Abstract
Human immunodeficiency virus type 1 (HIV-1) entry into the host cell involves a cascade of events and currently represents one of most attractive targets in the search for new antiviral drugs. The fusion-active gp41 core structure is a stable six-helix bundle (6-HB) folded by its trimeric N-terminal heptad repeat (NHR) and C-terminal heptad repeat (CHR). Peptides derived from the CHR region of HIV-1 gp41 are potent fusion inhibitors that target the NHR to block viral and cellular membrane fusion in a dominant negative fashion. However, all CHR peptides reported to date are derived primarily from residues 628 to 673 of gp41; little attention has been paid to the upstream sequence of the pocket binding domain (PBD) in the CHR. Here, we have identified a motif (621QIWNNMT627) located at the upstream region of the gp41 CHR, immediately adjacent to the PBD (628WMEWEREI635). Biophysical characterization demonstrated that this motif is critical for the stabilization of the gp41 6-HB core. The peptide CP621-652, containing the 621QIWNNMT627 motif, was able to interact with T21, a counterpart peptide derived from the NHR, to form a typical 6-HB structure with a high thermostability (thermal unfolding transition [Tm] value of 82°C). In contrast, the 6-HB formed by the peptides N36 and C34, which has been considered to be a core structure of the fusion-active gp41, had a Tm of 64°C. Different from T-20 (brand name Fuseon), which is the first and only HIV-1 fusion inhibitor approved for clinical use, CP621-652 could efficiently block 6-HB formation in a dose-dependent manner. Significantly, CP621-652 had potent inhibitory activity against HIV-1-mediated cell-cell fusion and infection, especially against T-20- and C34-resistant virus. Therefore, our works provide important information for understanding the core structure of the fusion-active gp41 and for designing novel anti-HIV peptides.
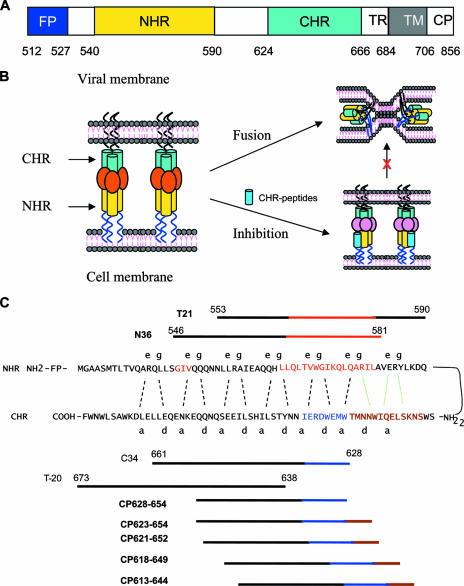
The entry of human immunodeficiency virus type 1 (HIV-1) into target cells is mediated by the attachment of its envelope (Env) glycoprotein to cell surface receptors. The Env glycoprotein, a type I transmembrane protein, is originally synthesized as a single, glycosylated, polyprotein precursor, gp160, which is believed to assemble a trimeric structure in the endoplasmic reticulum and is subsequently cleaved by a cellular protease to yield a surface subunit, gp120, and a transmembrane subunit, gp41 (23, 53). gp120 is responsible for virus binding to its cell receptor, CD4, and a coreceptor (CRR5 or CXCR4). gp41 mediates membrane fusion of the virus with the target cell (45). Like other type I transmembrane proteins, the gp41 molecule consists of extracellular, transmembrane, and cytoplasmic domains (Fig. 1A). Its extracellular domain (ectodomain) contains four major functional regions: a hydrophobic, glycine-rich fusion peptide (FP), an N-terminal heptad repeat (NHR) (or HR1), a C-terminal heptad repeat (CHR) (or HR2), and a tryptophan-rich region. Both the NHR and CHR contain 4-3 repeats of hydrophobic amino acids predicted to form coiled coils, but the exact boundary lines of the NHR and CHR regions could not be determined until 1995, when Lu et al. (36) isolated a stable, proteinase-resistant structure comprising two peptides designated N51 (amino acids [aa] 540 to 590) and C43 (aa 624 to 666) from the NHR and CHR regions by limited proteolysis of recombinant gp41 ectodomains. These two peptides associate to form a highly thermostable, helical, trimeric complex of heterodimers, suggesting that both peptides contain the full length of the 4-3 hydrophobic repeat sequences that can form an independent structural and functional domain with coiled-coil structure, which is relatively resistant to proteolytic enzymes. Therefore, their corresponding regions where N51 and C43 are derived were considered to be the NHR (aa 540 to 590) and CHR (aa 624 to 666) (36). The crystal structure of the complex formed by the NHR peptide containing aa 540 to 590 and the CHR peptide containing aa 624 to 665 was solved (51). Further digestion of the recombinant N51(L6)C43 polypeptide with proteinase K generated a stable subdomain formed by shorter NHR peptide N36 (aa 546 to 581) and CHR peptide C34 (aa 628 to 661) corresponding to the central regions of N51 and C43, respectively, which displays the salient feature of the stable core structure of the isolated gp41 (37). Crystallographic analysis showed that the complex comprising peptides N36 and C34 is a six-helix bundle (6-HB) consisting of three N36 helices forming a central parallel trimer and three C34 helices packing in an antiparallel manner into the hydrophobic grooves on the N trimer, representing the gp41 core domain (4, 5).
FIG. 1.
Structure and function of HIV-1 gp41. (A) Schematic view of the gp41 functional regions. The residue number of each region corresponds to its position in gp160 of HIV-1HXB2. TM, transmembrane domain; TR, tryptophan-rich region. (B) Model of gp41-mediated membrane fusion and inhibition. Upon gp120 binding to CD4 and a coreceptor on the target cell membrane, the FP of gp41 inserts into the target cell membrane, and the CHR and the NHR then form a 6-HB, which brings the viral and cellular membranes into close proximity for fusion. In the fusion-intermediate state, the C peptides (e.g., C34 and T-20) may bind to the viral NHR to block 6-HB formation, thus resulting in the inhibition of membrane fusion in a dominant negative fashion. (C) Interaction of the gp41 NHR and its downstream sequence with the CHR and its upstream sequence or with the C peptides. In the current fusion model, the CHR of gp41 folds back to interact with the NHR to form a hairpin structure. Three molecules of hairpins associate with each other to form a 6-HB. The dashed black lines between the NHR and the CHR indicate the interaction between the residues located at the e and g positions in the NHR and the a and d positions in the CHR, respectively. The interaction of the PBD (628WMEWEREI635) in the CHR (aa 628 to 635) (in blue) with the pocket-forming domain in the NHR (aa 565 to 581) (in red) is important for 6-HB formation. Both the C34 and T-20 peptides contain the sequences targeting the 547GIV549 motif (in brown), which is a major determinant of resistance to T-20 and C34 in the NHR. C34 contains the PBD sequence, while T-20 lacks it. Five designed peptides contain the motif 621QIWNNMT627 (in orange), the PBD, and the partial NHR-binding sequence but lack the GIV-binding sequence. The motif 621QIWNNMT627 may interact with the residues in the pocket-forming domain in the NHR and the downstream motif (AVERY) as shown by the dotted green lines.
Based on the above-described structural information, the current model for HIV-1-mediated membrane fusion was proposed (4, 5, 39). In brief, the sequential binding of gp120 to the primary receptor (CD4) and a coreceptor (CXCR4 or CCR5) triggers a series of conformational rearrangements in gp41, allowing the FP of gp41 to insert into the cell membrane, and the NHR and CHR associate to form a 6-HB core, bringing the viral and cellular membranes into proximity for fusion (Fig. 1B). The peptides derived from the gp41 CHR region, e.g., SJ-2176 (aa 630 to 659) (25), C34 (aa 628 to 661) (37), and T-20 (brand name of Fuzeon) (aa 638 to 673) (52), possess highly potent inhibitory activity against HIV-1 Env-mediated membrane fusion. T-20 is the first peptidic HIV fusion inhibitor approved by the U.S. FDA for clinical use. It has been believed that that these CHR peptides may bind to the viral gp41 NHR region to competitively block 6-HB formation, resulting in the inhibition of HIV entry (5, 34, 45).
Our previous studies indicated that the CHR region contains three functional domains: a pocket-binding domain (PBD) (aa 628 to 635), an NHR-binding domain (aa 628 to 666), and a lipid-binding or tryptophan-rich domain (aa 666 to 673) (7, 32). During the process of 6-HB formation, the PBD in the CHR binds with high affinity to a deep hydrophobic pocket in the NHR trimer, which has been proven to be important for 6-HB formation and has been proposed to be an attractive target for HIV fusion inhibitors (3, 5, 14). Both C34 and T-20 contain the NHR-binding domain, which may interact with the NHR domain, including the 547GIV549 motif, which is a major determinant of resistance to T-20 and C34 in the NHR (50). Although the sequences are largely overlapping, C34 contains the PBD at its N-terminal region, while T-20 lacks those residues; however, T-20 has a lipid-binding domain at its C terminus, which is critical for its antiviral activity (32).
Notably, most anti-HIV C peptides reported to date are derived primarily from the gp41 CHR region, while scant attention has been accorded to the upstream sequence of the CHR. In this study, we have identified a novel motif (621QIWNNMT627) located at the upstream region of the CHR, immediately adjacent to the PBD (628WMEWEREI635). Our data indicate that this motif is critical for the stabilization of the gp41 core structure. Peptides containing this motif, the PBD, and the partial CHR sequence but lacking the 547GIV549 motif-binding sequence could interact with the NHR peptides to form extremely stable 6-HB and exhibit highly potent antiviral activity against HIV-1 strains, especially those strains that are resistant to the CHR peptides, including T-20 and C34. Therefore, this newly identified motif, 621QIWNNMT627, and its binding site can serve as targets for designing novel HIV fusion inhibitors with improved or altered resistant profiles.
MATERIALS AND METHODS
Peptide synthesis.
A set of peptides derived from the NHR (N36 and T21) or the CHR (CP613-644, CP618-649, CP621-652, CP623-654, CP628-654, C34, and T-20) of HIV-1 gp41 (Fig. 1C) were synthesized by a standard solid-phase 9-fluorenylmethoxy carbonyl method using an Applied Biosystems model 433A peptide synthesizer. All peptides were acetylated at the N terminus and amidated at the C terminus. The peptides were purified to homogeneity (>95% purity) by high-performance liquid chromatography (HPLC) and identified by laser desorption mass spectrometry (PerSeptive Biosystems, Framingham, MA). The concentration of peptides was determined by UV absorbance and theoretically calculated molar extinction coefficients (ɛ) (280 nm) of 5,500 mol/liter−1·cm−1 and 1,490 mol/liter−1·cm−1 based on the number of tryptophan (Trp) residues and tyrosine (Tyr) residues (all the peptides tested contain Trp and/or Tyr), respectively.
CD spectroscopy.
Circular dichroism (CD) spectroscopy was performed as previously described (19). Briefly, an N peptide (e.g., T21 or N36) was incubated with a C peptide (e.g., CP621-652 or C34) at 37°C for 30 min (the final concentrations of N peptide and C peptide were 10 μM in 50 mM sodium phosphate and 150 mM NaCl [pH 7.2]). The isolated N and C peptides were also tested. The CD spectra of these peptides and peptide mixtures were acquired on Jasco spectropolarimeter (model J-715) at room temperature using a 5.0-nm bandwidth, a 0.1-nm resolution, a 0.1-cm path length, a 4.0-s response time, and a 50-nm/min scanning speed. The spectra were corrected by the subtraction of a blank corresponding to the solvent. The α-helical content was calculated from the CD signal by dividing the mean residue ellipticity at 222 nm by the value expected for 100% helix formation (−33,000 degrees cm2 dmol−1) as described previously (8, 47). Thermal denaturation was monitored at 222 nm by applying a thermal gradient of 2°C/min in the range of 4 to 98°C. To determine the reversibility, the peptide mixtures were cooled to 4°C and kept in the CD chamber at 4°C for 30 min, followed by the monitoring of thermal denaturation as described above. The reversibility was also measured by performing the reverse thermal melt from 98°C to 4°C. The melting curve was smoothened, and the midpoint of the thermal unfolding transition (Tm) values was calculated using Jasco software utilities as described previously (33). Thermal denaturation was analyzed by fitting the CD data to a simple two-state model (46).
N-PAGE assay.
Native polyacrylamide gel electrophoresis (N-PAGE) was carried out to determine the 6-HB formation between the N and C peptides as described previously (35). Briefly, an N peptide (N36 or T21) was mixed with a C peptide (C34 or CP621-652) at a final concentration of 40 μM and incubated at 37°C for 30 min. The mixture was loaded onto 10- by 1.0-cm precast 18% Tris-glycine gels (Invitrogen, Carlsbad, CA) at 25 μl/per well with an equal volume of Tris-glycine native sample buffer (Invitrogen). Gel electrophoresis was carried out with 125-V constant voltage at room temperature for 2 h. The gel was then stained with Coomassie blue and imaged with a FluorChem 8800 imaging system (Alpha Innotech Corp., San Leandro, CA).
Binding assays by size-exclusion chromatography.
Nonequilibrium binding analyses were performed using size-exclusion chromatography on a TSK-G 3000SWxl HPLC column (Tosoh Corporation, Japan) as described previously (12). An N peptide (T21) was mixed with a designed C peptide (CP621-652) at a molar ratio of 1:1 in 50 mM sodium phosphate-150 mM NaCl (pH 7.2) and incubated at 37°C for 30 min (the final concentration of the peptide was 0.20 mM). Thirty microliters of the mixture or peptide (0.2 mM, in 50 mM sodium phosphate-150 mM NaCl, at pH 7.2) was applied to the TSK-G 3000SWxl HPLC column equilibrated with 50 mM sodium phosphate-150 mM NaCl and eluted at 0.8 ml/min, and fractions were monitored at 214 nm.
Sedimentation equilibrium centrifugation.
Sedimentation equilibrium experiments were performed using an Optima XL-I analytical ultracentrifuge (Beckman Instruments, Palo Alto, CA) equipped with a standard two-channel cell in an An-60 Ti rotor (11). The concentration of the designed peptide was 25 μM in buffer consisting of 50 mM sodium phosphate-100 mM NaCl (pH 7.4), and the complex was composed of 12.5 μM N peptide (T21) and 12.5 μM C peptide (CP621-652). The samples were run at 25,000 or 33,000 rpm at 20°C for 24 h. Absorbance monitoring was performed at 280 nm. The apparent molecular weight (MWapp) was obtained by fitting the data to self-association using the sedimentation analysis software supplied by Beckman. The partial specific volumes used for T21 and CP621-652 were 0.738 and 0.743, respectively, as calculated from the mass average of the partial specific volumes of the individual amino acids.
Inhibition of HIV-1 gp41 6-HB formation by CP621-652.
The inhibitory activities of peptides (CP621-652, C34, and T-20) on 6-HB formation were measured by a modified enzyme-linked immunosorbent assay (ELISA)-based method as previously described (20). Briefly, a 96-well polystyrene plate (Costar; Corning Inc., Corning, NY) was coated with 6-HB-specific monoclonal antibody NC-1 immunoglobulin G (4 μg/ml in 0.1 M Tris, pH 8.8). A tested peptide (CP621-652, C34, or T-20) at graded concentrations was mixed with C34-biotin (0.25 μM) and incubated with N36 (0.25 μM) at room temperature for 30 min. The mixture was then added to the NC-1-coated plate, followed by incubation at room temperature for 30 min and washing with a washing buffer (phosphate-buffered saline [PBS] containing 0.1% Tween 20) three times. Streptavidin-labeled horseradish peroxidase (Invitrogen) and the substrate 3,3′,5,5′-tetramethylbenzidine (Sigma) were then added sequentially. The absorbance at 450 nm (A450) was measured using an ELISA reader (Ultra 384; Tecan, Research Triangle Park, NC). The percent inhibition by the peptides and the 50% inhibitory concentration (IC50) values were calculated as previously described (28).
Cell-cell fusion assay.
A dye transfer assay was used for the detection of HIV-1-mediated cell-cell fusion as previously described (27). Briefly, H9/HIV-1IIIB-infected cells were labeled with a fluorescent reagent, Calcein-AM (Molecular Probes, Inc., Eugene, OR) and then incubated with MT-2 cells (ratio of 1:5) in 96-well plates at 37°C for 2 h in the presence or absence of tested peptides. The fused and unfused calcein-labeled HIV-1-infected cells were counted under an inverted fluorescence microscope (Zeiss, Germany) with an eyepiece micrometer disc. The percent inhibition of cell-cell fusion and the IC50 values were calculated as described above using GraphPad Prism software (GraphPad Software Inc., San Diego, CA).
Inhibition of infection by T-20-resistant virus.
HIV-1 strain NL4-3 and its T-20-resistant mutant bearing V38E/N42S double mutations were obtained through the NIH AIDS reagent program (44). The inhibitory activities of peptides on infection by wild-type and T-20-resistant NL4-3 were determined as previously described (27). In brief, 1 × 104 MT-2 cells were infected with HIV-1 isolates at 100 50% tissue culture infective doses in 200 μl RPMI 1640 medium containing 10% fetal bovine serum in the presence or absence of the peptides at graded concentrations overnight. The culture supernatants were removed, and fresh media were added. On the fourth day postinfection, 100 μl of culture supernatants was collected from each well, mixed with equal volumes of 5% Triton X-100, and assayed for p24 antigen by ELISA.
RESULTS
Identification of a critical motif for the stabilization of the HIV-1 gp41 6-HB structure.
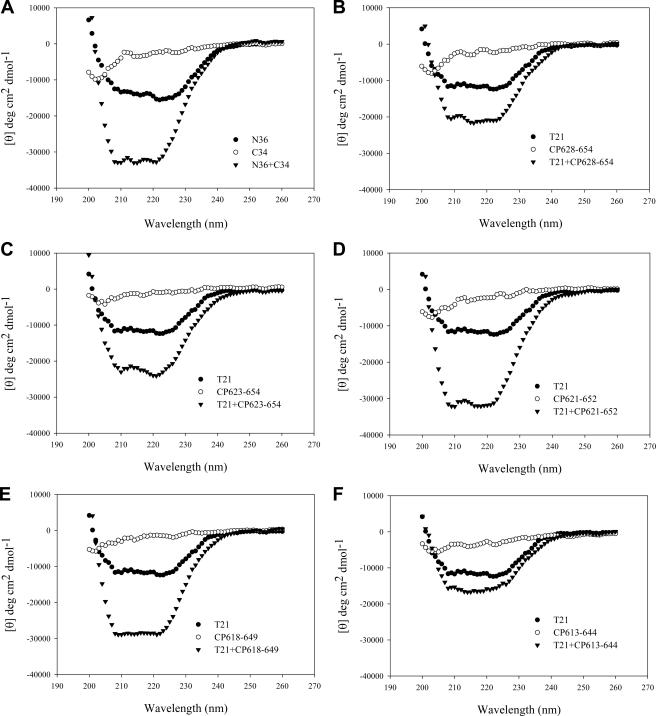
After analyzing the sequences that involved in the interactions between the NHR and the CHR (Fig. 1C), we predicted that the helical motif (WNNMT) preceding the PBD in the CHR may interact with the downstream helical sequence of the pocket in the NHR to form a highly stable fusion-active 6-HB structure. We therefore designed and synthesized a set of peptides to evaluate whether the WNNMT motif affects the α-helicity and stability of the gp41 6-HB structure. Four overlapping peptides (CP623-654, CP621-652, CP618-649, and CP613-644) were designed to contain various numbers of residues preceding the PBD, while a short peptide (CP628-654) was designed to lack these residues as a control. We used CD spectroscopy to analyze the interactions of NHR and CHR peptides. The counterpart peptides N36 and C34 were included in this study since they can form a thermostable 6-HB complex that represents a core structure of the fusion-active gp41 (4). As expected, the CD spectra of the isolated N peptides (N36 and T21) displayed about 40% α-helicity, while all C peptides showed random coil structures (Fig. 2). The equimolar mixture of N36 and C34 is a typical conformation of an α-helix, displaying the characteristic double minima at 208 and 222 nm (Fig. 2A). However, among the newly synthesized C peptides, only CP621-652 could significantly interact with N36 to form an α-helical structure (∼72% helicity); all others (CP613-644, CP618-649, CP623-654, and CP628-654) induced minor or no α-helicity (Table 1), possibly because they may not match well with N36 to bind each other (Fig. 1C). When T21 was used as a counterpart N peptide, all new C peptides could interact with T21 to form α-helical conformations except peptide CP613-644 (Fig. 2B to F). The combination of CP621-652 and T21 was fully helical (100%), while CP628-654, CP623-654, and CP618-649 induced about 64%, 72%, and 87% α-helicities, respectively.
FIG. 2.
The α-helical conformation of the complex formed by N and C peptides analyzed by CD spectroscopy. The final concentration of each peptide in PBS was 10 μM.
TABLE 1.
The α-helicity and thermostability of the complexes formed between NHR and CHR peptides determined by CD spectroscopy
| Peptide(s) | [θ]222 | Helix content (%) | Tm (°C) |
|---|---|---|---|
| N36 | −14,575 | 44 | Undetectable |
| T21 | −12,401 | 38 | Undetectable |
| N36 + CP613-644 | −18,982 | 58 | Undetectable |
| N36 + CP618-649 | −19,581 | 59 | 59 |
| N36 + CP621-652 | −23,782 | 72 | 65 |
| N36 + C34 | −32,550 | 100 | 64 |
| N36 + CP623-654 | −17,309 | 53 | 59 |
| N36 + CP628-654 | −9,642 | 29 | Undetectable |
| T21 + C34 | −28,463 | 86 | 65 |
| T21 + CP613-644 | −14,725 | 45 | Undetectable |
| T21 + CP618-649 | −28,587 | 87 | 78 |
| T21 + CP621-652 | −32,540 | 100 | 82 |
| T21 + CP623-654 | −23,678 | 72 | 76 |
| T21 + CP628-654 | −20,985 | 64 | 57 |
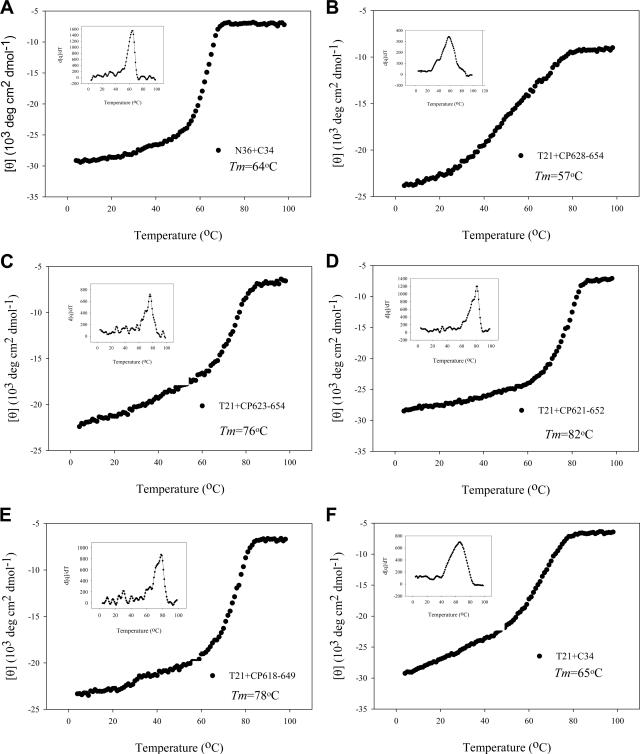
The stability of the peptide complex formed by N and C peptides was measured by thermal denaturation analyses. The signal of the peptide mixture at 222 nm was monitored when the temperature was slowly raised from 4°C to 98°C at a scan rate of 2°C/min. The melting curves for the peptide combination are shown in Fig. 3, and their Tm values were calculated (Table 1). As a control, the complex of N36 and C34 had a Tm of 64°C (Fig. 3A). Interestingly, while the complex of peptides CP628-654 and T21 had a Tm of 57°C (Fig. 3B), the complexes formed by the peptides containing the sequences preceding the PBD were highly stable (Fig. 3C to E). The Tm values for the CP623-654/T21, CP621-652/T21, and CP618-649/T21 complexes were 76°C, 82°C, and 78°C, respectively. Notably, although the C34/T21 complex was also highly helical (86%), it had a Tm (65°C) similar to that of the N36/C34 complex (Fig. 3F). Furthermore, these peptide pairs were unfolding reproducibly and reversibly while the reverse thermal melts were monitored from 98°C to 4°C or heated to 98°C again. These results suggest that the 621QIWNNMT627 motif in the C peptides is a key determinant for the stabilization of the α-helical complexes formed between the NHR and the CHR.
FIG. 3.
Thermostability of the complexes formed by N and C peptides. The unfolding temperature of each complex was scanned at 222 nm by CD spectroscopy, and their Tm values were calculated. Inserted is the first derivative of the curve against temperature, which was used to determine the Tm value.
Characterization of CP621-652 by N-PAGE and sedimentation equilibrium centrifugation.
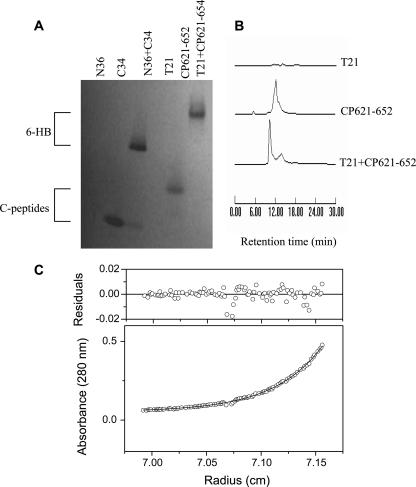
We previously developed an N-PAGE-based method to visualize the 6-HB formed by N36/C34 and used it to characterize the antiviral activities of HIV-1 fusion inhibitors (33, 35). In this study, N-PAGE was used to detect the complex formed by peptide CP621-652 and its counterpart peptide T21. As shown in Fig. 4A, the N peptide N36 shows no band in the native gel because it carries net positive charges and thus could migrate up and off the gel, but peptide C34 shows a specific band, consistent with our previous results (33). When C34 was mixed with N36, the specific bands corresponding to the 6-HB appeared. The migration rates of 6-HBs were dependent on their net charges and molecular sizes as well as the different shapes among the peptide combinations. Similarly, N peptide T21 migrated off the gel, whereas the peptide CP621-652 alone and its mixture with T21 gave a specific band, further confirming that CP621-652 and T21 formed a high order of complex. Their specific binding was further confirmed by size-exclusion HPLC (Fig. 4B).
FIG. 4.
Determination of 6-HBs formed by N and C peptides. (A) N-PAGE. (B) Size-exclusion HPLC analysis. (C) Molecular mass of the T21/CP621-652 complex determined by sedimentation equilibrium ultracentrifugation at concentrations of 25 μM in PBS buffer (pH 7.0) at a rotor speed of 33,000 rpm. The observed molecular mass is 24,250 Da (the calculated mass for a trimer is 25,530 Da).
The MWapp values for peptide CP621-652 and its complex with T21 were determined by sedimentation equilibrium ultracentrifugation at 20°C. The result showed that the MWapp of CP621-652 alone was 3,720, suggesting that it is a monomer in the solution. The MWapp of the mixture of CP621-652 and T21 was 24,250 (Fig. 4C). Compared to the expected molecular mass of 8,510 Da for the CP621-652/T21 heterodimer, we conclude that the CP621-652 and T21 peptides associate to form a 6-HB structure consisting of three molecules each of CP621-652 and T21.
Unlike T20, CP621-652 efficiently inhibited 6-HB formation as modeled by N36/C34.
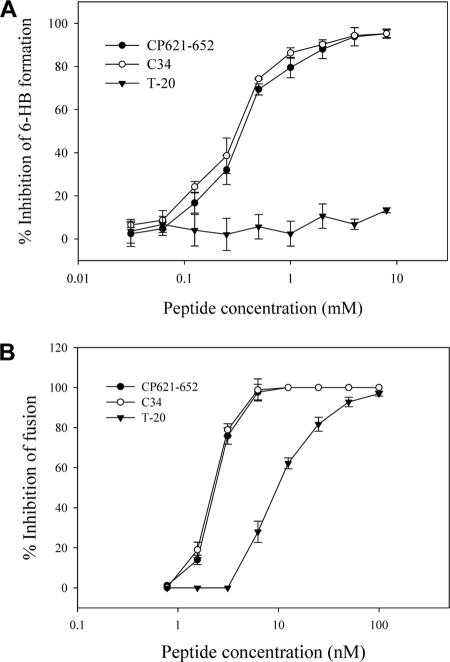
A model system of the gp41 6-HB was established by mixing the N and C peptides (N36 and C34) at equal molar concentrations (4). This model gp41 core structure can be detected by sandwich ELISA using a conformation-specific monoclonal antibody, NC-1 (24, 26). Using this system, we have tested whether peptide CP621-652 inhibits 6-HB formation, a proposed mechanism for anti-HIV peptides. As shown in Fig. 5A, CP621-652 could efficiently inhibit the 6-HB formation between N36 and C34 in a dose-dependent manner, comparable to that of C34 itself. However, T-20 had no this effect at a concentration as high as 8,000 nM, consistent with our previous data (33). This result suggests that CP621-652, different from T-20, can block 6-HB formation between the NHR and the CHR in a dominant negative fashion.
FIG. 5.
Inhibition of 6-HB formation and membrane fusion by CP621-652. (A) Unlike T-20, both CP621-652 and C34 can inhibit the 6-HB formation modeled by N36/C34 peptides in a dose-dependent manner. (B) CP621-652 can efficiently inhibit HIV-1IIIB-mediated cell-cell fusion.
Peptides containing the WNNMT motif exhibited potent inhibitory activities on HIV-1-mediated cell-cell fusion.
It is interesting to know whether the WNNMT motif-containing peptides have anti-HIV activity. First, their inhibitory activities on HIV-1-mediated cell-cell fusion were determined. Although the short peptide CP628-654 contained the most sequence of C34, it had no antiviral activity at a concentration of as high as 4,000 nM (Table 2). When five residues (WNNMT) were added to the N terminus of CP628-654, the resulting peptide, CP623-654, could efficiently inhibit HIV-1-mediated cell-cell fusion with an IC50 of 70.22 nM. Impressively, while CP623-654 shifted two residues upstream, peptide CP621-652 increased 17-fold in its anti-HIV potency (IC50 = 4.23 nM). However, a further shift upstream with three (CP618-649) or more (CP613-644) residues resulted in peptides with greatly decreased activity. As a control, C34 and T-20 could inhibit HIV-1-mediated cell-cell fusion with IC50 values of 3.65 nM and 26.43 nM, respectively. The representative data from the viral fusion inhibition experiments are shown in Fig. 5B. These results suggest that the heptad motif 621QIWNNMT627 is also a critical motif for anti-HIV peptides.
TABLE 2.
Inhibitory activities of CHR-derived peptides on HIV-1-mediated cell-cell fusion
| Peptide | Sequencea | Mean IC50 (nM) ± SD |
|---|---|---|
| CP613-644 | SWSNKSLEQIWNNMTWMEWDREINNYTSLIHS | 1,962.11 ± 29.72 |
| CP618-649 | SLEQIWNNMTWMEWDREINNYTSLIHSLIEES | 86.94 ± 1.63 |
| CP621-652 | QIWNNMTWMEWDREINNYTSLIHSLIEESQNQ | 4.23 ± 0.35 |
| CP623-654 | WNNMTWMEWDREINNYTSLIHSLIEESQNQQE | 70.22 ± 1.18 |
| CP628-654 | WMEWDREINNYTSLIHSLIEESQNQQE | >4,000.00 |
| C34 | WMEWDREINNYTSLIHSLIEESQNQQEKNEQELL | 3.65 ± 0.39 |
| T20 | YTSLIHSLIEESQNQQEKNEQELLELDKWASLWNWF | 26.43 ± 0.94 |
The residue numbers of each region corresponds to their positions in gp160 of HIV-1HXB2. The residues forming the PBD are in boldface type, and the residues preceding the PBD are in italic type.
Potent inhibition of CP621-652 against T-20- and C34-resistant virus.
Previous studies indicated that double mutations (V38E and N42S) in the NHR of HIV-1 strain NL4-3 confer genetic resistance to T-20 (44). Here, we sought to determine whether CP621-652 is able to inhibit the replication of HIV-1 strains that are resistant to T-20. As expected, T-20 could effectively inhibit the parental HIV-1 NL4-3 but failed to block infection by NL4-3 bearing V38E andN42S mutations at a concentration of as high as 2,000 nM (Table 3). This NL4-3 mutant was also resistant to C34 (22.97-fold change). However, CP621-652 was similarly effective against both wild-type (IC50 = 4.55 nM) and resistant (IC50 = 4.22 nM) viruses. As a control, peptide CP628-654, which lacks the 621QIWNNMT627 motif, had no inhibitory activity on these viruses. These results suggest that the peptide containing 621QIWNNMT627 is highly effective against HIV-1 strains, especially those that are resistant to T-20 and C34, therefore having a great potential to be developed as a novel peptidic anti-HIV drug.
TABLE 3.
Potent inhibition of CP621-652 against T-20- and C34-resistant virus
| Peptide | Mean IC50 (nM) ± SD
|
Fold change | |
|---|---|---|---|
| NL4-3 WT | NL 4-3 V38E + N42S | ||
| T-20 | 33.86 ± 1.12 | >2,000.00 | >59.17 |
| C34 | 3.60 ± 0.38 | 82.68 ± 2.21 | 22.97 |
| CP621-652 | 4.55 ± 0.33 | 4.22 ± 0.27 | 0.93 |
| CP628-654 | >2,000.00 | >2,000.00 | |
DISCUSSION
Structurally, the coiled-coil domain of HIV-1 fusion protein gp41 shares a characteristic 4-3 heptad repeat sequence, (abcdefg)n (4, 51). During viral fusion, the trimeric NHR and CHR fold into the fusion-active 6-HB structure, in which the N-terminal homotrimers are packed against each other through the interaction of residues at the a and d positions, and its residues at the e and g positions lie on the outside of the central coiled coil and create extensive interactions with the residues at the a and d positions of the C helices (Fig. 1). Previous studies indicated that the helical packing interactions in the HIV-1 gp41 6-HB core play an important role in viral infectivity (17, 31, 38). In particular, the hydrophobic interactions between the deep pocket in the C terminus of the NHR groove and three residues from the PBD of the CHR can determine the stability of the 6-HB (3, 5-7). In this study, we identified a motif, 621QIWNNMT627, located in the upstream region of the CHR, immediately adjacent to the PBD. This motif might be a key determinant for the stabilization of the gp41 6-HB core formed by the NHR and the CHR during the virus fusion process. Synthetic peptides containing this motif, the PDB, and NHR-binding sequence, e.g., CP621-652, could interact with the counterpart NHR peptide T21 to form a fully α-helical 6-HB with a higher stability (Tm = 82°C) than those formed between T21 and the overlapping peptides lacking the 621QIWNNMT627 motif, e.g., CP628-654 (Tm = 57°C) and C34 (Tm = 64°C) (Fig. 4 and Table 1). These results suggest that the peptide containing the 621QIWNNMT627 motif can bind with the NHR target to form a highly stable 6-HB core.
It was predicted previously that the 623WNNMT627 motif displays the α-helical conformation in the steric structure of the ectodomain of HIV-1 gp41 (1, 54). It is possible that these residues may interact with the downstream residues of the NHR pocket to stabilize the interaction of the PBD in the C helix with the pocket in N-trimeric helices (1, 2). The NHR peptide T21 contains the pocket downstream sequence 581AVERYLKDQ590, which has also been predicted to be in an α-helical conformation (1). The interaction between the PBD upstream and the pocket downstream might provide a high binding energy of peptides CP621-652 and T21. One can speculate that the interactions of these motifs may be together with the hydrophobic interactions within the pocket for CHR and NHR pairing during the membrane fusion process. Furthermore, the 623WNNMT627 motif has recently been identified as being a caveolin-1 binding site and has the ability to induce neutralizing antibodies (22). It is interesting to define whether these features are associated with the ability of CP621-652 to interact with the NHR peptide with a high affinity. Thus, it is necessary to further characterize the interacting interface of the NHR and the CHR. A crystallographic study on the 6-HB structure formed by CP621-652 and T21 is under the way, and hopefully, this study will provide more detailed information about their interaction. In addition, mutational analyses of the 621QIWNNMT627 motif may also reveal the residues responsible for the high-affinity interactions between these NHR and CHR peptides.
Application of the highly active antiretroviral therapy regimen, or “drug cocktail,” which is a combination of several antiretroviral drugs such as reverse transcriptase inhibitors and protease inhibitors, can dramatically reduced the morbidity and mortality of AIDS and increased the life expectancy of HIV-infected patients (41, 55). However, the need for new classes of anti-HIV drugs has become apparent because of increasing concern about the long-term toxic effects of existing drugs and the emergence of HIV-1 variants that are resistant to treatment (21, 43). HIV-1 entry inhibitors that target the Env of HIV-1 or its receptors have been considered to be the most promising treatment options (16, 39, 40). Enfuvirtide (T-20), which targets the gp41 NHR and blocks the fusion of viral and cellular membranes, is the first HIV-1 entry inhibitor approved for the treatment of HIV-1 in treatment-experienced patients (29, 30). Although T-20 has a pivotal role in optimizing antiretroviral drug combinations, it also has the problem of inducing drug resistance rapidly (9, 42). Therefore, it is essential to identify new targets in Env for designing novel HIV fusion inhibitors with improved or altered drug resistance profiles (13).
It was reported previously that the peptides containing the motifs that stabilize the helical structure have increased antiviral activity (3, 48), although more recent results have shown that this relationship may be complex (18, 49). Peptide CP621-652, containing the 621QIWNNMT627 motif, could effectively inhibit HIV-1-mediated cell-cell fusion (Table 2). In contrast, peptide CP628-654, lacking the 621QIWNNMT627 motif, had no or weak antiviral activity. Impressively, CP621-652 was also highly effective against both T-20- and C34-resistant HIV-1 strains (Tables 3). Previous studies suggested that both T-20 and C34 can target the “GIV” motif in the NHR and that the mutations in this site can confer resistance (44). However, CP621-652 targets primarily the site around the pocket region, and the “GIV” motif is not involved in its target, which may explain why this peptide can retain its potent inhibitory activity against T-20- and C34-resistant HIV-1 strains bearing V38E and N42S double mutations. Furthermore, our data presented here suggest that the sequence 581AVERYLKDQ590, located at the NHR pocket downstream, may serve as a target by the 621QIWNNMT627 motif. Interestingly, the sequence 581AVERYLKDQ590 also serves as a critical portion for the most immunodominant B-cell epitope and is highly conserved among all HIV-1 isolates (10). This feature implies that the 621QIWNNMT627 motif-containing peptides may have broad anti-HIV activity.
The antiviral activities of peptides are presumed to be related to the stability of the NHR/CHR bundle (3, 48). However, several recent studies demonstrated that some C peptides may have high Tm values with their NHR partners but have reduced antiviral activity (15, 49). We have recently shown that the engineered peptide C34 can form a 6-HB with N36 with much higher thermostability but that its anti-HIV activity has not been increased (20). In this study, we show that peptides CP613-644 and CP618-649, which also contain the 621QIWNNMT627 motif, have higher Tm values than C34 but that they have less anti-HIV activity. Notably, peptide C34 had an inhibitory activity similar to that of peptide CP621-652 against wild-type HIV-1-mediated fusion and infection, but it interacted with the N peptides (N36 and T21) with much lower Tm values (64°C and 65°C, respectively) than CP621-652. In addition, the anti-HIV activity of C34 could be abolished or severely impaired by the deletion of its C-terminal heptad residues (655KNEQEL661), as shown by peptide CP628-654, suggesting that the C-terminal residues of C34 play a critical role in its inhibitory activity. It is believed that the integrity of the NHR binding sequence is important for designing anti-HIV peptides rather than that the length of the peptides can simply cause the reduction of the inhibitory activity. As a control to this point, peptide CP613-644 is composed of 32 aa but is practically not active. Therefore, further studies need to be done to elucidate the relationship between antiviral activity and 6-HB stability.
In conclusion, the present studies not only provide important information for understanding the interhelical interactions in the fusion-active gp41 core structure but also suggest that peptide CP621-654 has potential to be further developed as a novel anti-HIV fusion inhibitor that can be used for the treatment of infection by HIV-1 strains that are resistant to anti-HIV peptides currently used in clinics or in preclinical studies (e.g., T-20, T-1249, and C34).
Acknowledgments
We thank Veronica Kuhlemann for her critical reading of the manuscript.
This work was supported by the 973 program (grant 2006CB504200) and the 863 program (grant 2006A09Z404) from the Chinese Ministry of Science and Technology.
Footnotes
Published ahead of print on 16 April 2008.
REFERENCES
- 1.Caffrey, M., M. Cai, J. Kaufman, S. J. Stahl, P. T. Wingfield, D. G. Covell, A. M. Gronenborn, and G. M. Clore. 1998. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 174572-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caffrey, M., M. Cai, J. Kaufman, S. J. Stahl, P. T. Wingfield, A. M. Gronenborn, and G. M. Clore. 1997. Determination of the secondary structure and global topology of the 44 kDa ectodomain of gp41 of the simian immunodeficiency virus by multidimensional nuclear magnetic resonance spectroscopy. J. Mol. Biol. 271819-826. [DOI] [PubMed] [Google Scholar]
- 3.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 9515613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89263-273. [DOI] [PubMed] [Google Scholar]
- 5.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93681-684. [DOI] [PubMed] [Google Scholar]
- 6.Chang, D. K., S. F. Cheng, and V. D. Trivedi. 1999. Biophysical characterization of the structure of the amino-terminal region of gp41 of HIV-1. Implications on viral fusion mechanism. J. Biol. Chem. 2745299-5309. [DOI] [PubMed] [Google Scholar]
- 7.Chang, D. K., and C. S. Hsu. 2007. Biophysical evidence of two docking sites of the carboxyl heptad repeat region within the amino heptad repeat region of gp41 of human immunodeficiency virus type 1. Antivir. Res. 7451-58. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Y. H., J. T. Yang, and K. H. Chau. 1974. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry 133350-3359. [DOI] [PubMed] [Google Scholar]
- 9.Chinnadurai, R., D. Rajan, J. Munch, and F. Kirchhoff. 2007. Human immunodeficiency virus type 1 variants resistant to first- and second-version fusion inhibitors and cytopathic in ex vivo human lymphoid tissue. J. Virol. 816563-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cumming, S. A., D. A. McPhee, W. J. Maskill, B. E. Kemp, R. R. Doherty, and I. D. Gust. 1990. Use of a conserved immunodominant epitope of HIV surface glycoprotein gp41 in the detection of early antibodies. AIDS 483-86. [DOI] [PubMed] [Google Scholar]
- 11.Dai, Q., F. J. Castellino, and M. Prorok. 2004. A single amino acid replacement results in the Ca2+-induced self-assembly of a helical conantokin-based peptide. Biochemistry 4313225-13232. [DOI] [PubMed] [Google Scholar]
- 12.Dai, Q., J. Zajicek, F. J. Castellino, and M. Prorok. 2003. Binding and orientation of conantokins in PL vesicles and aligned PL multilayers. Biochemistry 4212511-12521. [DOI] [PubMed] [Google Scholar]
- 13.Doms, R. W., and J. P. Moore. 2000. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151F9-F14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dwyer, J. J., A. Hasan, K. L. Wilson, J. M. White, T. J. Matthews, and M. K. Delmedico. 2003. The hydrophobic pocket contributes to the structural stability of the N-terminal coiled coil of HIV gp41 but is not required for six-helix bundle formation. Biochemistry 424945-4953. [DOI] [PubMed] [Google Scholar]
- 15.Dwyer, J. J., K. L. Wilson, D. K. Davison, S. A. Freel, J. E. Seedorff, S. A. Wring, N. A. Tvermoes, T. J. Matthews, M. L. Greenberg, and M. K. Delmedico. 2007. Design of helical, oligomeric HIV-1 fusion inhibitor peptides with potent activity against enfuvirtide-resistant virus. Proc. Natl. Acad. Sci. USA 10412772-12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Este, J. A., and A. Telenti. 2007. HIV entry inhibitors. Lancet 37081-88. [DOI] [PubMed] [Google Scholar]
- 17.Follis, K. E., S. J. Larson, M. Lu, and J. H. Nunberg. 2002. Genetic evidence that interhelical packing interactions in the gp41 core are critical for transition of the human immunodeficiency virus type 1 envelope glycoprotein to the fusion-active state. J. Virol. 767356-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallo, S. A., K. Sackett, S. S. Rawat, Y. Shai, and R. Blumenthal. 2004. The stability of the intact envelope glycoproteins is a major determinant of sensitivity of HIV/SIV to peptidic fusion inhibitors. J. Mol. Biol. 3409-14. [DOI] [PubMed] [Google Scholar]
- 19.He, Y., S. Liu, W. Jing, H. Lu, D. Cai, D. J. Chin, A. K. Debnath, F. Kirchhoff, and S. Jiang. 2007. Conserved residue Lys574 in the cavity of HIV-1 Gp41 coiled-coil domain is critical for six-helix bundle stability and virus entry. J. Biol. Chem. 28225631-25639. [DOI] [PubMed] [Google Scholar]
- 20.He, Y., Y. Xiao, H. Song, Q. Liang, D. Ju, X. Chen, H. Lu, W. Jing, S. Jiang, and L. Zhang. 2008. Design and evaluation of sifuvirtide: a novel HIV-1 fusion inhibitor. J. Biol. Chem. 28311126-11134. [DOI] [PubMed] [Google Scholar]
- 21.Hogg, R. S., D. R. Bangsberg, V. D. Lima, C. Alexander, S. Bonner, B. Yip, E. Wood, W. W. Dong, J. S. Montaner, and P. R. Harrigan. 2006. Emergence of drug resistance is associated with an increased risk of death among patients first starting HAART. PLoS Med. 3e356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hovanessian, A. G., J. P. Briand, E. A. Said, J. Svab, S. Ferris, H. Dali, S. Muller, C. Desgranges, and B. Krust. 2004. The caveolin-1 binding domain of HIV-1 glycoprotein gp41 is an efficient B cell epitope vaccine candidate against virus infection. Immunity 21617-627. [DOI] [PubMed] [Google Scholar]
- 23.Hunter, E., and R. Swanstrom. 1990. Retrovirus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 157187-253. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, S., K. Lin, and M. Lu. 1998. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 7210213-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365113. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, S., K. Lin, L. Zhang, and A. K. Debnath. 1999. A screening assay for antiviral compounds targeted to the HIV-1 gp41 core structure using a conformation-specific monoclonal antibody. J. Virol. Methods 8085-96. [DOI] [PubMed] [Google Scholar]
- 27.Jiang, S., H. Lu, S. Liu, Q. Zhao, Y. He, and A. K. Debnath. 2004. N-substituted pyrrole derivatives as novel human immunodeficiency virus type 1 entry inhibitors that interfere with the gp41 six-helix bundle formation and block virus fusion. Antimicrob. Agents Chemother. 484349-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, S. B., K. Lin, and A. R. Neurath. 1991. Enhancement of human immunodeficiency virus type 1 infection by antisera to peptides from the envelope glycoproteins gp120/gp41. J. Exp. Med. 1741557-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lalezari, J. P., J. J. Eron, M. Carlson, C. Cohen, E. DeJesus, R. C. Arduino, J. E. Gallant, P. Volberding, R. L. Murphy, F. Valentine, E. L. Nelson, P. R. Sista, A. Dusek, and J. M. Kilby. 2003. A phase II clinical study of the long-term safety and antiviral activity of enfuvirtide-based antiretroviral therapy. AIDS 17691-698. [DOI] [PubMed] [Google Scholar]
- 30.Lalezari, J. P., K. Henry, M. O'Hearn, J. S. Montaner, P. J. Piliero, B. Trottier, S. Walmsley, C. Cohen, D. R. Kuritzkes, J. J. Eron, Jr., J. Chung, R. DeMasi, L. Donatacci, C. Drobnes, J. Delehanty, and M. Salgo. 2003. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N. Engl. J. Med. 3482175-2185. [DOI] [PubMed] [Google Scholar]
- 31.Liu, J., S. Wang, J. A. Hoxie, C. C. LaBranche, and M. Lu. 2002. Mutations that destabilize the gp41 core are determinants for stabilizing the simian immunodeficiency virus-CPmac envelope glycoprotein complex. J. Biol. Chem. 27712891-12900. [DOI] [PubMed] [Google Scholar]
- 32.Liu, S., W. Jing, B. Cheung, H. Lu, J. Sun, X. Yan, J. Niu, J. Farmar, S. Wu, and S. Jiang. 2007. HIV gp41 C-terminal heptad repeat contains multifunctional domains. Relation to mechanisms of action of anti-HIV peptides. J. Biol. Chem. 2829612-9620. [DOI] [PubMed] [Google Scholar]
- 33.Liu, S., H. Lu, J. Niu, Y. Xu, S. Wu, and S. Jiang. 2005. Different from the HIV fusion inhibitor C34, the anti-HIV drug Fuzeon (T-20) inhibits HIV-1 entry by targeting multiple sites in gp41 and gp120. J. Biol. Chem. 28011259-11273. [DOI] [PubMed] [Google Scholar]
- 34.Liu, S., S. Wu, and S. Jiang. 2007. HIV entry inhibitors targeting gp41: from polypeptides to small-molecule compounds. Curr. Pharm. Des. 13143-162. [DOI] [PubMed] [Google Scholar]
- 35.Liu, S., Q. Zhao, and S. Jiang. 2003. Determination of the HIV-1 gp41 fusogenic core conformation modeled by synthetic peptides: applicable for identification of HIV-1 fusion inhibitors. Peptides 241303-1313. [DOI] [PubMed] [Google Scholar]
- 36.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 21075-1082. [DOI] [PubMed] [Google Scholar]
- 37.Lu, M., and P. S. Kim. 1997. A trimeric structural subdomain of the HIV-1 transmembrane glycoprotein. J. Biomol. Struct. Dyn. 15465-471. [DOI] [PubMed] [Google Scholar]
- 38.Lu, M., M. O. Stoller, S. Wang, J. Liu, M. B. Fagan, and J. H. Nunberg. 2001. Structural and functional analysis of interhelical interactions in the human immunodeficiency virus type 1 gp41 envelope glycoprotein by alanine-scanning mutagenesis. J. Virol. 7511146-11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore, J. P., and R. W. Doms. 2003. The entry of entry inhibitors: a fusion of science and medicine. Proc. Natl. Acad. Sci. USA 10010598-10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore, J. P., and M. Stevenson. 2000. New targets for inhibitors of HIV-1 replication. Nat. Rev. Mol. Cell Biol. 140-49. [DOI] [PubMed] [Google Scholar]
- 41.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, S. D. Holmberg, et al. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338853-860. [DOI] [PubMed] [Google Scholar]
- 42.Peuchant, O., S. Capdepont, J. M. Ragnaud, V. Aurillac-Lavignolle, R. Thiebaut, H. Fleury, and B. Masquelier. 2007. Primary resistance to enfuvirtide (T20) in recently HIV-1 infected, antiretroviral-naive patients from the ANRS Aquitaine Cohort. Antivir. Ther. 12559-562. [DOI] [PubMed] [Google Scholar]
- 43.Pillay, D., S. Taylor, and D. D. Richman. 2000. Incidence and impact of resistance against approved antiretroviral drugs. Rev. Med. Virol. 10231-253. [DOI] [PubMed] [Google Scholar]
- 44.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roux, K. H., and K. A. Taylor. 2007. AIDS virus envelope spike structure. Curr. Opin. Struct. Biol. 17244-252. [DOI] [PubMed] [Google Scholar]
- 46.Santoro, M. M., and D. W. Bolen. 1988. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry 278063-8068. [DOI] [PubMed] [Google Scholar]
- 47.Shu, W., J. Liu, H. Ji, L. Radigen, S. Jiang, and M. Lu. 2000. Helical interactions in the HIV-1 gp41 core reveal structural basis for the inhibitory activity of gp41 peptides. Biochemistry 391634-1642. [DOI] [PubMed] [Google Scholar]
- 48.Sia, S. K., P. A. Carr, A. G. Cochran, V. N. Malashkevich, and P. S. Kim. 2002. Short constrained peptides that inhibit HIV-1 entry. Proc. Natl. Acad. Sci. USA 9914664-14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steger, H. K., and M. J. Root. 2006. Kinetic dependence to HIV-1 entry inhibition. J. Biol. Chem. 28125813-25821. [DOI] [PubMed] [Google Scholar]
- 50.Trivedi, V. D., S. F. Cheng, C. W. Wu, R. Karthikeyan, C. J. Chen, and D. K. Chang. 2003. The LLSGIV stretch of the N-terminal region of HIV-1 gp41 is critical for binding to a model peptide, T20. Protein Eng. 16311-317. [DOI] [PubMed] [Google Scholar]
- 51.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387426-430. [DOI] [PubMed] [Google Scholar]
- 52.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA 8910537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 2801884-1888. [DOI] [PubMed] [Google Scholar]
- 54.Yang, Z. N., T. C. Mueser, J. Kaufman, S. J. Stahl, P. T. Wingfield, and C. C. Hyde. 1999. The crystal structure of the SIV gp41 ectodomain at 1.47 Å resolution. J. Struct. Biol. 126131-144. [DOI] [PubMed] [Google Scholar]
- 55.Yeni, P. 2006. Update on HAART in HIV. J. Hepatol. 44S100-S103. [DOI] [PubMed] [Google Scholar]