both the rpoB/C and the endpoint strain set hierarchy in terms of the endpoint strains the mutations were identified from (i.e., D > A > E > C). However, the order within the glpK hierarchy differs significantly from the consensus order identified in the other experiments (Fig. 3). This result suggests that the adaptive value of the acquired rpoB/C mutations largely determine the fitness of the endpoint strains, overshadowing the effects of the glpK mutations.
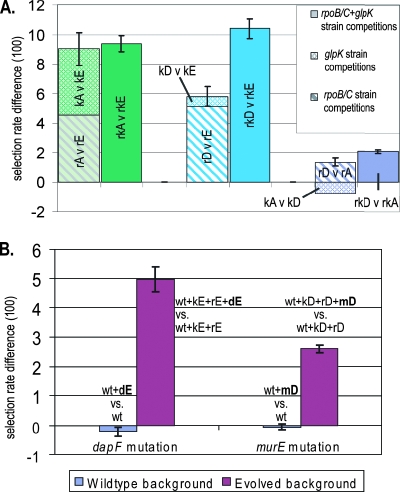
We additionally analyzed the selection rates from allele replacement strain competitions for evidence of epistatic interactions between coacquired glpK and rpoB/C mutations in the three endpoints that carried mutations to both genes. If there are no epistatic interactions, we expect the selection rate between two rpoB/C+glpK double allele replacement strains to approximately equal the sum of the selection rates of the same glpK and rpoB/C mutations when competed separately. This analysis is summarized in Fig. 4. The selection rates between allele replacement strains carrying GA's and GE's mutations are approximately additive, suggesting the pairs of mutations in neither of these strains have synergistic or antagonistic interactions. In contrast, when the mutations from GA or GE are similarly competed against GD's mutations, the fitness difference between the rpoB/C+glpK double mutant strains is significantly greater than the sum of the fitness differences found when the same rpoB/C mutations and glpK mutations are competed separately. This indicates epistatic interactions between the mutations in at least one strain—either synergism between glpK and rpoB/C mutations in the double-mutant of greater fitness (rkD in both cases), or antagonism between the mutations in the less-fit double-mutant (rkA or rkE) in each competition. If the latter case is true, the additive outcome observed from competitions between strains rkA and rkE could be produced if the antagonistic interactions between the glpK and rpoC mutations in strain GA and GE are approximately equal.
FIG. 4.
Epistatic interactions suggested by additive analysis. Epistatic interactions can be inferred when mutations cause different selection rate increases in different genetic backgrounds. (A) Each pair of bars shows the selection rates from competitions between mutations from the same endpoint strains. The selection rates represented in the left column result from competitions between allele replacement strains harboring single mutations; the horizontally striped bar represents the selection rate difference between the rpoB/C mutations of two endpoint strains (Fig. 3B), and the cross-hatched bar represents the selection rate difference between the glpK mutations from the same two endpoint strains (Fig. 3C). The solid-colored right-hand bar represents the selection rate difference found from competitions between allele replacement strains that carried both of a given endpoint strain's glpK and rpoB/C mutations (Fig. 3D). Significant discrepancies between the heights of each pair of bars indicate epistatic interactions between the glpK and rpoB/C mutations of one or both of the involved endpoint strains. An asterisk indicates the value does not agree well with estimates extrapolated from the rest of the set and may be questionable (see Discussion). (B) The adaptive values of the murE and dapF mutations increase in the evolved genetic background relative to wild type. Competitions were performed in both the wild-type and the evolved genetic background between strains that differed only by the murE or dapF mutation. Both mutations had effectively no adaptive value in the wild-type genetic background but were clearly adaptive in the presence of their coacquired rpoB/C and glpK mutations.
The GD and GE strains both acquired an additional mutation apart from the glpK and rpoB/C mutations. These mutations were in the murE and dapF genes involved in peptidoglycan synthesis (Fig. 1), and they were the last mutations these strains acquired (9). The growth rates of allele replacement strains harboring these mutations suggested that they have little to no adaptive value as single mutations to wild type, although they increased the growth rate when added to strains carrying their coacquired glpK and rpoB/C mutations. To test for potential epistatic relationships, we performed competition experiments between strains that differed only by the dapF or murE mutations in both the wild-type and the endpoint genetic backgrounds (Fig. 4B), which confirmed that both the dapF and the murE mutations considerably increase the relative fitness of strains with the coacquired glpK+rpoB/C mutations but have little impact on the wild type.
DISCUSSION
Previous experimental adaptive evolution studies have demonstrated strong parallelism between independent endpoints. Viral adaptation experiments have produced independent endpoints in which the majority of adaptive mutations are at identical loci (25). Although bacterial laboratory evolution studies have found far fewer exact allele changes in common between replicate evolutions, specific genes have been frequently identified that acquired adaptive mutations in all or most of the replicate evolved strains (9, 26, 29). This parallelism is presumed to be due to the strains following slightly divergent adaptive paths toward the same improved phenotype. By competing the endpoint strains and allele replacement strains, we have measured the relative effectiveness of the four apparently parallel adaptive paths taken by these strains.
The relative fitness phenotype hierarchy among the glpK and rpoB/C mutations will be used to help interpret results from assays of mutant enzyme activity. The fact that the majority of the replicate evolutions repeatedly acquire mutations to the same genes suggests that the mutations are selected to alter the same functional attribute. If so, fitness gains may be largely proportional to the mutation's effect on that function (although this may also be complicated by pleiotropic and epistatic interactions) (21). However, mutations can have multiple effects on enzyme activity, especially in proteins with complex functions like the β- and β′-RNA polymerase subunits. The relative fitness hierarchy can provide a criterion for identifying the activity changes that correspond to increasing the fitness phenotype. We have not yet been able to identify a specific functional attribute among the glpK or rpoB/C mutations that correlates to their relative fitness effects, let alone how such changes translate into increased fitness for the organism. Efforts to profile the activity of the mutated β- and β′-RNA polymerase subunits (encoded by rpoB and rpoC, respectively) are under way and indicate significant kinetic differences between wild-type and mutated polymerases (R. Landick, unpublished data). The mutant GlpK enzymes have increased Vmax and Ki-FBP (9), but neither of these parameters correlates to relative fitness among the corresponding glpK allele replacement strains, suggesting that increased fitness is due to another parameter or combination of parameters that we have not yet measured. It is interesting that both the glpK mutation with the greatest fitness impact (kC, 184 g→t) and the glpK loci most frequently mutated across 50 or more additional glycerol evolution experiments (kA, 218th nucleotide) (11) are located in the subunit interaction domain involved in GlpK's transition from an active dimer form to an inactive tetramer (15), suggesting that the dynamic equilibrium between these forms is an effective adaptive target. The other glpK mutations are located in fructose-1,6-bisphosphate binding regions, which are also involved in dimer-tetramer transitions, as well as in an uncharacterized region (9).
Although little is known about the underlying biochemistry of the rpoB/C mutations, it appears that their impact largely determines endpoint fitness. The order of the rpoB/C mutations in terms of increased fitness correlates to the rank order among the rpoB/C+glpK double-mutant strains and the endpoint strains. This, and the fact that the allele replacement strains harboring rpoB/C mutations have higher growth rates than strains harboring their coacquired glpK mutations (9), indicates that they contribute more to the fitness of the endpoint strains. This conclusion is further supported by the evolutionary trajectory of each strain, which show that the rpoB/C mutations were the first acquired in strains GA, GD, and GE (1, 9, 19, 20). Interestingly, the rpoB/C mutations did not become fixed until the lineages also acquired the glpK mutations.
The role of the rpoB/C mutations is believed to be linked to stabilizing the initially large-scale changes in the transcription profile observed in each evolution's population, which eventually stabilized and largely returned to the preperturbation expression state by the end of the adaptation period (5). The rpoB/C mutations may accomplish this by influencing catabolite repression (9) or the stringent response to minimal media. The biological effects of the rpoB/C mutations on global transcriptional activity are currently being investigated to clarify how the adaptive benefits of these mutations are realized.
All four sets of competitions had internally consistent outcomes, and their hierarchies correlate to the order of increasing growth rates among the strains (9), as predicted for selection under continuous exponential growth (14). The competition experiments performed here also resolved the relative fitness relationships between strains with indistinguishable growth rates (14).
The competition experiments also produce strong evidence that, except in the case of the strain GA, every causal mutation acquired by the studied endpoint strains was identified by resequencing during the previous study. We performed competitions against a common competitor against each endpoint strain and an allele replacement strain harboring all of the endpoint's known mutations (except for the large duplication in strain GC). No significant fitness differences were identified when the competitions were performed simultaneously with GD and GE strains, but this was not the case with GA and GC strains (Table 2). Resequencing of GA by using Solexa technology has since discovered an additional mutation in an uncharacterized gene (T. Conrad, unpublished results). The fitness inequality found between GC and its reconstruction is attributed to the duplicated region in the evolved strain.
TABLE 2.
Test for fitness differences between evolved strains and allele-replacement strains that carry all its identified mutationsa
| Competition | S (102) | Difference in S | SE | P | Outcome |
|---|---|---|---|---|---|
| GD-GA | 3.050 | 0.0042 | 0.0021 | 0.0446 | GD = rGD |
| rGD-GA | 2.625 | ||||
| GE-GD | -7.334 | 0.0048 | 0.0077 | 0.5400 | GE = rGE |
| rGE-GD | -7.128 | ||||
| GC-rGA | -6.702 | 0.0753 | 0.0041 | 7.06E-35 | GC > rGC |
| rGC-rGA | -14.351 | ||||
| GA-GC | 9.135 | -0.0245 | 0.0040 | 1.18E-08 | GA > rGA |
| rGA-GC | 6.702 |
S, selection rate. These experiments were done to verify whether all causative mutations had been found. Allele-replacement “reconstruction” strains are labeled rGX, i.e., for reconstruction of evolved strain GX.
In analyzing the competition outcomes, we discovered that selection rates are not universally transitive within each competition set (i.e., if GX > GY > GZ, the selection rates are transitive when the selection rate of GX versus GZ equals the sum of the selection rates from GX versus GY and from GY versus GZ). We determined that all major discrepancies could be attributed to the selection rates from the GA versus GC and from the wt+kA versus wt+kD competitions. The magnitudes of these selection rates appear to be over- and underestimated, respectively, in the competition assays, based on analyzing the transitivity of the endpoint strain (Fig. 2A) and the glpK allelic replacement strain (Fig. 2c) competition sets. The reason why these selection rate measurements appear to have systematic errors has not been determined. Possible error sources could be interactions between the strains in the form of cross-feeding or minor day-to-day differences in culture conditions such as medium preparation that specifically affect one of the strains. Despite these considerations, we are confident that the indicated direction of the fitness relationships are still valid in both cases since they are still statistically significant (i.e., the selection rates are significantly different from zero) and are supported by the outcomes of the rest of the competitions.
We identified several epistatic interactions between coacquired adaptive mutations. At least one endpoint's glpK and rpoB/C mutations have epistatic interactions, as indicated by Fig. 4A. The origin of this interaction is not yet known, but we have recently observed that some of the glpK-single knockin strains have slightly depressed (∼10 to 15%) growth rates on glucose minimal medium compared to the wild type (data not shown). We now suspect that the GlpK mutations subtly effect the enzyme's interactions with EIIAGlc; previous studies have suggested that interactions between these proteins influence glucose-stimulated inducer exclusion and catabolite repression (2, 23). If this is the case, these effects could mediate the interactions between the effects of the glpK and rpoB/C mutations responsible for the observed epistasis.
In addition, the murE and dapF mutations both synergistically interact with at least one of the mutations previously acquired by their respective endpoint strains (Fig. 4B). Because the DapF and MurE enzymes have no known relationship to glycerol metabolism or RNA polymerase/transcription regulation, it seems likely that the mutations are adaptive due to indirect consequences of the previous mutations on general cellular properties. We suspect the rpoB/C mutations largely facilitate most of the synergistic interactions, since it has been proposed that synergism is an emergent property of complex regulatory networks (24). In addition, while antagonistic interactions are known to be relatively common in organisms with small genomes (3, 21), there has previously been little evidence that synergistic interactions occur frequently enough to effect the evolution of organisms such as E. coli (24).
Measuring the fitness phenotype differences between single mutations may be applicable to the development of adaptation models. Fitness landscapes of individual enzymes have recently been mapped and successfully used to alter enzyme function by predicting the most effective sets of mutations (16, 28). However, these models require prior knowledge of the fitness value of each mutation since their fitness effects are not easily predicted and additionally assume that the effects of multiple mutations combine in an additive, nonepistatic manner (18, 28). Developing predictive models of adaptation on a genome scale promises to be much more complicated, since there are many more components and levels of interaction and, consequently, a non-negligible frequency of epistatic interactions (3). Studying the biochemical basis of epistatic interactions could become a valuable strategy for discovering subtle but important functional interactions between distinct cellular components and systems. Regardless, developing models of specific adaptation processes will likely require accurate fitness effect measurements of many single mutations.
This study delineated the relative fitness among four fully resequenced, parallel-evolved E. coli endpoint strains by head-to-head competitions. Competition sets among endpoint strains and constituent-mutation allele replacement strains were all internally consistent, confirm the distinct fitness phenotype of all of the strains, and indicate the range of fitness gains that can be expected from adaptation to this particular challenge. The relative fitness of the first acquired mutation, to the rpoB/C gene in three strains, largely determines the relative fitness among the endpoints. The adaptive benefits of two mutations, to genes murE and dapF, have little to no benefit in the wild-type background and are epistatically dependent on coacquired mutations, although the nature of their dependence is not yet known. The selection rates during each competition were measured with high sensitivity, suggesting this method has great potential for examining the fitness contributions of specific mutations and identifying epistatic interactions.
Supplementary Material
Acknowledgments
We gratefully acknowledge Trina Patel for her help during the development of this study, Christiane Honisch (Sequenom, Inc., San Diego, CA) for sharing her invaluable technical insights and expertise, Tom Conrad for his assistance growing cultures, and Peter Andolfatto (UCSD Department of Biology) and Chris Herring (Mascoma, Inc., Lebanon, NH) for insightful discussions.
Footnotes
Published ahead of print on 16 May 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Betancourt, A. J., and J. P. Bollback. 2006. Fitness effects of beneficial mutations: the mutational landscape model in experimental evolution. Curr. Opin. Genet. Dev. 16618-623. [DOI] [PubMed] [Google Scholar]
- 2.Chagneau, C., M. Heyde, S. Alonso, R. Portalier, and P. Laloi. 2001. External-pH-dependent expression of the maltose regulon and ompF gene in Escherichia coli is affected by the level of glycerol kinase, encoded by glpK. J. Bacteriol. 1835675-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elena, S. F., and R. E. Lenski. 1997. Test of synergistic interactions among deleterious mutations in bacteria. Nature 390395-398. [DOI] [PubMed] [Google Scholar]
- 4.Fitzmaurice, G. M., N. M. Laird, and J. H. Ware. 2004. Applied longitudinal analysis, vol. John Wiley & Sons, Inc., New York, NY.
- 5.Fong, S. S., A. R. Joyce, and B. O. Palsson. 2005. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome Res. 151365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fong, S. S., J. Y. Marciniak, and B. O. Palsson. 2003. Description and interpretation of adaptive evolution of Escherichia coli K-12 MG1655 by using a genome-scale in silico metabolic model. J. Bacteriol. 1856400-6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegreness, M., N. Shoresh, D. Hartl, and R. Kishony. 2006. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 3111615-1617. [DOI] [PubMed] [Google Scholar]
- 8.Herring, C. D., J. D. Glasner, and F. R. Blattner. 2003. Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene 311153-163. [DOI] [PubMed] [Google Scholar]
- 9.Herring, C. D., A. Raghunathan, C. Honisch, T. Patel, M. K. Applebee, A. R. Joyce, T. J. Albert, F. R. Blattner, D. van den Boom, C. R. Cantor, and B. O. Palsson. 2006. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat. Genet. 381406-1412. [DOI] [PubMed] [Google Scholar]
- 10.Imhof, M., and C. Schlotterer. 2001. Fitness effects of advantageous mutations in evolving Escherichia coli populations. Proc. Natl. Acad. Sci. USA 981113-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joyce, A. R. 2007. Modeling and analysis of the Escherichia coli transcriptional regulatory network: an assessment of its properties, plasticity, and role in adaptive evolution. Ph.D. thesis. University of California-San Diego, La Jolla.
- 12.Kauffman, S. A., and E. D. Weinberger. 1989. The NK model of rugged fitness landscapes and its application to maturation of the immune response. J. Theor. Biol. 141211-245. [DOI] [PubMed] [Google Scholar]
- 13.Knight, C. G., N. Zitzmann, S. Prabhakar, R. Antrobus, R. Dwek, H. Hebestreit, and P. B. Rainey. 2006. Unraveling adaptive evolution: how a single point mutation affects the protein coregulation network. Nat. Genet. 381015-1022. [DOI] [PubMed] [Google Scholar]
- 14.Lenski, R. E., M. R. Rose, S. C. Simpson, and S. C. Tadler. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Naturalist 1381315-1341. [Google Scholar]
- 15.Liu, W. Z., R. Faber, M. Feese, S. J. Remington, and D. W. Pettigrew. 1994. Escherichia coli glycerol kinase: role of a tetramer interface in regulation by fructose 1,6-bisphosphate and phosphotransferase system regulatory protein IIIglc. Biochemistry 3310120-10126. [DOI] [PubMed] [Google Scholar]
- 16.Lunzer, M., S. P. Miller, R. Felsheim, and A. M. Dean. 2005. The biochemical architecture of an ancient adaptive landscape. Science 310499-501. [DOI] [PubMed] [Google Scholar]
- 17.Maharjan, R., S. Seeto, L. Notley-McRobb, and T. Ferenci. 2006. Clonal adaptive radiation in a constant environment. Science 313514-517. [DOI] [PubMed] [Google Scholar]
- 18.Mildvan, A. S. 2004. Inverse thinking about double mutants of enzymes. Biochemistry 4314517-14520. [DOI] [PubMed] [Google Scholar]
- 19.Orr, H. A. 2005. The genetic theory of adaptation: a brief history. Nat. Rev. Genet. 6119-127. [DOI] [PubMed] [Google Scholar]
- 20.Orr, H. A. 2002. The population genetics of adaptation: the adaptation of DNA sequences. Evolution 561317-1330. [DOI] [PubMed] [Google Scholar]
- 21.Pepin, K. M., M. A. Samuel, and H. A. Wichman. 2006. Variable pleiotropic effects from mutations at the same locus hamper prediction of fitness from a fitness component. Genetics 1722047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinheiro, J. C., and D. M. Bates. 2002. Mixed-effects models in S and S-plus, vol. Springer, New York, NY.
- 23.Rohwer, J. M., R. Bader, H. V. Westerhoff, and P. W. Postma. 1998. Limits to inducer exclusion: inhibition of the bacterial phosphotransferase system by glycerol kinase. Mol. Microbiol. 29641-652. [DOI] [PubMed] [Google Scholar]
- 24.Sanjuan, R., and S. F. Elena. 2006. Epistasis correlates to genomic complexity. Proc. Natl. Acad. Sci. USA 10314402-14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wichman, H. A., M. R. Badgett, L. A. Scott, C. M. Boulianne, and J. J. Bull. 1999. Different trajectories of parallel evolution during viral adaptation. Science 285422-424. [DOI] [PubMed] [Google Scholar]
- 26.Woods, R., D. Schneider, C. L. Winkworth, M. A. Riley, and R. E. Lenski. 2006. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc. Natl. Acad. Sci. USA 1039107-9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright, S. 1931. Evolution in Mendelian populations. Genetics 1697-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshikuni, Y., and J. D. Keasling. 2007. Pathway engineering by designed divergent evolution. Curr. Opin. Chem. Biol. 11233-239. [DOI] [PubMed] [Google Scholar]
- 29.Zhong, S., A. Khodursky, D. E. Dykhuizen, and A. M. Dean. 2004. Evolutionary genomics of ecological specialization. Proc. Natl. Acad. Sci. USA 10111719-11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.