Abstract
Studies on the herpes simplex virus type 1 UL25-null mutant KUL25NS have shown that the capsid-associated UL25 protein is required at a late stage in the encapsidation of viral DNA. Our previous work on UL25 with the UL25 temperature-sensitive (ts) mutant ts1204 also implicated UL25 in a role at very early times in the virus growth cycle, possibly at the stage of penetration of the host cell. We have reexamined this mutant and discovered that it had an additional ts mutation elsewhere in the genome. The ts1204 UL25 mutation was transferred into wild-type (wt) virus DNA, and the UL25 mutant ts1249 was isolated and characterized to clarify the function of UL25 at the initial stages of virus infection. Indirect immunofluorescence assays and in situ hybridization analysis of virus-infected cells revealed that the mutant ts1249 was not impaired in penetration of the host cell but had an uncoating defect at the nonpermissive temperature. When ts1249-infected cells were incubated initially at the permissive temperature to allow uncoating of the viral genome and subsequently transferred to the restrictive temperature, a DNA-packaging defect was evident. The results suggested that ts1249, like KUL25NS, had a block at a late stage of DNA packaging and that the packaged genome was shorter than the full-length genome. Examination of ts1249 capsids produced at the nonpermissive temperature revealed that, in comparison with wt capsids, they contained reduced amounts of UL25 protein, thereby providing a possible explanation for the failure of ts1249 to package full-length viral DNA.
All herpesviruses have a characteristic morphology and a common assembly pathway. The viral DNA is enclosed in an icosahedral capsid, which in turn is surrounded by another protein layer, the tegument, most of which lacks icosahedral symmetry. The outer layer of the virion consists of a lipid membrane containing the viral glycoproteins that are required for fusion of the envelope with the host cell membrane. Studies with pseudorabies virus (PRV) have shown that upon entry of the virus into a permissive cell most of the tegument layer rapidly dissociates from the capsid, but at least two tegument proteins, UL36 and UL37, and the capsid-associated DNA-packaging protein UL25 remain attached to the capsid (18, 22, 28). Recent results obtained using herpes simplex virus type 1 (HSV-1) have revealed that proteasomal degradation of virion and/or host proteins in the cytoplasm is an essential step in the uncoating of the viral genome (11), and evidence suggesting that proteolytic cleavage of UL36 is important for DNA release has been obtained (21). After the capsid is transported along the microtubule to the microtubule-organizing center, it migrates by an unknown mechanism to the edge of the cell nucleus, where it interacts with the nuclear pore, releasing the viral genome into the nucleus by a process dependent on importin-β and Ran-GTP (3, 29, 38, 51).
DNA replication, capsid assembly, and DNA packaging all take place in the nucleus, initially within discrete regions referred to as replication compartments (10, 46). Cleavage of the replicated concatemeric viral DNA into monomeric units is tightly coupled to encapsidation of the DNA, and in the absence of capsid assembly, no viral DNA cleavage is observed (12). The replicated viral DNA is inserted into the cavity of the preformed spherical-shaped icosahedral procapsid, where it forms a highly compact, ordered structure (4). During DNA packaging, the scaffolding proteins inside the procapsid are cleaved by the viral serine protease and removed, and the capsid shell becomes a more stable, angularized structure (14, 44, 47, 58). In addition to DNA-containing, angularized capsids (C capsids), two other angularized capsid forms are observed in the nucleus, both of which are considered dead-end products. These are A capsids, which lack viral DNA and scaffolding proteins, and B capsids, which are devoid of viral DNA but contain the cleaved scaffolding proteins. The C capsid acquires an envelope by budding into the inner leaflet of the nuclear membrane (50). This layer is lost when the enveloped particle fuses with the outer nuclear membrane and the capsid is released into the cytoplasm, where it gains the tegument and subsequently an outer envelope (49; reviewed in references 33 and 52).
Of the seven HSV-1 DNA-packaging proteins, only UL6, UL17, and UL25 are found in significant amounts in the mature DNA-containing capsid (17, 32, 39, 48, 57). UL6 is present at the unique vertex on the capsid, forming the portal through which the DNA enters the capsid, whereas UL25 and UL17 are located at multiple sites on the capsid (6, 7, 36, 37, 56). Unlike UL17, UL25 is found in much smaller amounts in procapsids than in angularized B capsids, and therefore, it is likely that UL25 is added after procapsid formation (48). This conclusion is supported by the observation that UL25 binds in vitro to A or B capsids lacking UL25 (35). In the absence of UL17, small amounts of UL25 are present on B capsids in vivo, indicating that UL17 is required for the attachment of UL25 to capsids (56). Wild-type (wt) HSV-1 C capsids contain greater amounts of both UL17 and UL25 than B capsids, in keeping with the idea that UL25 and some of UL17 attach to capsids at a late stage in capsid maturation and that the presence of viral DNA inside the capsid exposes more binding sites on the capsid surface (57, 59). By use of cryoelectron microscopy and image reconstruction, extra mass on the surface of the capsid, adjacent to the pentons at the vertices, was identified on wt C capsids but not on wt and UL25-null mutant A capsids. It was proposed that this C-capsid-specific component was composed of a heterodimer of UL25 and UL17 (59).
In contrast to the other DNA-packaging proteins, UL25 is not absolutely essential for the cleavage of concatemeric viral DNA and initiation of DNA packaging but appears to be important at a later stage of HSV-1 genome encapsidation (32, 53). Subsequent studies of PRV and bovine herpesvirus 1 UL25 deletion mutants are in agreement with this conclusion (13, 24). Interestingly, the defect of the PRV UL25-null mutant can be partially reversed by growing the mutant on a cell line expressing the wt HSV-1 protein (25).
Since UL25 is present at multiple sites on the exterior surface of the capsid, UL25 may be required for stabilizing the capsid shell and possibly the packaging complex at the portal during and after DNA packaging. The findings that the UL25-null mutant KUL25NS stably packages only small amounts of viral DNA in nonpermissive cells and that most of the viral DNA encapsidated is of less than genome length are consistent with this proposal (53).
The first evidence that the UL25 gene product was important for DNA packaging was obtained from the analysis of two HSV-1 mutants, ts1204 and ts1208, that had temperature-sensitive (ts) mutations in the UL25 gene (1). ts1204 had two phenotypic defects at the nonpermissive temperature (NPT), a very early block in virus infection and another in the assembly of functional capsids. ts1208 had a defect in only the latter function. Initially, the early defect in ts1204 infection at the NPT was thought to be in penetration. This conclusion was based on the finding that high multiplicities of infection (MOI) with the mutant at the NPT prevented subsequent infection by wt HSV-1 but not HSV-2 and that the early defect could be overcome by polyethylene glycol, a membrane-fusing agent. Although we have been able to confirm the first observation, we have been unable to reproduce the latter finding. Since more-recent work has shown that UL25 is associated with the capsids in the cell nucleus, the defects in ts1204 have been reassessed. We discovered that ts1204 contained more than one ts mutation and constructed a new mutant, ts1249, possessing the UL25 ts mutation only. In this paper, we describe the characterization of this mutant to gain insight about the role of UL25 early in infection. In addition, ts1208 has also been analyzed further, and the nucleotide changes responsible for the defects in both ts1204 and ts1208 have been determined.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero) cells were grown in Dulbecco's modified Eagle's medium supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml), and 10% fetal calf serum. BHK21 clone 13 (BHK) cells were cultured in Glasgow minimal essential medium supplemented with 10% tryptose phosphate broth, 10% newborn calf serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) (ETC). The wt HSV-1 virus used was strain 17syn+. ts mutants ts1204 and ts1208 (derived from this strain), the disabled strain 17 mutant in1383, and a UL28-null mutant from strain KOS (gCB) have all been described previously (1, 42, 54).
Plasmids.
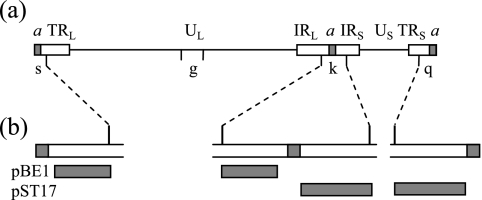
The amplicon pSA1 has a 200-bp packaging signal spanning the junction between two tandem a sequences derived from HSV-1 strain 17 and a copy of the HSV-1 oris origin of replication in the plasmid vector pAT153 (53). The BamHI g probe used in Southern blot hybridization was purified from BamHI-digested pGX37, which contains the HSV-1 strain 17 BamHI g genomic fragment (corresponding to HSV-1 nucleotides 52589 to 60363) inserted into the BamHI site of pAT153 (Fig. 1). Plasmids pBE1, containing HSV-1 sequences specific for IRL/TRL, and pST17, containing HSV-1 sequences specific for IRS/TRS, are described in detail in reference 53, and the locations of their HSV-1 sequences are shown in Fig. 1.
FIG. 1.
(a) Structure of the HSV-1 genome showing the unique regions (UL and US), the repeated regions (TRL, IRL, TRS, and IRS), and the a sequence, which contains the cis-acting packaging signals (shaded box). The positions of the BamHI fragments k, q, s, and g are shown. (b) Expanded section of BamHI fragments s, k, and q showing the HSV-1 regions present in the plasmids pBE1 and pST17.
Metabolic labeling of cells.
Vero cells and viruses were prewarmed to 42°C for 10 min prior to infection at the NPT. Cell monolayers (2 × 105 cells/15.6-mm-diameter well) were either mock infected or infected with 20 PFU of virus/cell. At 6 h postinfection (p.i.), the cells were washed twice with medium lacking methionine and fetal calf serum and incubated overnight in medium containing one-fifth the normal concentration of methionine, 2% fetal calf serum, and 20 μCi [35S]methionine/well.
Purification of viral capsids.
Viral capsids were purified from BHK cells, essentially as described by Preston and McDougall (45).
Western blotting.
Capsid proteins or virus-infected cell proteins, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), were blotted onto a nitrocellulose membrane and screened with antibodies, using the enhanced chemiluminescence method as described by Thurlow et al. (56). Protein bands were quantified by densitometric analysis of digital images by using Quantity One software (Bio-Rad Laboratories). The amount of each capsid preparation was standardized using the capsid shell protein VP19C, which was assumed to be present in the same copy number in both ts1249 and wt HSV-1 capsids.
Antibodies.
Mouse monoclonal antibodies (MAb) specific for UL6 (MAb 175), UL17 (MAb 203), UL25 (MAb 166), ICP0 (MAb 11060), VP5 (MAb DM165), and VP19C (MAb 2231) and the rabbit polyclonal antibodies specific for VP23 (R186) and UL25 (335) have all been described previously (16, 23, 30, 57). The actin-specific MAb AC-40 was purchased from Sigma.
Sequence analysis.
DNA was sequenced by the M13 chain terminator method by using a Sequenase kit (Amersham Biosciences) or sequenced commercially by Geneservice Ltd.
Analysis of replicated and packaged amplicon DNA.
Vero cell monolayers (106 cells/60-mm-diameter dish) were transfected with 2 μg pSA1, using Lipofectamine (12 μl) and Plus (8 μl) reagents (Invitrogen), and incubated at 36.5°C as described previously (45). Fifteen minutes prior to virus infection, 100 μg/ml cycloheximide was added to each sample. At 12 h after the addition of DNA to the cells, the cells were infected with 10 PFU of virus/cell, and incubation was continued at 36.5°C for 2 h in the presence of 100 μg cycloheximide/ml. This concentration of cycloheximide was used throughout. Cells were washed three times with tissue culture medium to remove the inhibitor, and samples were transferred to the NPT of 38.5°C or 39.5°C or the permissive temperature (PT) of 32°C. At 20 h p.i., the cells were harvested and total cellular DNA and DNase-resistant DNA were extracted from cells. An aliquot of DNA was digested with DpnI, together with EcoRI or BamHI, to remove the input plasmid DNA, and the fragments were separated on agarose gels as described by Stow (53). The DNA fragments were transferred onto a Hybond XL membrane (GE Healthcare) and hybridized to 32P-labeled pAT153 or a cloned HSV-1 BamHI g fragment, as described by Jamieson et al. (20). The DNA was radiolabeled with [α-32P]dCTP and [α-32P]dGTP by random primer extension, using a kit from Roche. Phosphorimages of Southern blots were obtained using a personal phosphorimager (Bio-Rad), and the relative amounts of radioactivity in DNA bands were determined using Quantity One software (Bio-Rad).
Analysis of total and encapsidated viral DNA.
Prior to virus infection, cycloheximide was added to the medium of BHK cell monolayers (106 cells in 60-mm-diameter petri dishes). Fifteen minutes later, the medium was removed and the cell monolayers were infected with 10 PFU of virus per cell, and the virus adsorbed to cells for 2 h at 36.5°C in the presence of cycloheximide. Cells were washed with 0.1 M glycine-0.14 M NaCl (pH 3.0) to inactivate virus bound to the cell surface as described by Stow (53). After this treatment, the cells were washed three times with medium prewarmed to 39°C and incubated at the NPT (38.5°C) or the PT (32°C) in the absence of cycloheximide. At 20 h p.i., the cells were harvested, total cellular DNA and DNase-resistant DNA were extracted from cells and digested with BamHI, and Southern blot analysis was carried out as described above.
Assay of VP16 activity in cells.
The multiply defective HSV-1 virus in1383 (42), carrying the lacZ reporter gene under the control of the ICP0 immediate-early promoter, was central in the design of an assay to determine whether VP16 was released from incoming ts1249 capsids in cells at the NPT. Vero cells were either mock infected or infected with 5 PFU of in1383/cell in the presence of cycloheximide at 38.5°C. After 30 min at the NPT, cells were mock infected or were infected with purified ts1249 virions or virions of a control virus at an MOI of 10 PFU/cell in the presence of cycloheximide. The virions were prewarmed to 42°C for 10 min immediately prior to infection. After incubation for 5.5 h at the NPT in the presence of cycloheximide, the cells were washed three times with medium containing 1 μg/ml of the RNA synthesis inhibitor actinomycin D to remove the cycloheximide block, and incubation was continued at the NPT in the presence of actinomycin D. The samples were harvested at 9 h and 20 min after the addition of the second virus. Cells were washed once with ice-cold phosphate-buffered saline (PBS) prior to the addition of 100 μl 10 mM Tris-HCl (pH 7.5), 2 mM MgCl2, 10 mM NaCl, and 0.1% NP-40. Dishes were stored overnight at −20°C. Samples were thawed, and cells were scraped off into the supernatant. The samples were centrifuged at 14,500 × g for 2 min at 4°C and assayed for β-galactosidase activity essentially as described by Preston and Nicholl (41).
Confocal immunofluorescence microscopy.
Prior to infection, viruses were warmed to 42°C for 10 min and cells were incubated at 38.5°C for 1 h. The prewarmed Vero cells on 13-mm-diameter coverslips (7 × 104 cells per well) were infected with 10 PFU of prewarmed, purified virions per cell at 38.5°C in the presence of cycloheximide. After 1 h at 39°C, the virus inoculum was removed and the cells were washed twice with prewarmed tissue culture medium containing cycloheximide. Incubation of virus-infected cells was continued at 39°C in the presence of cycloheximide for various times up to 4 h. Immunofluorescence was performed as described by Preston and McDougall (45), using MAb DM165, specific for VP5, and for some experiments 335, a polyclonal rabbit antibody to UL25 protein, as the primary antibodies. The secondary antibodies used were fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (IgG) (FITC-GAM) (Sigma) and Cy5-conjugated goat anti-rabbit IgG (Cy5-GAR) (Sigma). In experiments using only MAb DM165, after the cells were treated with secondary antibodies, they were incubated with propidium iodide (Sigma-Aldrich) at a concentration of 1 μg/ml in PBS containing 1% fetal calf serum. Stained cells were examined under a Zeiss LSM 510 confocal microscope, using a 63× oil immersion lens (NA 1.4). For samples containing FITC and Cy5 conjugates, lasers with excitation lines at 488 nm and 633 nm were used. For samples containing the FITC conjugate and propidium iodide, lasers with excitation lines of 488 nm and 543 nm were activated.
Fluorescence in situ hybridization.
Vero cells on 13-mm-diameter coverslips were infected with 50 PFU of prewarmed virus/cell. A high concentration of virus was used in these experiments because at lower levels the method was less sensitive. After 1 h at 38.5°C, the virus inoculum was removed, the cells were washed three times with medium warmed to 42°C, and incubation was continued at 38.5°C. At 5 h p.i., the cells were washed twice with PBS containing 1% fetal calf serum prior to fixation with precooled 95% ethanol-5% acetic acid for 5 min at −20°C. The fixed cells were washed three times with PBS-1% fetal calf serum and stored at 4°C until required. The probe used for in situ hybridization was a cosmid, cos56, containing HSV-1 strain 17 sequences from bp 79442 to 115152 (9, 31). The DNA was labeled by nick translation with Cy3-dCTP as described previously (15), and the cells were subsequently treated with DAPI (4′,6′-diamidino-2-phenylindole) to stain cell nuclei. Cells were examined under a Zeiss LSM 510 confocal microscope with 405- and 543-nm laser lines, with each channel scanned separately.
Reversibility of the ts1249 uncoating defect.
BHK cells in 35-mm dishes, prewarmed for 1 h at 38.5°C, were infected with virus (100 to 200 PFU per dish) that had been prewarmed at 42°C for 10 min and were incubated at 38.5°C. At 1 h after virus infection, the cells were washed once with tissue culture medium and overlaid with medium containing 3% human serum, 2% calf serum, and antibiotics. The cells were transferred to the PT at various times after virus infection and stained with Giemsa after 3 days at 32°C, and the number of virus plaques was recorded.
Sequence alignment.
The multiple-sequence alignment of the UL25nt domains (HSV-1 UL25 amino acid residues 134 to 580) from 38 herpesvirus members, described by Bowman et al. (5), was used. The alignment was generated with the program CLUSTALW (55).
RESULTS
Sequence analysis of the ts1204 and ts1208 mutations.
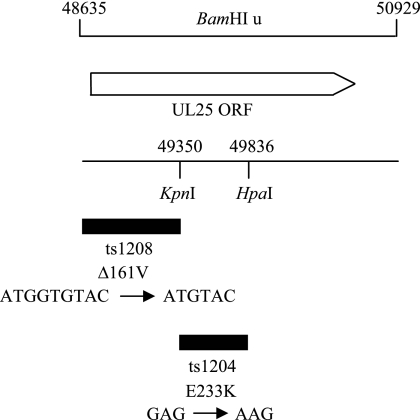
Previous marker rescue studies using cloned wt HSV-1 DNA had mapped each of the mutations to a small region within UL25 (1). These regions of the cloned wt HSV-1 DNA and the corresponding portions in the cloned mutant DNAs were sequenced (Fig. 2). The ts mutation in the ts1204 UL25 gene was caused by a single base pair change leading to the replacement of glutamic acid residue 233 with a lysine. Unexpectedly, ts1208 had an in-frame deletion of three base pairs within the UL25 gene, resulting in a ts protein that lacked valine 161. No other differences from the sequence of wt HSV-1 (31) were present.
FIG. 2.
Identification of ts1204 and ts1208 UL25 mutations. The solid black box indicates the location of the ts mutation within the BamHI u genomic fragment obtained previously by marker rescue experiments. The numbers refer to the positions of the restriction endonuclease sites in the HSV-1 strain 17 genome. The precise base pair change determined by DNA sequence analysis and the effect of the mutation on the UL25 amino acid sequence are shown below the marker rescue data. ORF, open reading frame.
Construction of ts1249.
A ts+ marker rescuant of ts1204, isolated from cells transfected with ts1204 viral DNA and a cloned fragment containing the wt UL25 gene, behaved like wt HSV-1 at 38.5°C. On the basis of this finding, it was concluded that ts1204 had a single ts lesion (1). It was subsequently discovered that at a higher NPT of 39.5°C the marker rescuant formed plaques at low efficiency compared with wt HSV-1 and therefore had more than one ts defect. The cloned ts1204 genomic EcoRI f fragment containing the UL25 gene was cotransfected with wt HSV-1 DNA into cells, and ts virus isolates that failed to recombine with ts1204 were plaque purified. One of the isolates, ts1249, was selected for further study. Sequence analysis confirmed that ts1249 contained the original UL25 mutation that was present in ts1204 (data not shown). ts+ marker rescuants were generated by recombining the cloned wt genomic BamHI u fragment into the ts1249 DNA. One of these, ts+1249MR, and a spontaneous ts+ revertant of ts1249, ts+1249rev, were each shown by sequence analysis to contain the wt sequence in place of the ts1249 mutation (data not shown). Both produced plaques at 39.5°C and the PT of 32°C with efficiency similar to that of wt HSV-1, suggesting that ts1249 had a single ts lesion (Table 1). ts+ marker rescuants of ts1208, produced in the same way as ts1249 marker rescuants, also behaved like wt HSV-1 at both temperatures, indicating that this mutant had a single ts mutation (Table 1).
TABLE 1.
Comparison of the efficiencies of plaque formation (e.o.p.) at the NPT of 38.5°C or 39.5°C and the PT of 32°Ca
| Virus | Efficiency of plaque formation at NPT indicated and PT of 32°C
|
||
|---|---|---|---|
| 38.5°C | 39.5°C | 39.5°C | |
| ts1249 | 8.5 × 10−5 | 1.0 × 10−4 | |
| ts+1249rev | 1.1 | 0.9 | |
| ts+1249MR | 1.5 | 1.1 | |
| ts1204 | 2.4 × 10−5 | <1.2 × 10−5 | |
| ts+1204rev | 1.1 | 3.0 × 10−2a | |
| Strain 17 wt | 1.6 | 1.2 | |
| ts1208 | 4.3 × 10−4 | ||
| ts+1208MR | 0.9 | ||
| Strain 17 wt | 0.5 | ||
Plaques were very small compared with wt HSV-1 plaques at 39.5°C.
Virus-infected cell polypeptides.
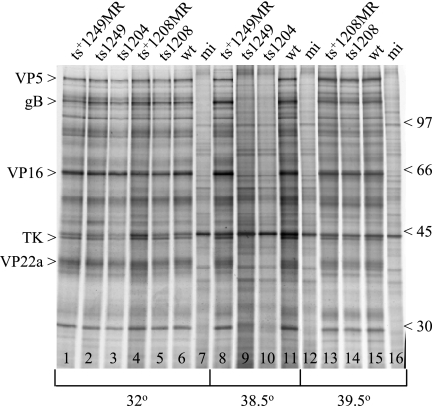
To confirm that ts1249, like ts1204 (1), has a very early defect, the virus-infected cell polypeptides synthesized at the NPT and PT were radiolabeled with [35S]methionine and analyzed by SDS-PAGE. wt HSV-1-, ts+1249MR-, and mock-infected cells were also included as controls. In addition, the polypeptide profiles of ts1208 and its marker rescuant were examined. Both ts1204 and ts1249 failed to synthesize any detectable viral polypeptides in cells infected at the NPT, and the profiles of the mutant-infected cell polypeptides resembled that of the mock-infected cell control (Fig. 3, lanes 9, 10, and 12). This result suggests that the ts lesions in the UL25 genes of ts1204 and ts1249 are responsible for the lack of viral protein synthesis in cells infected with either of these viruses at the NPT. The finding that ts+1249MR produced levels of viral polypeptides similar to those of wt HSV-1-infected cells at the NPT supported this conclusion (Fig. 3, lanes 8 and 11). The profile of the ts1208-infected cell polypeptides resembled those of ts1208MR- and wt HSV-1-infected cell polypeptides at the NPT (Fig. 3, lanes 13, 14, and 15), in agreement with previous published results (1).
FIG. 3.
Comparison of [35S]methionine-labeled mutant virus-infected cell polypeptides synthesized at the PT and NPT. Vero cells were mock infected (mi) or infected with wt HSV-1, ts1204, ts1249, ts+1249MR, ts1208, or ts+1208MR, and the virus-infected cells were incubated at 32°C, 38.5°C, or 39.5°C. At 5 h p.i., [35S]methionine was added to the cells and incubation continued for a further 15 h. Samples were analyzed on an SDS-10% polyacrylamide gel and the proteins detected by autoradiography. Selected virus polypeptides are labeled on the left-hand side, and on the right-hand side the positions of molecular mass markers (kDa) are indicated. TK, thymidine kinase.
Virus entry into cells.
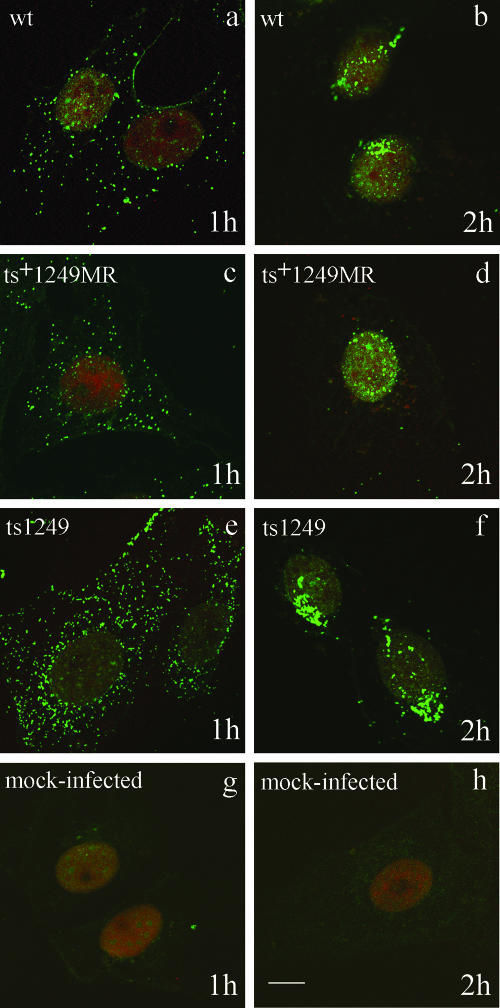
The failure of ts1249 to synthesize viral proteins at the NPT could be due to a defect in attachment to or penetration of the host cell or an uncoating defect. To determine whether ts1249 was able to enter cells at the NPT, an indirect immunofluorescent assay was used to detect incoming capsids (51). Vero cells were infected at 38.5°C with ts1249, wt HSV-1, or ts+1249MR or were mock infected and incubated in the presence of cycloheximide to block de novo protein synthesis. At 1, 2, and 4 h p.i., the cells were fixed, permeabilized, and probed with MAb specific for the major capsid protein VP5. At each time point, all the virus-infected cell samples had similar patterns of distribution of capsids. At 1 h p.i., most of the virus-infected cells contained capsids that were dispersed all over the cell (Fig. 4a, c, and e). By 2 h, in many of the cells the capsids were concentrated together, often adjacent to or over the nuclei (Fig. 4b, d, and f). At 4 h p.i., the pattern of immunofluorescence was similar to that observed at 2 h, with most of the cells containing capsids concentrated near or over the nuclei (data not shown). The results suggest that ts1249 is not impaired in entry into the cell or transport within the cytoplasm to the edge of the nucleus. Greater numbers of capsids were apparent in ts1249-infected cells than in cells infected with wt HSV-1 or ts+1249MR, suggesting that the ts1249 virion preparation exhibited a higher particle/PFU ratio than the control viruses. This, however, was not a consistent feature of ts1249 virus stocks.
FIG. 4.
Localization of incoming capsids in the cell. Vero cells were either mock infected or infected with wt HSV-1, ts+1249 MR, or ts1249 in the presence of cycloheximide and incubated for 1 (a, c, e, g) or 2 (b, d, f, h) h at 38.5°C. Capsids were detected by indirect immunofluorescence with MAb DM165, specific for VP5, and the secondary IgG antibody FITC-GAM. The nuclei were stained with propidium iodide. The cells were examined by confocal microscopy and digital images taken. Bar, 10 μm.
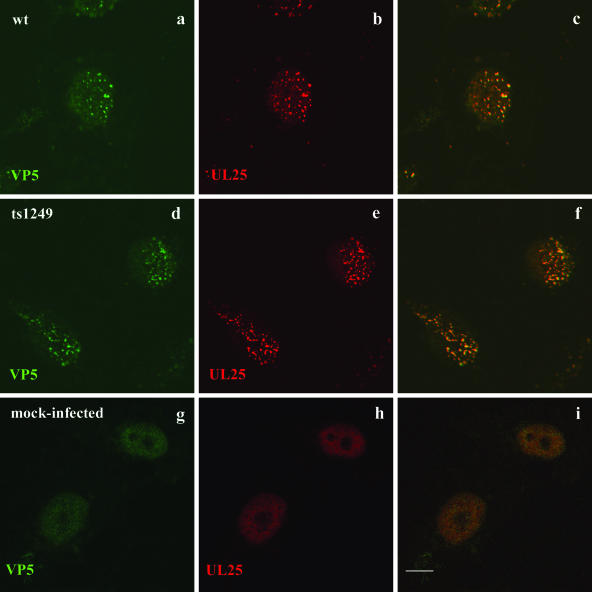
To determine whether UL25 remained attached to the incoming capsids, a similar experiment, in which cells were incubated for 4 h p.i. at the NPT, fixed, permeabilized, and probed with VP5- and UL25-specific antibodies, was performed. In cells infected with either wt HSV-1 or ts1249, UL25 colocalized with VP5, suggesting that UL25 remained bound to the capsids (Fig. 5). This finding is consistent with the findings of an earlier study using PRV (22). Although incoming ts1249 capsids migrated toward the cell nuclei, further experiments are required to determine whether the mutant capsids associated with the nuclear pores.
FIG. 5.
Colocalization of UL25 with VP5 in incoming ts1249 capsids at the NPT. Vero cells were either mock infected or infected with wt HSV-1 or ts1249 in the presence of cycloheximide and incubated for 4 h at 38.5°C. Capsids were detected in the cell by indirect immunofluorescence with MAb DM165, specific for VP5, and the secondary IgG antibody FITC-GAM (a, d, e), and UL25 was identified with rabbit antibody 335 and the secondary IgG antibody Cy5-GAR (b, e, h). Each set of three images shows FITC staining of VP5 (left), Cy5 staining of UL25 (middle), and a merged image of the two for the same field of infected cells (right). The cells were examined by confocal microscopy and digital images taken. Bar, 10 μm.
Release of viral DNA into the cell nucleus.
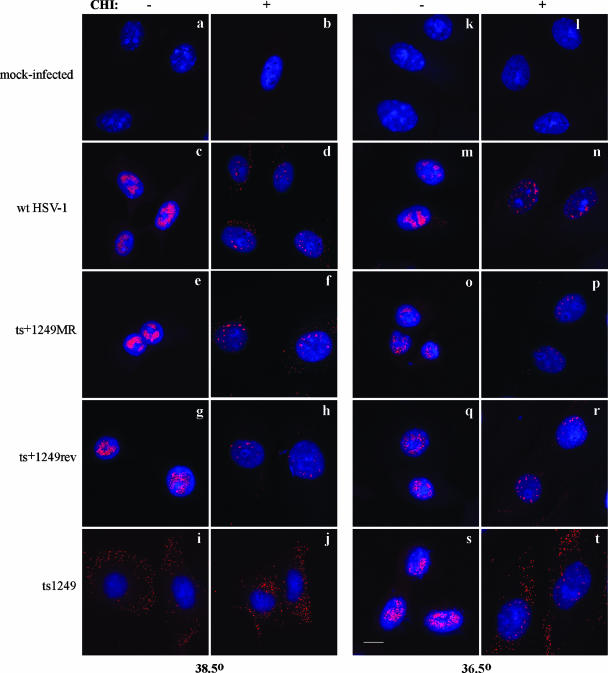
To confirm that ts1249 had an uncoating defect, a fluorescent in situ hybridization experiment was carried out to locate the HSV-1 DNA in the cell. Vero cells were infected with ts1249, wt HSV-1, ts+1249rev, or ts+1249MR or were mock infected and incubated at 38.5°C or 36.5°C in the presence or absence of cycloheximide for 5 h, fixed, and probed with Cy3-labeled cosmid DNA containing HSV-1 DNA sequences (Fig. 6). A PT of 36.5°C was used in these experiments instead of 32°C because the rate of viral DNA synthesis was more comparable to that at 38.5°C. At 36.5°C, the mutant grew almost as well as at the lower temperature of 32°C (data not shown). In wt HSV-1-, ts+1249MR-, or ts+1249rev-infected cells incubated at the NPT in the absence of cycloheximide, viral DNA was concentrated in the cell nuclei, the site of viral DNA replication (Fig. 6c, e, and g). In ts1249-infected cells treated under the same conditions as wt HSV-1-, ts+1249MR-, or ts+1249rev-infected cells, viral DNA was not observed in most of the nuclei, consistent with the suggestion that ts1249 had an uncoating defect (Fig. 6i). Viral DNA was also detected in the nuclei of wt HSV-1-, ts+1249MR-, or ts+1249rev-infected cells treated with the protein synthesis inhibitor but in smaller amounts than in cells grown in the absence of the drug because viral DNA replication cannot occur (Fig. 6d, f, and h). Low levels of viral DNA were also found in the cytoplasm, with the genomes generally being dispersed. The pattern of fluorescence in ts1249-infected cells grown in the presence of cycloheximide was very similar to that of mutant-infected cells grown in the absence of the drug (Fig. 6i and j). The viral DNA formed speckles of fluorescence distributed throughout the cytoplasm and was only very occasionally detected in the cell nuclei.
FIG. 6.
Localization of incoming virion DNA in the cell by in situ hybridization. Vero cells were either mock infected or infected with wt HSV-1, ts+1249MR, ts+1249rev, or ts1249 in the presence or absence of cycloheximide (CHI) and incubated for 5 h at 38.5°C or 36.5°C as indicated. HSV-1 DNA was detected by in situ hybridization and confocal microscopy. Bar, 10 μm.
The finding that input ts1249 DNA was dispersed throughout the cytoplasm was in contrast to results from immunofluorescence experiments, in which incoming capsids were found concentrated around or close to the cell nuclei at 4 h p.i., in agreement with earlier studies with wt HSV-1 (51). It should be noted that the cells in the two experiments were treated differently in fixation and subsequent procedures, which could account for the difference. It is also possible that viral DNA was lost from some of the capsids, although preliminary electron microscopic experiments suggest that most mutant capsids retained their DNA (data not shown).
At 36.5°C, much of ts1249 input DNA remained in the cytoplasm in cells grown in the presence of cycloheximide, although some genomes appeared to be transported into the nucleus. Consistent with this, in the absence of cycloheximide, ts1249 DNA replication was detected in most of the nuclei (Fig. 6s and t). It should be noted that at 36.5°C ts1249 had a plating efficiency similar to that at 32°C and the yield from cells infected with 10 PFU/cell of mutant virus at 36.5°C was similar to the yield obtained at 32°C, indicating that 36.5°C is permissive for replication of ts1249 (data not shown).
VP16 activity in cells infected at the NPT by ts1249.
A possible explanation for the failure of ts1249 to uncoat its genome at the NPT is that some of the tegument proteins remain attached to the incoming capsid, blocking the release of the genome into the nucleus at the nuclear pore. VP16 is a major tegument protein that, in addition to its structural role, is required for the transcription of immediate-early viral genes. To determine whether VP16 was released from incoming ts1249 capsids, an experiment was carried out using the highly disabled HSV-1 mutant in1383 containing a reporter gene carrying lacZ under the control of the immediate-early promoter of ICP0 in place of the thymidine kinase gene (42). This virus has three other defects. It has a ts mutation (tsK) in the ICP4 gene, which makes the protein nonfunctional at 38.5°C, and an insertion in the VP16 gene, which abolishes the transactivating function of VP16 but does not affect its structural role as a tegument protein. in1383 also contains a nonfunctional ICP0 gene in which sequences encoding the zinc ring finger in ICP0 have been deleted, generating a frameshift mutation. As a consequence of these mutations, in1383 is severely impaired in viral mRNA synthesis. β-Galactosidase activity expressed from mRNA transcribed from the lacZ reporter gene in the in1383 genome is minimal but is increased substantially when another virus supplies VP16.
Cells were infected at the NPT with purified virions of ts1249, ts+1249MR, or wt HSV-1 or were mock-infected in the presence or absence of in1383 and incubated at the NPT in the presence of cycloheximide to allow the accumulation of immediate-early mRNAs from input viral genomes released into the cell nuclei. After 5.5 h, the medium containing cycloheximide was removed and replaced with medium containing the RNA synthesis inhibitor actinomycin D for 4 h to permit translation of the immediate-early transcripts. Cells were harvested, and extracts were assayed for β-galactosidase activity.
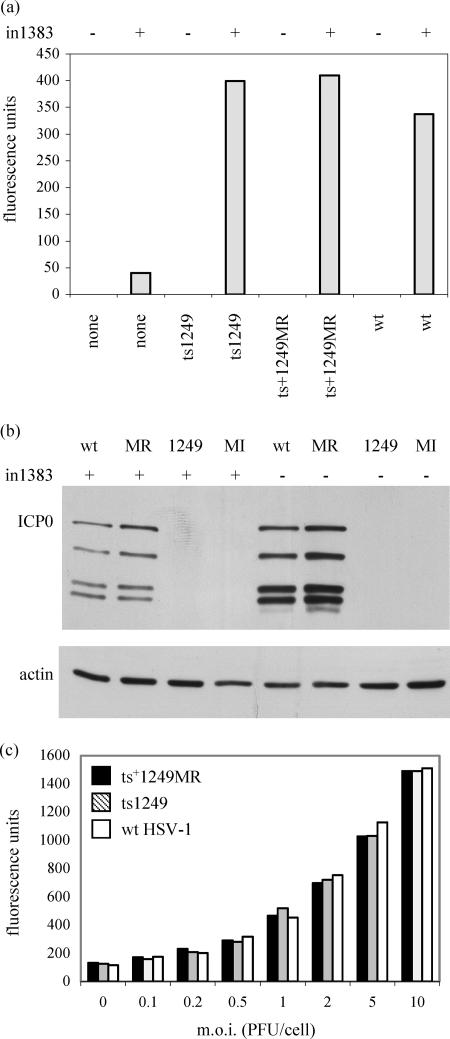
No β-galactosidase was detected in the absence of in1383, and only very low levels were found in cells infected with in1383 alone. In contrast, extracts of cells that had been coinfected with in1383 and ts1249 expressed high levels of β-galactosidase, comparable to the amounts detected in cells coinfected with in1383 and wt HSV-1 or ts+1249MR, suggesting that VP16 was released from the capsids of input ts1249 (Fig. 7a). Extracts were also screened for the presence of ICP0 by using a Western blot assay. In the presence or absence of in1383, wt HSV-1 and ts+1249MR produced high levels of ICP0, much of which was present as proteolytic fragments, which are typically observed in Vero cells. By contrast, ICP0 was not detected in cells infected with in1383 alone, ts1249 alone, or in1383 plus ts1249 (Fig. 7b). Thus, although VP16 supplied by ts1249 activated the ICP0 promoter in in1383 driving β-galactosidase expression, it failed to express ICP0 from its own genome, supporting the finding that incoming ts1249 capsids at the NPT are unable to release the viral DNA into cell nuclei.
FIG. 7.
(a) β-Galactosidase activity in cell extracts. Cells were mock infected (−) or infected with in1383 (+), followed by superinfection with ts1249, ts+1249MR, or wt HSV-1 virions as indicated. The activities shown are expressed in arbitrary fluorescence units and represent the means of readings obtained for duplicate samples. (b) Western blot analysis of protein extracts from cells infected with ts1249 (1249), ts+1249MR (MR), or wt HSV-1 (wt) or mock infected (MI) cells in the presence or absence of in1383 infecting virus, screened with ICP0 or actin antibodies. (c) β-Galactosidase activity in extracts from cells infected with in1383 and different MOI of ts1249, ts+1249MR, or wt HSV-1 virions.
To establish that the efficiency of VP16 release from ts1249 virions infecting the cells was similar to that of wt HSV-1 and ts+1249MR virions, the experiment was repeated using a range of MOI of these viruses (Fig. 7c). At each MOI, cells infected with ts1249, ts+1249MR, or wt HSV-1 virions expressed similar levels of β-galactosidase activity, and there was a strong correlation between the expression level and the input multiplicity. This result provides strong evidence that ts1249 releases VP16 as efficiently as wt HSV-1 and ts+1249MR.
Reversibility of the uncoating phenotype of ts1249.
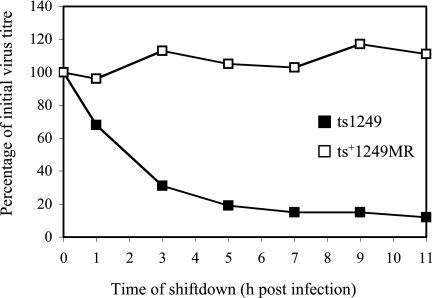
BHK cells were infected at the NPT with approximately 150 PFU of ts1249 or the marker rescuant per dish and were transferred to the PT of 32°C at various times after infection to determine whether the effect of the ts1249 uncoating defect could be overcome by lowering the incubation temperature. The ability of ts1249 to form plaques at the PT declined with increasing time at the NPT, in contrast to ts+1249MR, which gave similar numbers of plaques upon downshift of virus-infected cells, irrespective of the time of downshift (Fig. 8).
FIG. 8.
Reversibility of the ts1249 uncoating defect. BHK cells were infected with either ts1249 or ts+1249MR (MR) at the NPT and transferred to the PT at the times indicated. After 3 days at 32°C, the virus plaques were counted. The values shown are the means of values obtained for duplicate samples.
Packaging of viral DNA at the NPT.
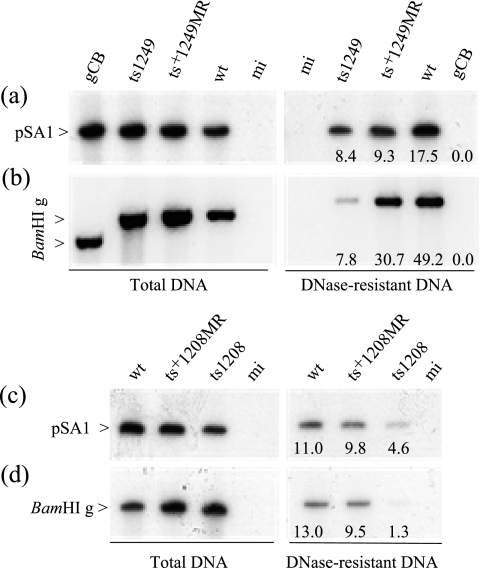
Previous work showed that ts1204-infected cells incubated for 8 h at the NPT after virus adsorption for 1 h at the PT contained fewer capsids in the cell nuclei than wt HSV-1-infected cells, and DNA-containing capsids were not observed, suggesting that ts1204 had a defect in the assembly of mature capsids (1). Similarly, no C capsids were detected in ts1208-infected cells at the NPT. Initially, ts1249 and ts1208 were screened for the ability to amplify and package the plasmid pSA1, containing an HSV-1 origin of replication (oris) and the minimal cleavage/packaging signal Uc-DR1-Ub sequence (53). The UL28-null mutant gCB, which does not package any viral or amplicon DNA under restrictive conditions, was also included as a negative control. Cells were transfected with the plasmid and subsequently infected with ts1249, wt HSV-1, or ts+1249MR in the presence of cycloheximide for 2 h at 36.5°C to allow for uncoating of the viral genome prior to incubation at the NPT of 38.5°C in the absence of the drug. This step was omitted for cells infected with ts1208, ts+1208MR, or wt control virus. Instead, these virus-infected cell samples were incubated only at the NPT of 39.5°C. Total cell DNA and DNase-resistant DNA were extracted from the cells at 20 h p.i., and a portion was treated with DpnI and EcoRI and screened by Southern blot analysis for the presence of the plasmid, using 32P-labeled vector pAT153 as a probe (Fig. 9a and c). In contrast to the UL28-null mutant, ts1249 packaged the replicated amplicon DNA at the NPT (Fig. 9a). Similarly, ts1208 packaged the amplicon but not as efficiently as ts1249 (Fig. 9c). A sample of each DNA was also digested with DpnI and BamHI and probed with 32P-labeled genomic fragment BamHI g, which maps in the long unique region of the viral genome to examine the packaging of the viral genome (Fig. 9b and d). ts1249 packaged the viral DNA less efficiently than the amplicon DNA, as had been reported previously for the UL25-null mutant KUL25NS (53). Likewise, at 39.5°C ts1208 had a lower efficiency of packaging than wt HSV-1, although at this temperature wt HSV-1 encapsidated a smaller proportion of viral DNA than at 38.5°C.
FIG. 9.
Amplification and packaging of the plasmid pSA1 in Vero cells by ts1249 and ts1208. One set of cell monolayers was transfected with pSA1, and cells subsequently were infected with wt HSV-1, gCB, ts+1249MR, or ts1249 or were mock infected (mi) in the presence of cycloheximide at 36.5°C. After 2 h, the cycloheximide block was removed and the samples were transferred to 38.5°C. The second set of cell monolayers was transfected with pSA1, and cells were subsequently infected with ts1208, ts+1208MR, or wt HSV-1 or were mock infected. These samples were incubated only at 39.5°C in the absence of cycloheximide. At 20 h p.i., the cells were harvested and total cellular DNA and DNase-resistant DNA were prepared and digested with DpnI and EcoRI (a, c) or BamHI (b, d). Southern blot analysis was carried out, using 32P-labeled plasmid vector pAT153 (a, c) or the 32P-labeled, cloned HSV-1 genomic fragment BamHI g (b, d) as a probe. The numbers at the bottom of the lanes indicate the percentage of radioactivity present in the band in the DNase-treated sample relative to that in the band in the total DNA sample. Note that gCB BamHI g is smaller than that of wt HSV-1 strain 17.
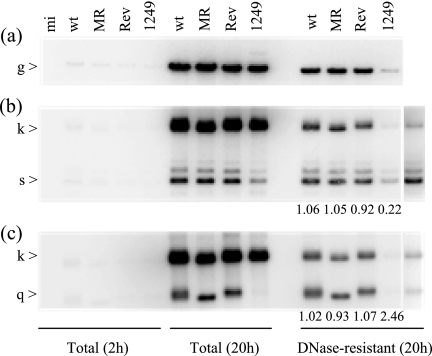
Previous characterization of the UL25-null mutant KUL25NS suggested that the mutant had a reduced capacity to encapsidate full-length viral DNA in nonpermissive cells (53). The packaging of viral DNA by ts1249 was examined in more detail to determine whether this mutant had a phenotype similar to that of KUL25NS at the NPT. ts1208 was not included in these experiments, because the level of viral DNA packaged at 39.5°C was so low. Cells were infected with ts1249, wt HSV-1, ts+1249rev, or ts+1249MR in duplicate in the presence of cycloheximide. After 2 h at 36.5°C, the cells were washed three times with medium to remove the cycloheximide block. One set of plates was transferred to 38.5°C, and the total cell DNA and DNase-resistant DNA were extracted from these cells at 20 h p.i. The duplicate set of samples was harvested after incubation for 2 h at 36.5°C, immediately after removal of the cycloheximide block. The DNAs were digested with BamHI and screened by Southern blot analysis, initially using 32P-labeled cloned genomic fragment BamHI g as a probe. Reduced amounts of BamHI g were present in packaged DNA from ts1249-infected cells incubated at the NPT compared to those for the control viruses (Fig. 10a), consistent with the experiment whose results are shown in Fig. 9. By contrast, cells infected at the PT of 32°C with ts1249 had levels of packaged viral DNA similar to those for wt HSV-1, ts+1249rev, and ts+1249MR grown at the same temperature (data not shown). To ascertain whether ts1249 also had a late defect in packaging, the DNA samples were also probed with 32P-labeled pBE1 and with 32P-labeled pST17, which hybridize to the long-repeat and short-repeat sequences, respectively. At the NPT, ts1249-packaged DNA contained reduced amounts of the S-terminal fragment q, compared to those of the junction fragment k, whereas the L-terminal fragment s was overrepresented, indicating that most of the DNA packaged was not full-length (Fig. 10b and c).
FIG. 10.
Analysis of ts1249 total and DNase-resistant viral DNAs at the NPT. Duplicate samples of cells were infected with wt HSV-1, ts+1249MR (MR), ts+1249rev (Rev), or ts1249 or were mock infected (mi) in the presence of cycloheximide at 36.5°C. After 2 h, the cycloheximide block was removed and the samples were transferred to 38.5°C. One set of samples was harvested at 4 h p.i. and the other set at 20 h p.i. Total cellular DNA and DNase-resistant DNA were prepared and digested with BamHI. Southern blot analysis was carried out, using 32P-labeled, cloned HSV-1 genomic fragment BamHI g (a), pBE1 (b), or pST17 (c) as a probe. The far right-hand lanes in panels b and c are the ts1249 lanes after longer exposure. The numbers at the bottom of the lanes indicate the ratio of radioactivity present in the joint fragment k to that in the terminal s or q fragment in the DNase-treated sample.
Association of UL25 protein with ts1249 B capsids.
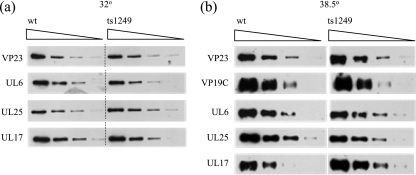
The pattern of encapsidation of viral DNA by ts1249 at the NPT was very similar to that observed previously for the UL25-null mutant. One possibility for the similar phenotypes of the two mutants is that at the NPT the ts1249 UL25 protein was impaired in its ability to bind to capsids. The amount of UL25 protein present in ts1249 B capsids produced at 38.5°C was compared to the levels in wt B capsids generated at the same temperature. Cells were infected with ts1249 and were incubated for 2 h at 36.5°C to enable release of ts1249 DNA from incoming capsids prior to incubation at 38.5°C. At 20 h p.i., the cells were harvested and the capsids purified. As a control, purified B capsids from wt HSV-1 and ts1249-infected cells grown at the PT of 32°C were also prepared. Capsid proteins were separated by SDS-PAGE, and Western blot analysis was carried out, using VP23 polyclonal antibody as a probe to determine the relative amount of protein in each capsid preparation. The concentration of each capsid preparation was adjusted so that they all had similar levels of VP23. Samples were serially twofold diluted, and the separated proteins were blotted onto nitrocellulose and probed with antibodies to UL6, UL25, and UL17 (Fig. 11a and b). In addition, the polypeptides from capsids formed at the NPT were probed with VP19C MAb. ts1249 B capsids isolated from cells grown at the PT of 32°C had levels of UL6, UL25, and UL17 similar to those for wt B capsids produced under the same conditions (Fig. 11a). The ts1249 B capsids extracted from cells grown at the NPT also had amounts of UL6 similar to the levels present in wt HSV-1 B capsids produced at the same temperature but had twofold less UL25 and, unexpectedly, had slightly enhanced levels of UL17, approximately twofold greater than those in wt B capsids (Fig. 11b). The relative amounts of each packaging protein present in B capsids assembled in cells at the NPT were determined by densitometric analysis of the digital images of the exposed film of the Western blots, assuming equal levels of VP19C protein in each capsid preparation (Table 2).
FIG. 11.
(a) Association of UL25 with ts1249 B capsids produced at the PT. Cells were infected with wt HSV-1 and ts1249 at 36.5°C for 2 h and transferred to 32°C for 18 h. Serial twofold dilutions of purified B capsids were prepared and equalized on the basis of their VP23 content by Western blotting. The polypeptides from the equalized capsids were resolved by SDS-PAGE, transferred onto nitrocellulose, and probed sequentially with UL6, UL17, and UL25 MAbs as indicated on the left side. (b) Association of UL25 with ts1249 B capsids produced at the NPT. Capsids were treated in the same way as those produced at the PT and the equalized capsids screened sequentially with UL6, UL17, UL25, and VP19C MAbs.
TABLE 2.
Quantification of DNA-packaging proteins in ts1249 B capsids produced at the NPTa
| Capsid protein | Relative amt
|
||
|---|---|---|---|
| wt | ts1249 (1) | ts1249 (2) | |
| VP19C | 1.0 | 1.0 | 1.0 |
| UL6 | 1.0 | 1.1 | 0.9 |
| UL25 | 1.0 | 0.5 | 0.5 |
| UL17 | 1.0 | 2.2 | 2.5 |
The relative amount of protein was determined by densitometric analysis of digital images of exposed films of Western blots of B capsid proteins. wt and ts1249 B capsids produced at the NPT were equalized on the basis of their VP19C levels, and the amounts of the packaging protein in two different ts1249 capsid preparations relative to the level in the wt HSV-1 B capsid sample were calculated.
DISCUSSION
The structure of the N-terminally truncated UL25 protein (amino acid residues 134 to 580) has been determined (5). The protein has numerous flexible loops radiating from a box-shaped core and a distinctive electrostatic pattern, with one face of the protein containing a large cluster of positively charged residues and the opposite face having a high concentration of negatively charged residues. The flexible architecture of UL25 makes it ideally suited to adjust to conformational changes in the capsid that occur during DNA encapsidation and to interact with protein partners at different stages in virion assembly (5). Work with PRV has suggested that UL25, as well as having a role in DNA packaging, may be important for egress of capsids from the cell nuclei (24). Recently, evidence has been obtained for HSV-1 and PRV that UL25 is required for the attachment of the tegument to the capsid through its interaction with the large tegument protein UL36 (8, 26). The characterization of the UL25 ts mutant ts1249 has shown that UL25 is also involved at a very early stage in virus infection, in uncoating the viral genome.
Analysis of incoming ts1249 capsids by immunofluorescence revealed that ts1249 was able to enter the cell at the NPT, and this result was supported by the finding that cells coinfected with ts1249 and the defective virus in1383 at the NPT expressed high levels of β-galactosidase activity due to the release of functional ts1249 VP16 into the nuclei. The observations that cells infected with ts1249 at the NPT produced very low levels of viral proteins and that most of the input viral DNA remained in the cytoplasm are consistent with an uncoating defect, although the precise role of UL25 in this process is uncertain. The ts UL25 protein might directly prevent the release of the viral DNA from the capsid or release the DNA prematurely. Alternatively, it might interfere with other key processes, such as the association of the capsid with the nuclear pore complex or the dissociation of viral proteins from the capsid. Two ts mutants of UL36 have also been implicated in uncoating, and one of them, tsB7, has features similar to those of ts1249 inasmuch as incoming capsids at the NPT accumulate around the nuclei of virus-infected cells (3, 43). UL36 is thought to be important for the transport of capsids along the microtubules to the microtubule organizer, although direct evidence for this function is lacking.
The uncoating defect in ts1249-infected cells at the NPT was reversible at early times by transferring the cells to 36.5°C or lower temperatures. The decline in reversibility with time could be the result of cellular defense mechanisms reducing virus viability, or alternatively, the ts UL25 protein could become locked in an irreversible conformation. Although the ts1249 protein functioned at 36.5°C, it was clear from in situ hybridization experiments that ts1249 showed some impairment in the release of viral DNA into the cell nuclei, consistent with the observation that ts1249 formed slightly smaller plaques than wt HSV-1 at this temperature and also at 32°C. Nonetheless, virus stocks with high titers were readily obtained at both 36.5°C and 32°C, suggesting that the ts mutant had only a slight growth disadvantage at these temperatures.
The phenotype of ts1249 at the NPT upon release of the viral DNA from the incoming capsids was similar to that of the UL25-null mutant KUL25NS under restrictive conditions (53). Like the null mutant, ts1249 initiated packaging but was impaired at a late stage in the process. Furthermore, both sets of data supported a model in which viral DNA is packaged from the L segment end. Taking into account the proposed location of UL25 on the capsid and the phenotype of the UL25-null mutant, we favor the idea that UL25 is required for capsid stabilization during late stages of DNA packaging and that if it is also located around the portal, it may additionally stabilize the packing machinery. This stabilization may need to be reversed during uncoating to allow the viral DNA to be released from the capsid.
In contrast to what was found for the UL25-null mutant capsids, however, some UL25 was present in purified ts1249 B capsids produced at the NPT, about twofold less than the level observed in wt HSV-1 B capsids, suggesting that the ts protein was able to bind to capsids, although less efficiently than wt UL25. Preliminary immunogold analysis of ts1249 capsids by use of UL25 antibody supported this finding (data not shown), and Western blot analysis of UL25 in wt HSV-1- and ts1249-infected cells at the NPT excluded the possibility that the mutant UL25 was less stable than the wt protein (data not shown). Recent work by Trus et al. (59) suggested that UL25 on C capsids interacted with three different proteins (VP5, present in the peripentonal hexon; UL17; and VP19C), and it is likely that one or more of these interactions is impaired in the association of the ts UL25 protein with the capsid at the NPT. ts1249 UL25 was also retained on incoming capsids at the NPT, although we do not have any information as to whether the amounts present on the capsid were similar to those on wt HSV-1 capsids. It is conceivable that UL36 could stabilize the interaction of ts1249 UL25 with the capsid at the NPT.
Both the ts1208 and the ts1249 mutations map within the UL25 protein region whose structure is known (5). The ts1249 mutation E233K lies toward the end of an alpha helix close to a loop. Given the predicted flexible architecture of UL25, the change in the charge of residue 233 in ts1249 UL25 by the replacement of an acidic amino acid with a basic residue may be responsible for altering the conformation of the protein on the capsid at the NPT and could affect the strength of interaction with one or more of its binding partners. This glutamic acid residue is highly conserved among the alphaherpesvirus and betaherpesvirus UL25 orthologues analyzed but is not found in gammaherpesvirus counterparts (Table 3). Valine residue 161, absent in ts1208 UL25 protein, is also located toward the end of an alpha helix. It is conserved in 11 of the 16 alphaherpesviruses, and all but one had a small hydrophobic amino acid at this position (Table 3). The UL25 proteins of both the beta- and the gammaherpesviruses also had a small, hydrophobic amino acid at this position. Interestingly, all 14 gammaherpesviruses had a leucine present at this site.
TABLE 3.
Conservation of amino acid sequence at the sites of the ts1249 and ts1208 mutations among the alpha-, beta-, and gammaherpesviruses
| Mutant | Virus | Sequencea |
|---|---|---|
| ts1249 | Alphaherpesviruses | |
| GaHV-2 | SSLECAILCLYA | |
| GaHV-3 | SSLESAVLCLYA | |
| MeHV-1 | SSFECAVLCLYA | |
| CeHV-7 | SAFECAVLCLYL | |
| VZV | SAFECAVLCLYL | |
| BoHV-1 | SAFECAVLCLHL | |
| BoHV-5 | SAFECAVLCLHL | |
| EHV-4 | SAFEAAVLCLHL | |
| EHV-1 | SAFEAAVLCLHL | |
| SuHV-1 | SALESAALCLHL | |
| HSV-2 | SAFECAVLCLYL | |
| HSV-1 | SAFECAVLCLYL | |
| CeHV-1 | SALECAVLCLYL | |
| BoHV-2 | SAFECAVICLYM | |
| GaHV-1 | SGFEATVLCLLH | |
| PsHV-1 | SAFEAAVMCLFQ | |
| Betaherpesviruses | ||
| HHV-7 | SDLEAGLCLLTA | |
| HHV-6B | SDLEAALCILAA | |
| HHV-6A | SDLEAALCILAA | |
| HCMV | SDLEAAACLLAA | |
| CCMV | SDLEAAMCLLAA | |
| RhCMV | SDLEAAMCLLAA | |
| TuHV-1 | SDVEAAACLLAA | |
| RCMV | TDLEVAVVLLAA | |
| Gammaherpesviruses | ||
| PLHV-1 | SDLCAGLTIINA | |
| PLHV-2 | SDFCAGLTIINA | |
| PLHV-3 | SDFCAGLTIINA | |
| AlHV-1 | SDFNAALCILNA | |
| SaHV-2 | SDTSSALVILNA | |
| AtHV-3 | SDTNAALVILNA | |
| BoHV-4 | SDTNAALCIING | |
| CeHV-17 | SDTQAALCLLNG | |
| HHV-8 | TDTQAALCLVNA | |
| EHV-2 | SDVNAALCLLNG | |
| MuHV-4 | SDTMSALCLIAA | |
| EBV | PDLQAALILSVA | |
| CeHV-15 | PDLQAALCLSVG | |
| CaHV-3 | PDIHSAMCLSVG | |
| ts1208 | Alphaherpesviruses | |
| GaHV-2 | VDLLNLVYANRN | |
| GaHV-3 | VDLLNIIYASRG | |
| MeHV-1 | VDLLNIIYASRG | |
| CeHV-7 | IDFLTLVYVERS | |
| VZV | VDFITLVYLGRA | |
| BoHV-1 | ADLLAMVYTARA | |
| BoHV-5 | ADLLAMVYTARA | |
| EHV-4 | VDLLTTVFVGRA | |
| EHV-1 | VDLLATVFVSRA | |
| SuHV-1 | LDFLAMVYAARG | |
| HSV-2 | VDLLHMVYAGRG | |
| HSV-1 | VDLLHMVYAGRG | |
| CeHV-1 | VDMLRMVYAGRA | |
| BoHV-2 | VDLLHMVYAGRG | |
| GaHV-1 | VDFLTTSFAIAG | |
| PsHV-1 | VNFVTMAFAASG | |
| Betaherpesviruses | ||
| HHV-7 | PEMIKTFYNNTQ | |
| HHV-6B | GEMINTFFNNAQ | |
| HHV-6A | GEMINTFFNNAQ | |
| HCMV | GEVVNTMFENAS | |
| CCMV | GELVSTMFENAS | |
| RhCMV | GELINTMFENAS | |
| TuHV-1 | GELIETMFGSAQ | |
| RCMV | GEVVDTLYNVGQ | |
| Gammaherpesviruses | ||
| PLHV-1 | EEFIASLYTSPN | |
| PLHV-2 | EEFLSSLYASPN | |
| PLHV-3 | EEFLASLYTSPN | |
| AlHV-1 | REFLSGLYATSA | |
| SaHV-2 | IELVSSLYTNQQ | |
| AtHV-3 | LELVSSLYTNQQ | |
| BoHV-4 | LEFLASLYTQQN | |
| CeHV-17 | LELLPSLYMNQN | |
| HHV-8 | GELMPTLYMNQN | |
| EHV-2 | TEFISGLYTRQS | |
| MuHV-4 | QKIWPMLYLHQQ | |
| EBV | QTLLGNLYGNIN | |
| CeHV-15 | RTLLGNLYGNVH | |
| CaHV-3 | QTFLRNLYATPT |
The amino acid in bold is the site where the ts mutation lies.
The early stage of virion morphogenesis in which the viral DNA is packaged into a protein shell has remarkable similarities to the process in tailed, double-stranded-DNA bacteriophage from the family Caudovirales. With the discoveries that the HSV-1 major capsid protein VP5 has the same structural organization in the capsid floor as members of Caudovirales and that the HSV-1 portal complex has a morphology similar to the structure of bacteriophage portals, evidence is accumulating for the existence of a common ancestor. Although there have been speculations that various bacteriophage capsid-stabilizing proteins may be analogous to UL25, including lambda gpD and gpW and T4 Hoc and Soc, these have different distributions to the predicted location of UL25 on the capsid (2, 5, 19, 27, 40, 60). Given the likely flexible architecture of UL25 and its position on the outside of the capsid, it is probable that UL25 is either a novel herpesviral protein or one that has evolved considerably to adapt to its eukaryotic environment, taking on new roles. To date, UL25 is the only herpesvirus DNA-packaging protein that is known to be important in initiating virus infection, although it is likely that the portal protein is also involved in this process (34).
Acknowledgments
We thank F. Homa for providing the UL28-null mutant gCB, M. Murphy for technical assistance, R. Everett for advice on in situ hybridization, and F. Rixon for polyclonal antibody to VP23 and for electron microscopic analysis of ts1249 capsids. We are grateful to D. McGeoch and R. Everett for critically reading the manuscript.
Footnotes
Published ahead of print on 30 April 2008.
REFERENCES
- 1.Addison, C., F. J. Rixon, J. W. Palfreyman, M. O'Hara, and V. G. Preston. 1984. Characterization of a herpes simplex virus type-1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology 138246-259. [DOI] [PubMed] [Google Scholar]
- 2.Baines, J. D., and S. K. Weller. 2005. Cleavage and packaging of herpes simplex virus 1 DNA, p. 135-150. In C. Catalano (ed.), Viral genome packaging machines. Kluwer Academic/Plenum Publishers, New York, NY.
- 3.Batterson, W., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in the release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booy, F. P., W. W. Newcomb, B. L. Trus, J. C. Brown, T. S. Baker, and A. C. Steven. 1991. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell 641007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman, B. R., R. L. Welschhans, H. Jayaram, N. D. Stow, V. G. Preston, and F. A. Quiocho. 2006. Structural characterization of the UL25 DNA-packaging protein from herpes simplex virus type 1. J. Virol. 802309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardone, G., D. C. Winkler, B. L. Trus, N. Cheng, J. E. Heuser, W. W. Newcomb, J. C. Brown, and A. C. Steven. 2007. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology 361426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, J. T., M. F. Schmid, F. J. Rixon, and W. Chiu. 2007. Electron cryotomography reveals the portal in the herpesvirus capsid. J. Virol. 812065-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coller, K. E., J. I.-H. Lee, A. Ueda, and G. A. Smith. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 8111790-11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham, C., and A. Davison. 1993. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology 197116-124. [DOI] [PubMed] [Google Scholar]
- 10.de Bruyn Kops, A., S. L. Uprichard, M. Chen, and D. M. Knipe. 1998. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in various viral functions. Virology 252162-178. [DOI] [PubMed] [Google Scholar]
- 11.Delboy, M. G., D. G. Roller, and A. V. Nicola. 2008. Cellular proteasome activity facilitates herpes simplex virus entry at a postpenetration step. J. Virol. 823381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai, P., N. A. Deluca, J. C. Glorioso, and S. Person. 1993. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J. Virol. 671357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desloges, N., and C. Simard. 2003. Implication of the product of the bovine herpesvirus type 1 UL25 gene in capsid assembly. J. Gen. Virol. 842485-2490. [DOI] [PubMed] [Google Scholar]
- 14.DiIanni, C. L., J. T. Stevens, M. Bolgar, D. R. O'Boyle II, S. P. Weinheimer, and R. J. Colonno. 1994. Identification of the serine residue at the active site of the herpes simplex virus type 1 protease. J. Biol. Chem. 26912672-12676. [PubMed] [Google Scholar]
- 15.Everett, R. D., and J. Murray. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 795078-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett, R. D., A. Orr, and M. Elliott. 1991. High level expression and purification of herpes simplex virus type 1 immediate early polypeptide Vmw110. Nucleic Acids Res. 196155-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goshima, F., D. Watanabe, H. Takakuwa, K. Wada, T. Daikoku, M. Yamada, and Y. Nishiyama. 2000. Herpes simplex virus UL17 protein is associated with B capsids and colocalizes with ICP35 and VP5 in infected cells. Arch. Virol. 145417-426. [DOI] [PubMed] [Google Scholar]
- 18.Granzow, H., B. G. Klupp, and T. Mettenleiter. 2005. Entry of pseudorabies virus: an imunogold-labeling study. J. Virol. 793200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishii, T., and M. Yanagida. 1977. The two dispensable structural proteins of the T4 phage capsid; their purification and properties, isolation and characterization of the defective mutants, and their binding with the defective heads in vitro. J. Mol. Biol. 109487-514. [DOI] [PubMed] [Google Scholar]
- 20.Jamieson, D. R. S., L. H. Robinson, J. I. Daksis, M. J. Nicholl, and C. M. Preston. 1995. Quiescent viral genomes in human fibroblasts after infection with herpes simplex virus type 1 Vmw65 mutants. J. Gen. Virol. 761417-1431. [DOI] [PubMed] [Google Scholar]
- 21.Jovasevic, V., L. Liang, and B. Roizman. 2008. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J. Virol. 823311-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaelin, K., S. Dezélée, M. J. Masse, F. Bras, and A. Flamand. 2000. The UL25 protein of pseudorabies virus associates with capsids and localizes to the nucleus and to microtubules. J. Virol. 74474-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirkitadze, M. D., P. N. Barlow, N. C. Price, S. M. Kelly, C. Boutell, F. J. Rixon, and D. M. McClelland. 1998. The herpes simplex virus triplex protein, VP23, exists as a molten globule. J. Virol. 7210066-10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klupp, B. G., H. Granzow, G. M. Keil, and T. Mettenleiter. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 806235-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn, J., T. Leege, B. G. Klupp, H. Granzow, W. Fuchs, and T. Mettenleiter. 2008. Partial functional complementation of a pseudorabies virus UL25 deletion mutant by herpes simplex virus 1 pUL25 indicates overlapping functions of alphaherpesvirus pUL25 proteins. J. Virol. 825725-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J. I.-H., G. W. G. Luxton, and G. A. Smith. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 8012086-12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leiman, P. G., S. Kanamaru, V. V. Mesyanzhinov, F. Arisaka, and M. G. Rossman. 2003. Structure and morphogenesis of bacteriophage T4. Cell. Mol. Life Sci. 602356-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luxton, G. W. G., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 1025832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lycke, E., B. Hamark, M. Johansson, A. Krotowil, J. Lycke, and B. Svennerholm. 1988. Herpes simplex infection of the human sensory neuron. An electron microscopic study. Arch. Virol. 10187-104. [DOI] [PubMed] [Google Scholar]
- 30.McClelland, D. A., J. D. Aitken, D. Bhella, D. McNab, J. Mitchell, S. M. Kelly, N. C. Price, and F. J. Rixon. 2002. pH reduction as a trigger for dissociation of herpes simplex virus type 1 scaffolds. J. Virol. 767407-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 691531-1574. [DOI] [PubMed] [Google Scholar]
- 32.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 721060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106167-180. [DOI] [PubMed] [Google Scholar]
- 34.Newcomb, W. W., F. P. Booy, and J. C. Brown. 2007. Uncoating the herpes simplex virus genome. J. Mol. Biol. 370633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newcomb, W. W., F. L. Homa, and J. C. Brown. 2006. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J. Virol. 806286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 7510923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 751427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ojala, P. M., B. Sodeik, M. W. Ebersold, U. Kutay, and A. Helenius. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 204922-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel, A. H., and J. B. Maclean. 1995. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology 206465-478. [DOI] [PubMed] [Google Scholar]
- 40.Perucchetti, R., W. Parris, A. Becker, and M. Gold. 1988. Late stages in lambda head morphogenesis: in vitro studies on the action of the bacteriophage lambda D-gene and W-gene products. Virology 165103-114. [DOI] [PubMed] [Google Scholar]
- 41.Preston, C. M., and M. J. Nicholl. 1997. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J. Virol. 717807-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preston, C. M., A. Rinaldi, and M. J. Nicholl. 1998. Herpes simplex virus type 1 immediate early gene expression is stimulated by inhibition of protein synthesis. J. Gen. Virol. 79117-124. [DOI] [PubMed] [Google Scholar]
- 43.Preston, V. G. 1990. Herpes simplex virus activates expression of a cellular gene by binding in a specific manner to the cell surface. Virology 176474-482. [DOI] [PubMed] [Google Scholar]
- 44.Preston, V. G., J. A. V. Coates, and F. J. Rixon. 1983. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J. Virol. 451056-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Preston, V. G., and I. M. McDougall. 2002. Regions of the herpes simplex virus scaffolding protein that are important for intermolecular self-interaction. J. Virol. 76673-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36857-868. [DOI] [PubMed] [Google Scholar]
- 47.Rixon, F. J., and D. McNab. 1999. Packaging-competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro-assembled procapsids. J. Virol. 735714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J. Virol. 755697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, J. D. 1980. An additional role for the outer nuclear membrane in the morphogenesis of herpes simplex virus. Intervirology 13312-316. [DOI] [PubMed] [Google Scholar]
- 51.Sodeik, B., M. W. Ebersold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1361007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, NY.
- 53.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 7510755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tengelsen, L. A., N. E. Pederson, P. R. Shaver, M. W. Wathen, and F. L. Homa. 1993. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J. Virol. 673470-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW: improving the sensitivity of progressive, multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thurlow, J. K., M. Murphy, N. D. Stow, and V. G. Preston. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 802118-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thurlow, J. K., F. J. Rixon, M. Murphy, P. Targett-Adams, M. Hughes, and V. G. Preston. 2005. The herpes simplex virus type 1 DNA packing protein UL17 is a virion protein that is present in both the capsid and tegument compartments. J. Virol. 79150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trus, B. L., F. P. Booy, W. W. Newcomb, J. C. Brown, F. L. Homa, D. R. Thomsen, and A. C. Steven. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263447-462. [DOI] [PubMed] [Google Scholar]
- 59.Trus, B. L., W. W. Newcomb, N. Cheng, C. Giovanni, L. Marekov, F. L. Homa, J. C. Brown, and A. C. Steven. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA filled HSV-1 capsids. Mol. Cell 26479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang, F., P. Forrer, Z. Dauter, J. F. Conway, N. Cheng, M. E. Cerritelli, A. C. Steven, A. Pluckthun, and Wlodawer. 2000. Novel fold and capsid-binding properties of the lambda-display platform protein gpD. Nat. Struct. Mol. Biol. 7230-237. [DOI] [PubMed] [Google Scholar]