Abstract
Sequencing of plant nuclear genomes reveals the widespread presence of integrated viral sequences known as endogenous pararetroviruses (EPRVs). Banana is one of the three plant species known to harbor infectious EPRVs. Musa balbisiana carries integrated copies of Banana streak virus (BSV), which are infectious by releasing virions in interspecific hybrids. Here, we analyze the organization of the EPRV of BSV Goldfinger (BSGfV) present in the wild diploid M. balbisiana cv. Pisang Klutuk Wulung (PKW) revealed by the study of Musa bacterial artificial chromosome resources and interspecific genetic cross. cv. PKW contains two similar EPRVs of BSGfV. Genotyping of these integrants and studies of their segregation pattern show an allelic insertion. Despite the fact that integrated BSGfV has undergone extensive rearrangement, both EPRVs contain the full-length viral genome. The high degree of sequence conservation between the integrated and episomal form of the virus indicates a recent integration event; however, only one allele is infectious. Analysis of BSGfV EPRV segregation among an F1 population from an interspecific genetic cross revealed that these EPRV sequences correspond to two alleles originating from a single integration event. We describe here for the first time the full genomic and genetic organization of the two EPRVs of BSGfV present in cv. PKW in response to the challenge facing both scientists and breeders to identify and generate genetic resources free from BSV. We discuss the consequences of this unique host-pathogen interaction in terms of genetic and genomic plant defenses versus strategies of infectious BSGfV EPRVs.
Plant pararetroviruses are nonenveloped viruses with a noncovalently closed circular double-stranded DNA of 7 to 8 kbp (11). After infection, open circular viral DNA is released into the nucleus of the cell, where it is converted into supercoiled DNA and associates with histones to form a minichromosome. The viral DNA is then transcribed into mRNA, as well as pregenomic RNA, which is used for DNA replication in the cytoplasm via reverse transcription (23). In contrast to retroviruses, integration of the pararetroviral genome into the host genome is not required for viral replication. Nevertheless, pararetroviral integrations within the host genome exist and are assumed to have originated from illegitimate recombination during the minichromosome phase (53). Such integrants, called endogenous pararetroviruses (EPRVs), range from small, incomplete fragments to larger sequences, and become part of the plant genome via integration in a germinal cell subsequently becoming fixed in the plant population by the evolutionary forces of natural selection and/or genetic drift.
EPRVs are widespread within the plant kingdom. Thus far, the genomes of bitter orange (Poncirus trifoliata), potato (Solanum tuberosum), rice (Oriza sativa), tomato (Lycopersicon sp.), petunia (Petunia sp.), tobacco (Nicotiana sp.), and banana (Musa sp.) have been shown to harbor such integrants (24, 53). In 1999, Jakowitsch et al. (26) described tobacco EPRVs as a novel class of dispersed repetitive elements. EPRV can reach up to a 1,000 copies in tobacco (17, 37). The widespread distribution of EPRVs among plants, and their scattering within the host genome thus results in a discernible impact on host genome shape, plasticity, and evolution.
A surprising discovery was that some EPRVs could release virions. The data on the existence of these infectious EPRVs came from observations of spontaneous viral infection in petunia, tobacco, and banana by Petunia vein clearing virus (PVCV) (42), Tobacco vein clearing virus (TVCV) (34), and Banana streak virus (BSV) (7), respectively. The de novo apparition of these viruses followed stresses, wounding, or tissue culture processes in environments free of vector insects, suggesting that these viruses could only be derived from integrated forms. In 2003, Richert-Poggeler et al. (41) showed that PVCV EPRV (denoted ePVCV) is infectious by demonstrating release of a complete viral DNA genome that contributes to the viral infection. It is important to note that EPRVs, just like their exogenous counterparts, can lead to epidemics and are therefore of considerable economic importance.
BSV is a plant bacilliform pararetrovirus belonging to the family Caulimoviridae and the genus Badnavirus (22). BSV is one of five described viruses of banana (genus Musa) and plantain. This virus causes streak mosaic disease, which had until recently never been considered a serious threat (10). However, in the last 15 years, numerous spontaneous outbreaks of the disease have occurred in all banana-producing areas among promising banana breeding lines and micropropagated interspecific Musa hybrids, all originating from virus-free parents. The origin of these outbreaks was correlated with the presence of EPRVs in the genome of the cultivars. This phenomenon has contributed to the widespread distribution of BSV within banana-producing areas (33). Two types of BSV-related EPRVs have been described thus far in banana. The first type is defined by noninfectious sequences with nonfunctional viral open reading frames (ORFs) containing premature stop codons, frameshift mutations, and/or incomplete viral genomes. Such BSV EPRVs are present in the two most common Musa species from which most cultivated banana is derived: Musa acuminata (denoted A) and Musa balbisiana (denoted B) (16). BSV EPRVs of the other, so-called infectious type contain the complete functional viral genome.
The first tentative description of an infectious BSV EPRV concerns the 5′ part of the integrated species BSV Obino l'Ewai (BSOlV EPRV) present in the genome of the plantain cv. Obino l'Ewai (AAB) (38). This BSV EPRV has a complex structure consisting of noncontiguous back-to-back viral sequences, interrupted by Musa sequences. Although this BSV integrant is not fully described, it contains the entire BSOlV genome at least once. The authors of that study suspected BSOlV EPRV to be pathogenic and hypothesized a mechanism involving two homologous recombination events to release an infectious BSV genome.
Four natural widespread BSV species have thus far been identified as integrants: Banana streak Obino l'Ewai virus (BSOlV), Banana streak Imové virus (BSImV), Banana streak Mysore virus, and Banana streak Goldfinger virus (BSGfV) (16). In banana, abiotic stresses such as micropropagation by in vitro culture processes (7) and genetic hybridization (30) are known to contribute to triggering the production of episomal BSV from EPRVs. Studies on the apparition of BSV after interspecific genetic crosses revealed that at least two factors are involved in BSV expression. The first is the ploidy of the B genome in Musa genotypes. M. balbisiana diploid genotypes (BB) such as cv. Pisang Klutuk Wulung (PKW) and cv. Pisang Batu, which are used as female parents, harbor infectious BSV EPRVs in their genome but are nevertheless resistant to any multiplication of BSV, whether from EPRV activation or from exogenous BSV infection (25, 32). In contrast, genotypes with haploid B genomes harboring BSV EPRVs, such as the triploid hybrids (AAB) arising from interspecific genetic crosses, as well as other natural triploids (AAB cv. Kelong Mekintou and Black Penkelon) (12) or newly created tetraploids (AAAB FHIA 21) (7), can express BSV after stresses and are susceptible to BSV infection. The second factor is a genetic factor called BSV expressed locus (BEL) identified in the triploid (AAB) progeny of interspecific genetic crosses between virus-free diploid M. balbisiana (BB) cv. PKW and tetraploid M. acuminata (AAAA) cv. IDN 110 4x parents (30). In that study the authors characterized the segregation of BSOlV appearance among AAB F1 progeny expressing the disease as a Mendelian monogenic allelic system, strongly regulated by BEL and conferring the role of carrier on the M. balbisiana diploid parent, cv. PKW.
Comparisons with other well-described infectious EPRVs, e.g., PVCV in petunia and TVCV in tobacco, has unfortunately not been very informative up to now in suggesting ways to efficiently manage BSV expression. EPRVs differ considerably in copy number per genome and structure, as well as in their mechanisms of regulation by the host plant. For instance, EPRV expression is repressed by DNA methylation in petunia (39) and tomato (52), whereas this is not the case for BSV EPRVs (M. L. Iskra-Caruana, unpublished data).
Of the three latter pathosystems, BSV/Musa remains the most critical in terms of economic impact. Bananas are the developing world's fourth most important food crop, and three major issues concern BSV EPRVs. First, the main method of propagating banana plantlets is micropropagation by in vitro culture, which can trigger activation of BSV EPRVs. Second, in tropical zones global warming is responsible for strong variations of water regime and thermal amplitude, a well-established activator stress for BSV EPRVs (6). Finally, the numerous infectious BSV EPRVs of different BSV species are restricted to the B genome used in Musa breeding programs as a source of genes of agronomic interest. This consequently reduces considerably the possibility of using genetics to control banana sigatoka leaf spots, the main fungal constraint for the banana crop industry.
Until now, the description of a complete BSV EPRV and a more detailed analysis of the mechanisms of the activation of infectious EPRV have been lacking. To further describe the genetic mechanisms of the regulation of BSV EPRV, we report here the full molecular organization of the pathogenic BSV Goldfinger species (BSGfV) EPRVs in the genome of the wild diploid (BB) M. balbisiana cv. PKW and demonstrate that its integration is the result of a single event.
MATERIALS AND METHODS
Hybridization of BAC clones with BSGfV probe.
Bacterial artificial chromosome (BAC) libraries were obtained from the M. balbisiana wild diploid cv. PKW (43) and two M. acuminata banana plants: the wild diploid cv. Calcutta 4 (AA) (58) and the triploid “Cavendish” subgroup cv. Petite Naine (AAA). These BAC libraries were constructed by partial digestion of genomic DNA with HindIII restriction enzyme and cloning into the pIndigoBAC-5 HindIII cloning-ready and pCC1BAC HindIII cloning-ready vector. BAC DNA was isolated, digested to completion with NotI, and separated, alongside the lambda ladder PFG marker (New England Biolabs, Pickering, Ont.), by PFGE on a 1% (wt/vol) agarose gel in 0.5X TBE under the following conditions: 6 V/cm, switch time 5 to 15 s, and an angle of 120°C for 5 h at 14°C. Clones of the BAC libraries were spotted onto high-density Hybond N+ filters (AP Biotech, Little Chalfont, United Kingdom) by using a Flexys robot. The filters were hybridized with two BSGfV probes: full-length (pCR-TOPO [6,001 bp]) and fragment BSGfV (pCR-TOPO [1,262 bp]), covering the entire viral genome.
Fingerprint: digestion by restriction enzymes and use of Southern blotting.
BAC DNA was digested with five different enzymes (HindIII, EcoRI, BamHI, PstI, and XhoI) to release the BAC fragments. The digested clones were separated on a 0.8% agarose gel in 1× Tris-acetate-EDTA at 60 V, run for 20 h. The separated fragments were denatured and transferred to nitrocellulose membrane Hybond-N+ (Amersham Pharmacia Biotech) (45). Southern hybridization was realized in high-stringency conditions using both full-length or fragments of virus genome probes (45). Filters with the digested BAC clones were hybridized with the two BSGfV probes (pCR-TOPO [1,262 bp] and pCR-TOPO [6,001 bp]).
Sequencing of BAC clones.
Selected BAC clones were sequenced by using the shotgun approach at the National Institute of Agrobiological Sciences. BAC shotgun sequencing was performed by using 2,000 shotgun (2-kb and 5- to 7-kb) clones of 10× coverage and a BigDye terminator kit (ABI) on ABI 3700 sequencers, assembled with Phred/phrap software (8, 9); contig gaps were filled by the primer-extension method when necessary. The GenBank accession numbers were AP009325 and AP009326 for MBP_71C19 and MBP_94I16, respectively.
Sequence annotation.
Each BAC sequence was processed through algorithms for predicting genes (FgenesH for monocot plants [44]; Softberry Software) and Genemark.hmm (35). The BLAST algorithm (1) was used for homology searches against nucleotide and protein databases. Information obtained by the different similarity searches and by the gene prediction programs was imported into the annotation platform Artemis (3) for further manual analysis. Dotter (51), REPuter (28), OligoRep (50; http://wwwmgs.bionet.nsc.ru/mgs/programs/oligorep/), and RepeatMasker (http://repeatmasker.org/) were used to search for repeated sequences. Gene structures and names were manually inspected and refined as necessary. Annotated gene models were scanned for Musa transposable element nucleotide sequences downloaded from GenBank. The BSV integrants sequences were manually annotated based on BSV sequences available in public databases.
Pairwise sequence comparison.
Sequences were aligned with the CLUSTAL W algorithm (56) implemented in BioEdit (18) and corrected manually. Insertion and deletion events were removed prior to nucleotide identity calculation.
Interspecific genetic crosses of cv. PKW with cv. IDN 110 4x.
The plant population used in the present study consisted of 165 F1 allotriploid hybrids (AAB) derived from interspecific genetic crosses between the virus-free wild diploid (BB) M. balbisiana female parent cv. PKW and the virus-free autotetraploid (AAAA) M. acuminata male parent cv. IDN 110 4x confirmed by immunosorbent electron microscopy and by immunocapture PCR (IC-PCR) (30). This genetic cross was fully described and characterized in Lheureux et al. (30). A total of 13% of the progeny was propagated in a vegetative manner to produce duplicates or triplicates of the original hybrid (235 hybrids). Leaf samples were stored at −80°C.
DNA extraction.
Total DNA was extracted by the method described in Gawel and Jarret (14) from leaf tissue of AAB progeny stored at −80°C. The quality and amount of DNA was visually estimated after separation of 5 μl of DNA extraction in a 0.8% agarose gel, staining with ethidium bromide, and visualizing the sample with a UV transluminator.
PCRs.
All PCRs were performed on 5 to 20 ng of template DNA using a common mix composed of 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 0.1 mM each deoxynucleoside triphosphate, 1.5 mM MgCl2, 400 nM of the forward and reverse primers, and 1 U of Taq DNA polymerase (Eurogentec, Seraing, Belgium) in a final volume of 25 μl. DNA was amplified after one cycle at 94°C for 4 min, 35 cycles of 94°C for 30 s, primer annealing at the temperature indicated for 30 s, 72°C for 1 min per kb, and a final extension at 72°C for 10 min. Amplicons were visualized after migration of 8 μl of PCR products on a 1.5% agarose gel in 0.5× TBE (45 mM Tris-borate, 1 mM EDTA [pH 8]). The gel was stained with ethidium bromide, and amplified bands were visualized under UV light.
EPRV genotyping.
For the PCR-restriction fragment length polymorphism (RFLP) DifGf-TaaI method, the primers DifGfF (5′-TTGCAGGAGCAGGAATTACA-3′) and DifGfR (5′-GGATGGAAGATGAGCTCTTTG-3′) (annealing temperature [Ta] = 60°C) amplify both ORF1 and ORF2 regions in BSGfV EPRVs (positions 702 to 1372 in BSGfV AY493509). PCR products (7 μl; 0.2 to 1.5 μg of DNA) were digested with 5 U of TaaI (Fermentas; restriction site 5′-AC,N′GT-3′) in 1× Tango buffer (Fermentas; 33 mM Tris-acetate [pH 7.9], 10 mM magnesium acetate, 66 mM potassium acetate, 0.1 mg of bovine serum albumin/ml) in a final volume of 10 μl. Incubations were performed at 65°C for 2 h. Digested DNA was loaded onto a 2.5% Nusieve (3:1) agarose (Lonza) gel stained with ethidium bromide, and the bands were visualized under UV light. In the multiplex PCR (VV3F/R-VV5F/R) method, the first set of primers (VV3F 5′-TTGCCAAGAATTCCTCCAAG-3′ and VV3R 5′-AAGTTCTTGTCGGCAAGGTG-3′, Ta = 60°C, positions 524 to 543 and 2888 to 2907 in BSGfV) hybridize with both alleles and yield an amplicon of 376 bp. The second set of primers (VV5F 5′-CCATGGAGGTTGACCTGTCT-3′ and VV5R 5′-ACCCCTCTGTCTTCCCAACT-3′, Ta = 60°C, positions 1896 to 1915 and 205 to 224 in BSGfV) hybridize with EPRV-9 only and yield a 628-bp amplification product. The multiplex PCR method generates a 1,012-bp product from the combination of primers VV5F and VV3R that hybridized elsewhere in both allelic EPRVs. In the PCR spe7/spe9bis method, we designed a set of PCR markers specific for Musa flanking regions of EPRV. Sequences of BAC MBP_71C19 and MBP_94I16 were aligned by using CLUSTAL W (56). Insertion and deletion events were detected manually and then used to design PCR primers. A first set of primers located upstream of the viral integration site is specific to EPRV-7 (spe7F [5′-TGGCTACTCGTTTGCCTTTT-3′] and spe7R [5′-CCGTAGCTCTTGTGGCTAGG-3′], Ta = 59°C). A second set is specific to EPRV-9 and is located downstream of the EPRV (spe9bisF [5′-TGATAGAAATACTAAAGATAGCTCATTACA-3′] and spe9bisR [5′-TTTTTGATTATTGCTTCTCTTTTT-3′], Ta = 50°C).
BSV genotyping: IC-multiplex-PCR-RFLP DifGf/Actin-TaaI.
For BSV genotyping, the immunocapture step consisted of coating sterile polypropylene thin-walled 0.2-ml microfuge tubes (Axygen, Union City, CA) for 4 h at 37°C with 25 μl of immunoglobulin G purified from the polyclonal antiserum raised against BSV species and Sugarcane bacilliform virus species, diluted at 2 μg/ml in carbonate coating buffer (15 mM sodium carbonate, 34 mM sodium bicarbonate [pH 9.6]). The tubes were then washed three times with 100 μl of PBT washing buffer (136 mM NaCl, 1.4 mM KH2PO4, 2.6 mM KCl, 8 mM NA2HPO4, 0.05% Tween 20 [pH 7.4]). Plant extracts were prepared by grinding 0.5-g leaf samples in 5 ml of grinding buffer (2% polyvinylpyrrolidone 40, 0.2% sodium sulfite, and 0.2% bovine serum albumin prepared in PBT) using a manual bead grinder and plastic grinding bags (Bio-Rad Phytodiagnostics, Marnes-la-Coquette, France). Portions (1 ml) of plant extracts were transferred to microfuge tubes and clarified by centrifugation at room temperature for 5 min at 7,000 rpm. Then, 25 μl of the supernatant was loaded into coated tubes, followed by incubation for 1 h 30 min at room temperature. The tubes were washed five times with 100 μl of PBT, three times with 100 μl of sterile water, and then dried briefly. Multiplex PCR was carried out directly in tubes using DifGfF and DifGfR primers described above and Actine1F (5′-TCCTTTCGCTCTATGCCAGT-3′) and Actine1R (5′-GCCCATCGGGAAGTTCATAG-3′) primers that amplify the Musa actin housekeeping gene. PCRs were performed as described above at a Ta of 58°C for 25 cycles. A nested PCR using the internal primers VV1F (5′-ACAGCTCCAGGAGATTGGAA-3′) and GfM2 (5′- GAGCTCTTTGAGTCGTCATTG-3′) was then performed at a Ta of 63°C on 2 μl of diluted PCR products (1:100 or 1:1,000). TaaI digestion was subsequently carried out as described above on VV1F-GfM2 PCR products.
RESULTS
Detection of BSGfV EPRVs in the Musa nuclear genome.
In order to exhaustively define the integration patterns of BSGfV in the banana nuclear genome, we used three BAC libraries derived from common cultivars representative of Musa cultivated species: the triploid M. acuminata Cavendish subgroup cv. Petite Naine (AAA), the wild diploid M. acuminata cv. Calcutta 4 (AA), and the wild diploid M. balbisiana cv. PKW (BB). We screened these three BAC libraries for integrated BSGfV by hybridizing with two viral probes covering the complete BSGfV genome. The M. acuminata libraries did not hybridize with either BSGfV probe. Of more than 36,864 BAC clones from the M. balbisiana library, 9 were found to contain BSGfV EPRVs (Table 1). Since the cv. PKW BAC library represents nine genome equivalents of M. balbisiana (43), the very small number of hits indicates low-copy integration.
TABLE 1.
Screen of Musa genomic BAC libraries by hybridization with BSGfV probes
| BSGfV probe (size [bp]) |
M. balbisiana cv. PKW (BB)
|
M. acuminata (no. of hits)
|
||
|---|---|---|---|---|
| No. of hits | BAC clones | cv. Calcutta 4 (AA) | cv. Petite Naine (AAA) | |
| pPCR-TOPO (6,001) | 9 | 30F18, 41K09, 48D15, 64H02, 71C19, 72M20, 73C24, 94I16, 96J15 | 0 | 0 |
| pPCR-TOPO (1,262) | 9 | 30F18, 41K09, 48D15, 64H02, 71C19, 72M20, 73C24, 94I16, 96J15 | 0 | 0 |
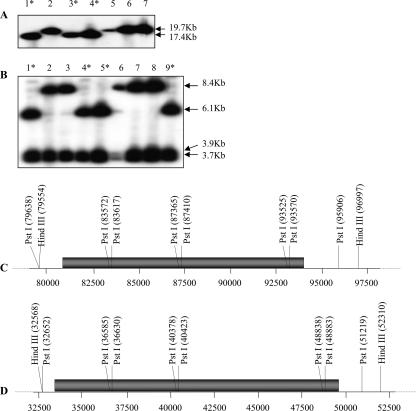
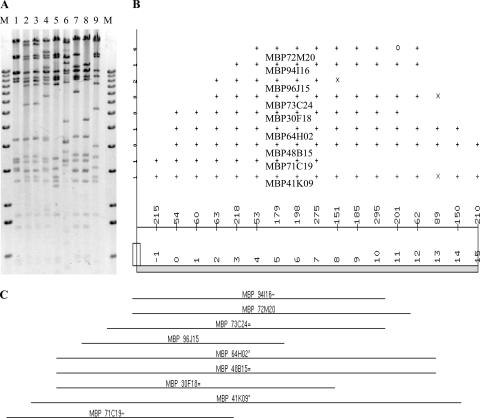
We analyzed the nine BAC clones by using an RFLP and a fingerprint approach in order to establish whether they correspond to different integration events. Two BSGfV probes were used for hybridization. Among the five restriction enzymes tested, HindIII and PstI yielded informative patterns (Fig. 1) and led to the identification of two classes of BSGfV EPRVs in the genome of cv. PKW. We performed physical mapping and contig building of the nine positive BAC clones from the XhoI fingerprint by using FPC software (Fig. 2). All BAC clones cluster in a single contig with a high confidence level (score = 0.991), suggesting a single locus of BSGfV integration in the cv. PKW genome. This result was subsequently confirmed by using a PstI fingerprint (data not shown). We removed incomplete EPRVs by discarding clones with BSGfV EPRV in the boundary regions. For this purpose, we sequenced the ends of each BAC clone (Table 2). Clones MBP_71C19 and MBP_94I16 were selected for sequencing since they were representative of each of the two classes and contained the full BSGfV EPRV present within the BAC. The corresponding GenBank accession numbers are AP009325 and AP009326, respectively.
FIG. 1.
Integration patterns of BSGfV EPRVs in cv. PKW. Fingerprint patterns obtained after digestion of BAC clones containing BSGfV EPRVs with HindIII (A) or PstI (B) and hybridization with two BSGfV probes covering the full-length viral genome are shown. Lanes 1 to 9 show the results obtained with BAC clones containing BSGfV inserts: lane 1, MBP 30_F18; lane 2, MBP 41_K09; lane 3, MBP 64_H02; lane 4, MBP 71_C19; lane 5, MBP 72_M20; lane 6, MBP 73_C24; lane 7, MBP 94_I16; lane 8, MBP 48_D15; and lane 9, MBP 96_J15. Asterisks indicate BAC clones with the same restriction pattern. Deduced restriction maps of BSGfV EPRV in BAC clones MBP 71_C19 (C) and MBP 94_I16 (D) are presented. Numbers refer to the position according to BAC annotation. Gray bars indicate the BSGfV EPRV.
FIG. 2.
Fingerprint contig building. (A) Restriction patterns of nine XhoI-digested BAC clones containing BSGfV integration analyzed with the software Image, version 3.10 (54). Lane M, 1-kb ladder (Invitrogen); lanes 1 to 9, BAC clones (the lane order is the same as in Fig. 1). (B) Consensus band map displayed in FPC version 4.7.9 (49) from the fingerprint analysis in panel A showing the ordering of clones and their fragments. At the top of the panel, the length of each clone is equal to the number of bands in the clone (total length of 17, ranging from −1 to 15). The points represent partially ordered groups: “+” indicates a match with the bottom band within the tolerance, “×” indicates a match within twice the tolerance, and “○” indicates no match. The middle portion of the panel indicates the consensus band numbers. The parameters used were as follows: tolerance, 7; cutoff, 10e-7. (C) Resulting contig of the nine BAC clones. The suffix symbols “*”, “=”, and “∼” represent the status of each clone: the “*” indicates a parent clone, which shares the same common bands with exact child clones (=) or a percentage of the common bands with approximate child clones (∼).
TABLE 2.
Sequencing ends of M. balbisiana cv. PKW BAC clones hybridizing with BSGfV probe
| BAC clone | Probe | Primer | Sequence length (bp) | BLASTN
|
BLASTX
|
||
|---|---|---|---|---|---|---|---|
| Result | E value | Result | E value | ||||
| 30-F18 | BSGfV | Forward | 519 | Oryza sativa genomic DNA, chromosome 1 | 1e.09 | WAK-like kinase (Arabidopsis thaliana) | 2e.40 |
| 30-F18 | BSGfV | Reverse | 372 | No significant similarity found | No significant similarity found | ||
| 64-H02 | BSGfV | Forward | 456 | No significant similarity found | No significant similarity found | ||
| 64-H02 | BSGfV | Reverse | 458 | No significant similarity found | No significant similarity found | ||
| 71-C19 | BSGfV | Forward | 644 | No significant similarity found | No significant similarity found | ||
| 71-C19 | BSGfV | Reverse | 436 | No significant similarity found | No significant similarity found | ||
| 96-J15 | BSGfV | Forward | 676 | No significant similarity found | No significant similarity found | ||
| 96-J15 | BSGfV | Reverse | 346 | No significant similarity found | No significant similarity found | ||
| 41-K09 | BSGfV | Forward | 922 | No significant similarity found | No significant similarity found | ||
| 41-K09 | BSGfV | Reverse | 734 | No significant similarity found | No significant similarity found | ||
| 48-D15 | BSGfV | Forward | 781 | No significant similarity found | Hypothetical protein At2g28370 (Arabidopsis thaliana) | 9e.07 | |
| 48-D15 | BSGfV | Reverse | NAa | NA | NA | NA | NA |
| 72-M20 | BSGfV | Forward | 536 | Calycanthus fertilis chloroplast genome | e.174 | ATPase α subunit (Calycanthus fertilis) | 8e.81 |
| 72-M20 | BSGfV | Reverse | NA | NA | NA | NA | NA |
| 73-C24 | BSGfV | Forward | 559 | No significant similarity found | No significant similarity found | ||
| 73-C24 | BSGfV | Reverse | 480 | Calycanthus fertilis chloroplast genome | 1e.15 | DNA-directed RNA polymerase common soapwort chloroplast | 2e.05 |
| 94-I16 | BSGfV | Forward | 576 | No significant similarity found | No significant similarity found | ||
| 94-I16 | BSGfV | Reverse | 449 | M. acuminata retrotransposon monkey | 8e.07 | No significant similarity found | |
NA, not available.
Structure of BSGfV EPRV-7 and EPRV-9.
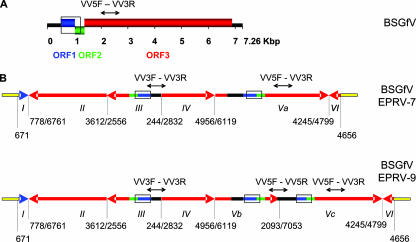
The sizes of the sequenced BAC clones MBP_71C19 and MBP_94I16 are 133,041 and 119,244 bp, respectively. Each carries one copy of the full BSGfV integrant. Figure 3 shows the annotation of EPRV-7 and EPRV-9, named according to their BAC number. The integrants are much longer than a single BSGfV genome (7.26 kb): 13.28 kb for EPRV-7 and 15.58 kb for EPRV-9. The integrant is composed only of viral sequences, with no Musa genome sequences embedded within it. BSGfV EPRVs exhibit a complex rearrangement of viral sequences in the same and opposite orientation relative to the organization of the BSGfV genome. EPRV-7 is comprised of six juxtaposed viral fragments (I to VI), while EPRV-9 carries an additional segment. Both EPRVs are strikingly similar to each other since the first four fragments (I to IV) and the last fragment (VI) display the same structure and size. Although the EPRVs appear as a succession of several fragmented, inversed, and partially repeated BSGfV genomes, Fig. 4 shows that most of the viral regions in EPRV-7 and EPRV-9 (69 and 72%, respectively) are duplicated and therefore present in two or three copies within each EPRV. Most importantly, the entire BSGfV genome (the ORFs and intergenic region) is present at least once in each EPRV.
FIG. 3.
BSGfV EPRV structures in cv. PKW. (A) Organization (linear view) of the BSGfV genome (GenBank AY3509). Blue, green, and red boxes indicate the three ORFs of the virus. The intergenic region is shown in black. (B) Structures of BSGfV EPRV-7 (top) and EPRV-9 (bottom) resulting from annotated BAC clones MBP_71C19 and MBP_94I16, respectively. Arrows indicate the orientation of fragments of the BSGfV genome integrated in the Musa genome, shown in yellow. Blue, green, and red (the same code as used in panel A) refer to the different ORFs. Roman numerals identify the fragment. Numbers below each EPRV indicate the position of the fragment in the BSGfV genome. Open boxes indicate the region used in EPRV genotyping by PCR-RFLP DifGf F/R, and black arrows above the fragments indicate the regions amplified by multiplex-PCR with VV3F/R-VV5F/R.
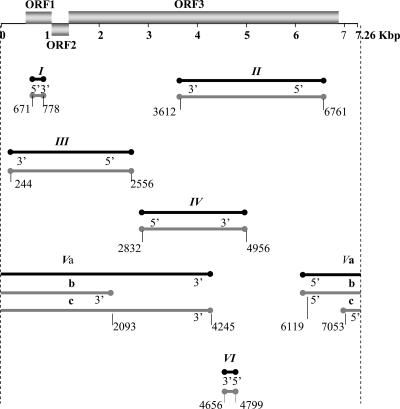
FIG. 4.
Positions of EPRV fragments in the BSGfV genome. The genome of BSGfV is represented as in Fig. 3 (top). Lines below the BSGfV genome represent all of the fragments of EPRV-7 (in black) and EPRV-9 (in gray), and the circles indicate the boundaries of each fragment. Fragment names are indicated above each line. Fragments are arranged relative to their position in the BSGfV genome. The annotations 5′ and 3′ indicate the orientation of the fragments relative to the BSGfV genome.
We then tested whether the high degree of similarity observed within and between EPRVs is confirmed by a similarity in nucleotide sequence. We performed a pairwise comparison of sequences within each EPRV. We chose a sequence of 1,847 bp present in two copies in EPRV-7 and in three copies in EPRV-9, thereby allowing intra- and inter-EPRV comparisons. This fragment is representative of the BSGfV genome since it contains part of the intergenic region, ORF1, ORF2, and the first 748 bp of ORF3 (positions 245 to 2090 of the BSGfV genome). The percentage of nucleotide sequence identity is very high within EPRV-7 (100%) and EPRV-9 (99.7 to 99.8%), as well as between EPRV-7 and EPRV-9 (99.8%) (Table 3). To support this result, we aligned the full sequence of EPRV-7 and EPRV-9 and compared the 13.1 kb of sequences they have in common. Only 28 substitutions differentiate EPRV-7 and EPRV-9, which thus share 99.8% nucleotide sequence identity.
TABLE 3.
Percentage of nucleotide identity within and between EPRVsa
| EPRV | % Nucleotide identity
|
||||
|---|---|---|---|---|---|
| 7 (FIII) | 7 (FVa) | 9 (FIII) | 9 (FVb) | 9 (FVc) | |
| 7 (FIII) | 100 | ||||
| 7 (FVa) | 100 | 100 | |||
| 9 (FIII) | 99.78 | 99.78 | 100 | ||
| 9 (FVb) | 99.78 | 99.78 | 99.67 | 100 | |
| 9 (FVc) | 99.78 | 99.78 | 99.78 | 99.67 | 100 |
Identity within EPRVs is indicated in boldface. A pairwise comparison was made on the same 1,847-bp sequence of BSGfV EPRV. The numbers 7 and 9 refer to EPRV-7 and EPRV-9, respectively. The specific EPRV fragment is indicated in parentheses.
Mutations accumulated in BSGfV EPRVs suggest that EPRV-7 is involved in release of functional BSGfV genomes.
We analyzed the type of mutations that have accumulated in EPRV-7 and EPRV-9 (Table 4). Each EPRV fragment was compared to its corresponding homologous region in the BSGfV genome. Despite the very close similarity (99.3% identity on average) with the BSGfV sequence, the few differences in the EPRV sequences are relevant in terms of functional BSGfV virus released after EPRV activation. EPRV-9 is indeed more distantly related to BSGfV than is EPRV-7 (on average 0.74% versus 0.62% divergence, respectively) and has accumulated 35 more mutations. We next analyzed the mutations found in the three ORFs of the EPRVs (Table 5). Within the coding regions, EPRV-9 has accumulated 18 more substitutions than EPRV-7. EPRV-9 accumulated 11 nonsynonymous substitutions compared to the BSGfV genome, whereas EPRV-7 has only 2. Finally, we found three null mutations in ORFs of EPRV-9, but none in EPRV-7. We observed two substitutions leading to premature stop codons in ORF3 (fragments II and IV) and one adenosine insertion responsible for a frameshift leading to a stop codon in ORF1 (fragment Vc). The quality of the chromatograms of the EPRV-9 sequence confirmed that the three null substitutions exist in the cv. PKW genome and are not due to sequencing errors (data not shown).
TABLE 4.
Number of mutations accumulated on each EPRV fragment compared to the known genome of BSGfV
| Fragment | Size (bp)a | EPRV-7
|
EPRV-9
|
||
|---|---|---|---|---|---|
| No. of mutations | Homology (%) with BSGfV | No. of mutations | Homology (%) with BSGfV | ||
| FI | 101 | 0 | 100 | 0 | 100 |
| FII | 3,152 | 28 | 99.11 | 32 | 98.98 |
| FIII | 2,318 | 11 | 99.53 | 16 | 99.31 |
| FIV | 2,124 | 17 | 99.20 | 19 | 99.11 |
| FVa | 5,445 (5,383) | 46 | 99.15 | ||
| FVb | 3,290 (3,228) | 38 | 98.82 | ||
| FVc | 4,456 (4,455) | 32 | 99.28 | ||
| FVI | 141 | 1 | 99.29 | 1 | 98.99 |
| Total | 103 | 138 | |||
| Mean | 99.38 | 99.26 | |||
The numbers in parentheses refer to the size of the aligned sequences without gaps.
TABLE 5.
Type of mutations accumulated in ORF1, ORF2, and ORF3 of both EPRVs relative to the ORFs of BSGfV genome
| Type | No. of mutations
|
|||
|---|---|---|---|---|
| Shared between ERPV-7 and EPRV-9 | Specific to EPRV-7 | Specific to EPRV-9 | Total | |
| Synonymous substitution | 32 | 1 | 7 | 40 |
| Non synonymous substitution | 15 | 2 | 11 | 28 |
| Premature stop codon | 0 | 0 | 2 | 2 |
| Insertion leading to frameshift | 0 | 0 | 1 | 1 |
| Total | 47 | 3 | 21 | 72 |
Thus, we found strong conservation of all ORFs in each fragment constituting ERPV-7 and significant alterations of the ORFs in EPRV-9. EPRV-7 is therefore more likely than EPRV-9 to be involved in the restitution of infectious episomal BSGfV.
Musa genomic environment of BSGfV integrants in cv. PKW.
As demonstrated above, EPRV-7 and EPRV-9 are very similar in both structure and nucleotide sequence. This observation could be explained either by a duplication of an ancestral ERPV to another locus in the Musa genome or divergence of two allelic EPRVs at the same locus. We first annotated and aligned the two BAC clones MBP_71C19 and MBP_94I16 (GenBank accession numbers AP009325 and AP009326, respectively). An 89.5-kb overlapping region between the two BAC clones with a very high sequence identity (99.7%) was found. The strongly conserved synteny of all genes in this overlapping area is shown in Table 6. These results are consistent with an allelic insertion of BSGfV in cv. PKW, where the two EPRVs are located on homologous chromosomes.
TABLE 6.
Comparative analysis of annotated EPRVsa
| Gene | Annotation | Putative gene position
|
No. of introns/no. of exons | |
|---|---|---|---|---|
| MBP_71C19 | MBP_94I16 | |||
| I | Hypothetical protein | 739-1212 | 1/2 | |
| II | Ribophorin I | 4062-10471 | 6/7 | |
| III | Glycosyl transferase | 11995-14863 | 2/3 | |
| IV | Epsin | 17264-24154 | 12/13 | |
| VI | Putative lysine decarboxylase | 29386-31885 | 6/7 | |
| VII | Pentatricopeptide (PPR) repeat protein | 34768-37419 | 2/3 | |
| 1 | Ty3/gypsy-like retrotransposon, pseudogene | 45714-52790 | 1-5536 | |
| 2 | Hypothetical protein | 55099-55341 | 8116-8358 | 1/2 |
| 3 | Phenylalanine ammonia-lyase | 55781-58032 | 8798-11049 | 1/2 |
| 4 | Hypothetical protein | 58333-59186 | 11349-12202 | 2/3 |
| 5 | Zonadhesin | 60143-67561 | 13159-20577 | 3/4 |
| 6 | Transcriptional regulator related to the mom pseudogene, 3′ part | 75953-78770 | 28967-31784 | 2/1 |
| 7 | Ty3/gypsy retrotransposon, pseudogene, 3′ part | 79531-80691 | 32546-33704 | |
| EPRV | BSGfV integration | 80691-93970 | 33704-49283 | |
| 8 | Ty3/gypsy retrotransposon, pseudogene, 5′ part | 93997-97330 | 49308-52643 | |
| 9 | Transcriptional regulator related to mom, pseudogene, 5′ part | 97779-100187 | 53095-55500 | 4/5 |
| 10 | Putative transposon, pseudogene | 101698-104653 | 57011-59852 | |
| 11 | Hypothetical protein | 107736-111174 | 63069-66507 | 7/8 |
| 12 | Hypothetical protein | 112684-117441 | 68017-72774 | 5/6 |
| 13 | Hypothetical protein | 118473-118806 | 73806-74139 | 2/3 |
| 14 | Auxin-responsive protein | 122614-123216 | 78063-78665 | 1/2 |
| 15 | Actin-depolymerizing factor | 125042-127808 | 80491-83257 | 1/2 |
| 16 | Hypothetical protein | 128615-129077 | 84064-84490 | 1/2 |
| 17 | myb transcriptional factor | 92254-93778 | 2/3 | |
| 18 | Putative wall-associated kinase | 97501-100128 | 2/3 | |
| 19 | Hypothetical protein | 102598-103133 | 2/3 | |
| 20 | Putative wall-associated kinase | 105488-108091 | 2/3 | |
| 21 | Hypothetical protein | 109438-110678 | 1/2 | |
| 22 | Putative wall-associated kinase | 113308-115900 | 2/3 | |
The synteny between BAC clones MBP_71C19 (AP009325) and MBP_94I16 (AP009326) is presented. Putative genes and their positions are given. The middle section of the table (separated by space above and below) represents the overlapping region between both BACs.
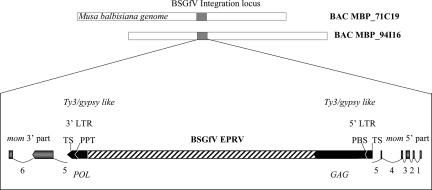
Next, we analyzed the genomic environment of BSGfV integrations in cv. PKW. It was noted that BSGfV integrated into a gene-rich region of the M. balbisiana genome. Surprisingly, a finer annotation of the insertion locus revealed that both BSGfV EPRV-7 and EPRV-9 had integrated in the middle of a Ty3/gypsy retrotransposon (Fig. 5). Although the retroelement ORFs show signs of degradation, we found two long terminal repeats (LTRs; sequence TGTTAG-CTAACA) with a target site (sequence GTGGC) at each side, the primer-binding site (sequence TGGTATCAGAC), and the polypurine track (sequence GAAGAGGACGGG). The 3′ LTR (393 bp) is longer than the 5′ LTR (351 bp) because of a 42-bp insertion in the middle of the LTR. This 42-bp sequence is identical to the 5′ LTR sequence positions 144 to 176. The retroelement itself is integrated in the fifth intron of a mom gene. The two split parts were called mom 3′ and 5′ part on the two Musa BAC clones. However, the N-terminal end of the corresponding protein is missing. The 5′ part of the mom gene might have been disrupted by another pseudogene or putative transposon found upstream.
FIG. 5.
Musa genomic environment of BSGfV EPRV. The orientation of the mom gene putative exons (gray arrows) and regions of the Ty3/gypsy-like retrotransposon (black arrows) are indicated. mom gene introns are numbered and indicated by thin lines. The 3′ and 5′ LTRs are indicated. TS, target site; PBS, primer binding site; PPT, polypurine tract. The regions of the GAG and POL polyproteins (Ty3/gypsy-encoded RT) in the retrotransposon are indicated.
Segregation of EPRV-7 and EPRV-9 in genetic crosses.
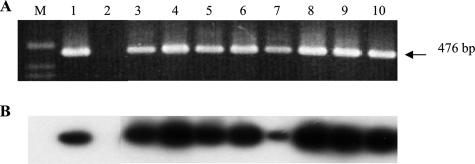
To confirm the allelic EPRV insertion in cv. PKW, we monitored the segregation of BSGfV EPRV-7 and EPRV-9 carried by M. balbisiana cv. PKW (BB) (female parent) in the triploid (AAB) F1 progeny of a genetic cross with M. acuminata cv. IDN 110 4x (AAAA) (male parent). First, we confirmed the absence of BSGfV EPRV within the genomic DNA of M. acuminata cv. IDN 110 4x by PCR amplification using primers specific to ORF3 of BSGfV (Gf F/R) (Fig. 6). As expected, a product of the predicted size (476 bp) is obtained only from total genomic DNA of both cv. PKW (BB) and the entire F1 population. These results were confirmed by Southern blot hybridization. We also confirmed by using a Multiplex-Immuno-Capture PCR (29) that both parents were virus free (data not shown).
FIG. 6.
PCR analysis of cv. PKW and cv. IDN 110 4x and their F1 progeny, using BSGfV specific primers. (A) Ethidium bromide-stained agarose gel analysis of PCR product (GfF [5′-ACGAACTATCACGACTTGTTCAAGC-3′] and GfR [5′-TCGGTGGAATAGTCCTGAGTCTTC-3′]). (B) Southern blot hybridization of the gel shown in panel A using complete genome probes of BSGfV. Lane M, 1-kb ladder; lane 1, cv. PKW; lane 2, IDN 110 4x; lanes 3 to 6, F1 plants showing no sign of banana streak disease; lanes 7 to 10, F1 plants showing symptoms and BSV particles by immunosorbent electron microscopy as described by Lheureux et al. (30). Hybridization was performed according to the method of Sambrook et al. (45) using the two BSGfV probes (pCR-TOPO [1,262 bp] and pCR-TOPO [6,001 bp]).
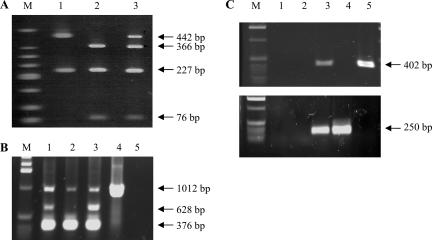
Next, we developed a set of specific molecular markers to genotype each EPRV. Since the two EPRVs are highly similar, we first developed a PCR-RFLP test that enables them to be distinguished due to a single-base substitution. The primer set DifGf F/R amplifies the same fragment of 670 bp containing ORF1 and ORF2, which is present as two copies in EPRV-7 and three copies in EPRV-9 (Fig. 3B). The PCR products from EPRV-7 and EPRV-9 carry different numbers of sites for the restriction endonuclease TaaI (two versus one). Consequently, the PCR-RFLP test can distinguish between EPRV-7 and EPRV-9 and also indicates whether both EPRVs are present in the same genome, as is the case in cv. PKW (Fig. 7A). We subsequently screened the AAB F1 population and observed strict segregation of EPRV-7 and EPRV-9, which were found in 52.82 and 47.18% of the hybrid population, respectively. No heterozygote pattern was observed among the 142 hybrids tested. This result confirmed a monogenic 50-50 segregation (df = 2, X2 = 0.75, P = 0.80) between the two BSGfV EPRVs.
FIG. 7.
Genotyping of BSGfV EPRV-7 and EPRV-9 (A, B, and C) and detection of recombinant EPRV (C). (A) PCR DifGf F/R-RFLP to genotype BSGfV EPRVs in cv. PKW. Endonuclease TaaI discriminates between EPRV-7 and EPRV-9; amplification products carry two versus one restriction sites, respectively. Lane M, DNA ladder (low molecular weight; Invitrogen). Digestion of PCR product DifGfF/R on clone MBP_94I16 carrying EPRV-9 (lane 1), on clone MBP_71C19 carrying EPRV-7 (lane 2), and on M. balbisiana cv. PKW carrying both EPRVs (lane 3) was performed. No amplification product was seen on M. acuminata cv. IDN 110 4x (data not shown). (B) Multiplex PCR with primers VV3 and VV5 for BSGfV EPRV genotyping. Primers VV3F/R amplify a 376-bp product in both EPRVs, primers VV5F/R amplify a 628-bp product in EPRV-9 only, and primers VV5F/R amplify a 1,012-bp product on both EPRVs and the BSGfV circular genome. Lane M, 1-kb ladder (Invitrogen). Lane 1, BAC MBP_94I16; lane 2, BAC MBP_71C19; lane 3, DNA of M. balbisiana cv. PKW; lane 4, DNA of M. acuminata infected by BSGfV; lane 5, PCR negative control. (C) PCR detection of recombination between the two BSGfV EPRVs. PCR results with Spe7F/R (top) specific to EPRV-7 and Spe9bisF/R (bottom) specific to EPRV-9. Lane M, 1-kb ladder (Invitrogen). Lane 1, negative PCR control (water); lane 2, M. acuminata cv. IDN 110 4x genomic DNA; lane 3, M. balbisiana cv. PKW genomic DNA; lane 4, BAC MBP_94I16; lane 5, BAC MBP_71C19.
In 2003, Lheureux et al. (30) demonstrated that a part of the progeny becomes infected de novo by BSV due to the activation of BSV EPRVs and that at least three different BSV species are expressed (data not shown), including the BSGfV studied here. Unfortunately, the PCR primer set DifGf F/R also recognizes the circular genome of BSGfV as a template for amplification. To avoid this cross-reaction, we developed a multiplex PCR using the primers VV3F/R and VV5F/R (Fig. 3B), which are highly specific for EPRV integrants, and able to differentiate EPRV-9 from EPRV-7. The VV5F/R primer set amplifies a 628-bp product with EPRV-9 only (Fig. 7B). The VV3F/R primer set amplifies a 376-bp fragment with both EPRVs and confirms EPRV amplification. In the multiplex PCR, the combination of primers VV5F and VV3R also amplifies a product of 1,012 bp from both EPRV and the circular BSGfV genome. We then screened the AAB population again and found exactly the same results as with the PCR-RFLP test.
Finally, in order to detect the possible recombination of BSGfV EPRVs in the hybrid progeny, we designed two additional PCR markers surrounding each of the two integration sites. The Spe7F/R primer set (Fig. 7C) amplifies a region located 28.2 kb upstream of EPRV-7, and the Spe9bisF/R primer set amplifies a region located 26.8 kb downstream of EPRV-9. None of the progeny showed a recombinant profile with either no amplification or both PCR products in the same individual. This latter genotyping method further confirmed the strict segregation of EPRV-7 and -9 in the progeny. Thus, three experimental approaches independently confirmed that the two BSGfV EPRVs, EPRV-7 and EPRV-9, are located on homologous chromosomes in the genome of M. balbisiana cv. PKW. We conclude that EPRV-7 and EPRV-9 are two alleles of the same locus in cv. PKW.
Which of the two EPRVs, EPRV-7 or EPRV-9, is infectious?
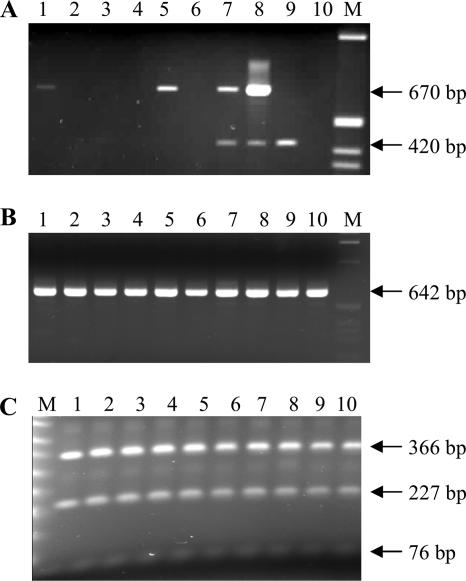
To demonstrate the infectious nature of EPRVs and determine which allele—EPRV-7 and EPRV-9—is infectious, we genotyped the BSGfV particles expressed in the AAB progeny. We developed an IC-multiplex PCR-RFLP method to genotype the molecular EPRV signature of BSGfV particles (Fig. 8). The IC step allows the capture of viral particles by a BSV polyclonal antiserum. Then, a single multiplex PCR specifically amplifies a 670-bp product from immunocaptured BSGfV with the DifGfF/R primers, whereas the primer set Act1F/R amplifying a 420-bp product from Musa housekeeping actin gene monitors the possible residual Musa genomic DNA containing BSV EPRV contaminations (Fig. 8A). A final nested PCR with internal primers increases the quantities of the PCR product (Fig. 8B), allowing an efficient digestion by TaaI endonuclease (Fig. 8C) to a final genotyping of BSGfV viral particles. We screened the 166 F1 hybrids by using this method. Seventeen hybrids were infected by BSGfV (Fig. 8A). There was no amplification of the actin gene, attesting to the amplification of episomal viral genome only. The molecular EPRV signature of viral particles recorded was always the same as for EPRV-7 (Fig. 8C); no viral particle carried the signature of EPRV-9. All 17 infected plants harbor the EPRV-7 allele.
FIG. 8.
Genotyping of BSGfV viral particles in infected hybrids. (A) IC-multiplex PCR allows specific detection of BSGfV particles (DifGfF/R, 670-bp product) and a monitoring of plant DNA contaminations (Actin1F/R, 420-bp product). Lane M, 1-kb ladder (Invitrogen); lanes 1 to 6, coated plant extracts; lanes 7 to 10, plant total DNA. Lanes 1 to 3 show results for F1 AAB hybrids; lanes 4 to 6 show results for the IC control (lane 4, M. balbisiana cv. PKW; lane 5, M. acuminata cv. Grande Naine infected by BSGfV; lane 6, M. acuminata cv. Grande Naine BSGfV-free). Lanes 7 to 10 show results for the PCR control (lane 7, M. balbisiana cv. PKW; lane 8, M. acuminata cv. Grande Naine infected by BSGfV; lane 9, M. acuminata cv. Grande Naine BSGfV-free; and lane 10, water control). (B) Nested PCR using the internal primers VV1F/GfM2 (642-bp product) and increasing PCR product quantity from diluted DifGfF/R PCR product of infected hybrids. Lanes 1 to 10 show the results for AAB F1 hybrids infected with BSGfV; lane M, 1-kb ladder (Invitrogen). (C) TaaI RFLP test (described in Fig. 7A) to genotype the molecular EPRV signature of the viral BSGfV particle. Lanes 1 to 10 show the results for AAB F1 hybrids infected with BSGfV; lane M shows the results for the 50-bp ladder (NEB).
These results demonstrate that allele EPRV-7 only is infectious and is able to release infectious BSGfV, causing systemic infection.
DISCUSSION
Using a high-resolution hybridization method, we demonstrated that BSGfV sequences are integrated only in the genome of M. balbisiana cultivar cv. PKW and are absent from two other common cultivars of M. acuminata tested. This result reinforces previous observations that species of the BSV clade sensu stricto, to which BSGfV belongs, are integrated mainly in the B genome (15, 16), whereas a minority of BSV species, e.g., BSCavV (Iskra-Caruana et al., unpublished) and BSAcVNV (31) are thus far reported as being specific to the A genome.
Only two BSGfV EPRVs, EPRV-7 and EPRV-9, exist in the nuclear genome of cv. PKW, and their integration is unique among the EPRVs described thus far. First, despite the fact that the viral genome appears fragmented, inverted, and partially repeated, surprisingly each EPRV contains the full-length genome of BSGfV. The EPRVs also each contain all of the genetic information needed for “reconstruction” of a functional BSGfV genome very similar to that of the infectious BSGfV virus, with EPRV-7 being the most conserved and showing no evidence of ORF degradation. In the progeny, all hybrids infected with BSGfV harbor EPRV-7, and all BSGfV particles in these hybrids showed an EPRV-7 signature. We therefore demonstrate that EPRV-7 is the infectious EPRV in our pathosystem. Furthermore, EPRV-7 and EPRV-9 are highly similar in general structure and nucleotide sequence and share a common surrounding genomic environment, as detected by contig building from BAC fingerprints, as well as sequencing of BACs carrying the two types of BSGfV EPRVs. This situation either could be due to duplication of an ancestral EPRV in a different locus of the genome or could have originated from a divergence of two EPRVs located on homologous chromosomes. By examining EPRV segregation in interspecific crosses, we demonstrated that EPRV-7 and EPRV-9 are two alleles of the same locus. This integration is therefore the consequence of a single integration event with no subsequent copy number increase. Not only do the two alleles share great similarity of sequence and structure but also only a few mutations differentiate them from the BSGfV genome, thus indicating the integration event to be relatively recent. This situation differs greatly from other previously studied cases of EPRVs. First, the only described integration of infectious EPRV is one of the many ePVCV in the Petunia hybrida genome. This integrant is a tandem direct repeat (i.e., in the same orientation) of the full PVCV genome (41). Second, EPRV sequences of PVCV, TVCV, and several BSV-like species found in petunia, tobacco, and banana (M. acuminata and M. balbisiana), respectively, are highly decayed and are found as numerous small fragments of badnaviral genome, dispersed within the host genome and usually referred to as “dead sequences” (20, 27). Finally, all EPRVs described thus far, whether they contain the full viral genome or not, reach a high copy number in their host genome through a dynamic process of accumulation and elimination (17). EPRV copy number ranges from dozens to several hundreds—as, for example, with BSOlV EPRV in the cultivar Obino l'Ewai (AAB) (21), ePVCV in petunia (41), or LycEPRV in tomato (52)—to thousands for NsEPRV in tobacco (37) and NtoEPRV in Nicotiana tomentosiformis (17).
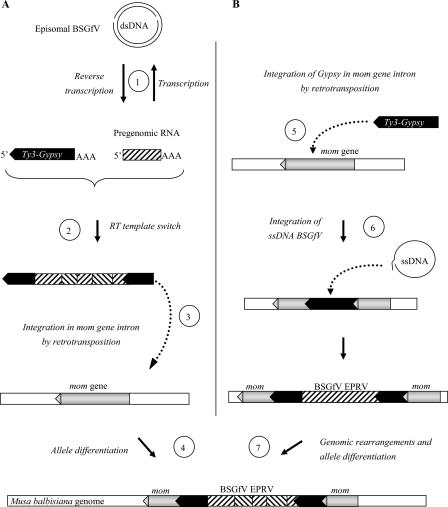
Endogenous viral sequences are a common constituent of many plant genomes (53). Integration generally results from an active mechanism, e.g., retroviral integrases, but this does not apply to pararetroviruses. Indeed, despite the fact that the petunia vein clearing pararetrovirus polyprotein contains two motifs resembling the catalytic domain motifs of integrase (42), no further sequence homology to putative integrase domains of retroelements could be found (20), and no experimental data confirm this function. Instead, plant pararetroviruses are thought to integrate in the host genome via accidental illegitimate recombination during the minichromosome phase. We propose two scenarios to explain the integration process and the final EPRV structure observed, taking into account both the BSGfV insertion locus in Musa chromosomes and the complex structure of EPRVs. One possible scenario (Fig. 9A) assumes recombination at the RNA level between the pregenomic viral RNA resulting from BSGfV infection and the RNA of a retrotranscribing Ty3/gypsy retrotransposon existing in the Musa genome. RNA recombination may originate from a template switch (5, 55) by the Ty3/gypsy reverse transcriptase (RT). An RT template switch between several chimeric pregenomic RNAs could also explain the rearrangements of viral sequences and thus the complex EPRV structure. If the chimeric RNA molecule produced retained its ability to fulfill the retrotranscription process, it could have integrated into the host genome to form the BSGfV integration observed today. In our model, integration of a chimeric Ty3/gypsy-BSGfV transposable element occurred in the fifth intron of the mom gene. Transposition of retrotransposons in gene introns is a frequent phenomenon observed, for example, in mammalian (47) and rice genomes (59). A second possible scenario (Fig. 9B) proposes integration of the Ty3/gypsy retrotransposon into the mom gene intron as a first event, predating integration of BSGfV DNA into the Ty3/gypsy element itself. It is generally acknowledged that integration of viral DNA occurs in the nucleus during viral replication and results from illegitimate recombination after a double-strand break repair. The presence of gaps in the open circular form of pararetroviral DNA may facilitate this mechanism (26). Furthermore, the Ty3/gypsy retrotransposon belongs to the Metaviridae, a family phylogenetically close to the family Caulimoviridae (19). In 2005, Puchta (40) showed that sequence homology, or microhomology, enhances recombination, in this case between badnaviruses and the retrotransposon responsible for integration of the viral genome. EPRVs are found preferentially in heterochromatin rather than euchromatin (52), often colocalizing with retrotransposon sequences, particularly with members of the family Metaviridae (Ty3/gypsy retrotransposons). This general feature was also observed for ePVCV (41), NsEPRV (26), NtoEPRV (17), LycEPRV (52), and BSOlV EPRV (38). However, the exact genomic location of BSGfV EPRV in cv. PKW remains unknown, and in situ hybridization will be required to answer this question.
FIG. 9.
Scenarios for BSGfV integration in the M. balbisiana nuclear genome. (A) Hypothesis 1: use of Ty3/gypsy retroelement for BSGfV integration. Viral RT leads to a pregenomic viral RNA (step 1) during episomal BSGfV infection. A template switch of retrotransposon RT between pregenomic viral RNA and a replicating Ty3/gypsy RNA may form a chimeric RNA (step 2). This step leads to the rearrangements of BSGfV EPRV: fragmentation, inversion, and duplication. Complete retrotransposition of the Ty3/gypsy element leads to its integration (step 3) into the genome of M. balbisiana (within intron 5 of the mom gene). Subsequent genomic change such as recombination may lead to the structural differences observed between the two alleles 7 and 9. (B) Hypothesis 2: Ty3/gypsy retroelement integrated in the fifth intron of the mom gene (step 5) during retrotransposition. Double-strand break repair could account for illegitimate recombination with the single-stranded DNA template generated during BSGfV reverse transcription and integration of the BSGfV genome (step 6). A subsequent genomic change, e.g., recombination, may have lead to both the fragmentation, inversion, and duplication observed in BSGfV EPRV and the structural differences between the alleles 7 and 9 (step 7).
EPRVs are also suspected to be beneficial by inducing viral resistance in the host. Species carrying EPRVs are frequently resistant to the corresponding virus (TVCV, diploids [BB] M. balbisiana), and Maori et al. (36) hypothesized that Israeli acute paralysis virus (dicistrovirus) integration into the honeybee (Apis mellifera) genome could explain bee resistance to this virus. This hypothesis could explain why infectious EPRVs are maintained by natural selection in plants as long as they bring a homology-dependent resistance (gene silencing). In line with this, moderate transcription and subsequent production of small RNAs complementary to ePVCV and LycEPRVs has recently been proved in the genomes of petunia and tomato, respectively (39, 52). Unfortunately, no BSOlV EPRV transcription or small interfering RNAs have been found thus far in cv. PKW (Iskra-Caruana et al., unpublished). Nevertheless, BSGfV EPRV is surrounded by the two LTR sequences of the Ty3/gypsy retrotransposon. LTRs contain promoters that might facilitate the expression of BSGfV EPRV. The RNA transcript from EPRV might undergo subsequent recombination, for instance, using endogenous Musa RT, thereby becoming pathogenic. In this respect, further studies on BSV EPRV expression and the levels of methylation will be required.
Among parasites, EPRVs are unusual pathogens. Each partner interacts at the genetic and genomic level and is engaged in an arms race. Probably in response to their potential harmful effects, natural selection has favored several host defenses against EPRV activation. First, the fragmentation, duplication, and inversion of EPRV sequences potentially decrease the probability that an EPRV can induce the production of a functional and infectious BSGfV genome. Maintaining such disorganization of integrated BSGfV genomes could be an evolving situation of host protection to hamper EPRV activation. Second, DNA and histone methylation are thought to explain the transcriptional silencing observed in ePVCV and LycEPRVs (39, 52). Although cv. PKW appears resistant to both EPRV expression and BSV infection despite harboring infectious EPRV, regulation of EPRV expression by DNA methylation has not been demonstrated, at least for BSOlV in cv. PKW (Iskra-Caruana et al., unpublished). We assume from our results that BSGfV integration in cv. PKW is a recent event from an evolutionary point of view. BSV integration is perhaps too recent for a resistance to BSV based on RNA interference-mediated silencing from expressed EPRVs, like that observed in other plants, to have evolved in Musa plants. From the pathogen point of view, three factors might be linked with the activation of BSGfV EPRV. First, because BSGfV integration is recent, BSGfV EPRVs have not yet evolved into “dead sequences.” The few mutations accumulated within BSGfV EPRV sequences were not numerous enough to result in the decay of viral ORFs in the case of EPRV-7. It is generally acknowledged referring to hypothesis developed from the partial BSOlV EPRV described in the AAB cv. Obino l'Ewai (38) that homologous recombination in the plant genome plays a role in the reconstruction of a BSGfV genome from functional ORFs in EPRVs (13, 24, 46), but the link is not yet firmly established. Next, a strong activation of retroelement transposition due to a release of epigenetic silencing is observed in response to UV exposure, temperature, radiation, wounding, cell culture, and polyploidization (4, 48). Musa hybrids are triploids (AAB), propagated by in vitro culture, and undergo subsequent environmental variation in the field. These stresses can explain why activation was restricted strictly to interspecific hybrids in our study, despite the fact that the genome of M. balbisiana cv. PKW carries infectious EPRVs. Because EPRVs are often found near or embedded in Metaviridae elements, a burst of retroelement transposition might facilitate EPRV transcription and therefore the activation of infectious BSGfV EPRVs. Such EPRV activation in hybrids is also observed for PVCV in P. hybrida and TVCV in Nicotiana tabacum. Lastly, BSGfV and the Ty3/gypsy retrotransposon are found in the fifth intron of a mom Musa gene. Integration of BSGfV and a retrotransposon in the mom gene intron might have disturbed its expression in cv. PKW. This could result in a loss of function of the mom gene, explaining why it subsequently became a pseudogene with decay in its coding sequence. Astonishingly, the artificial disruption of the Arabidopsis thaliana ortholog of the mom gene reactivates the transcription of previously silent genes (2) and repetitive sequences (57). It is therefore tempting to speculate that, as in A. thaliana, mom gene disruption by BSGfV EPRV and the Ty3/gypsy retrotransposon reconstitutes the expression of previously repressed genes. mom gene disruption might facilitate the expression of BSGfV EPRV itself, but also other BSV EPRVs present in the cv. PKW genome, thereby increasing the probability of their activation.
BSV sequences found integrated in the genome of the host banana (genus Musa) are of great concern since several BSV species integrated in M. balbisiana are infectious. Here, we report for the first time the full molecular organization and functional analysis of one such sequence present in the genome of the diploid M. balbisiana cv. PKW (BB). This viral sequence corresponds to the BSV species Goldfinger infectious in interspecific hybrids obtained by genetic crosses involving cv. PKW. Knowledge of the molecular organization of BSV EPRVs in the Musa genome is of crucial interest to researchers and plant breeders in order to overcome problems caused by their presence in banana plant genomes. Actually, the main difficulty comes from the fact that cv. PKW and M. balbisiana genotypes in general harbor at least three other integrated BSV species—BSOlV, BSImV, and Banana streak Mysore virus, each of them with several EPRVs—and that BSOlV and BSImV EPRVs are also infectious (Iskra-Caruana, unpublished). Identifying genetic resources free from BSV EPRVs and producing recombinant Musa genotypes having lost the set of infectious EPRV corresponding to the three BSV species are the challenges currently facing both scientists and breeders. Details of EPRV activation processes, including recombination at the plant DNA level and viral and host factors involved in the production of infectious BSGfV genomes in hybrids, need to be further characterized.
Acknowledgments
Construction of the BAC libraries was supported by CIRAD, Academy of Sciences of the Czech Republic (project A6038201), the International Atomic Energy Agency (research contract 12230/RBF), and the French Ministry of Foreign Affairs (COCOP project 17/01). The study was undertaken as a part of the Global Programme for Musa Improvement (PROMUSA) coordinated by BIOVERSITY (previously named INIBAP). P.G. is supported by a CIRAD/Région Languedoc-Roussillon Ph.D. grant.
We are very grateful to Liying Zhang for providing the two clones of BSGfV; Franc-Christophe Baurens and Stéphanie Sidibe Bocs for help with the Image and FPC softwares; and Kozue Kamiya, Hiroyuki Kanamori, and Takuji Sasaki for performing the sequencing of the two BAC clones (MBP_71C19 and MBP_94I16).
Footnotes
Published ahead of print on 16 April 2008.
REFERENCES
- 1.Altschul, S., W. Gish, W. Miller, E. Myers, and D. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amedeo, P., Y. Habu, K. Afsar, O. M. Scheid, and J. Paszkowski. 2000. Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405203-206. [DOI] [PubMed] [Google Scholar]
- 3.Berriman, M., and K. Rutherford. 2003. Viewing and annotating sequence data with Artemis. Brief Bioinform. 4124-132. [DOI] [PubMed] [Google Scholar]
- 4.Capy, P., G. Gasperi, C. Biemont, and C. Bazin. 2000. Stress and transposable elements: co-evolution or useful parasites? Heredity 85101-106. [DOI] [PubMed] [Google Scholar]
- 5.Cocquet, J., A. Chong, G. L. Zhang, and R. A. Veitia. 2006. Reverse transcriptase template switching and false alternative transcripts. Genomics 88127-131. [DOI] [PubMed] [Google Scholar]
- 6.Dahal, G., D. A. Hughes, G. Thottappilly, and B. E. L. Lockhart. 1998. Effect of temperature on symptom expression and expression and reliability of banana streak badnavirus detection in naturally infected plantain and banana (Musa spp). Plant Dis. 8216-21. [DOI] [PubMed] [Google Scholar]
- 7.Dallot, S., P. Acuna, C. Rivera, P. Ramirez, F. Cote, B. E. L. Lockhart, and M. L. Caruana. 2001. Evidence that the proliferation stage of micropropagation procedure is determinant in the expression of Banana streak virus integrated into the genome of the FHIA 21 hybrid (Musa AAAB). Arch. Virol. 1462179-2190. [DOI] [PubMed] [Google Scholar]
- 8.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8186-194. [PubMed] [Google Scholar]
- 9.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome. Res. 8175-185. [DOI] [PubMed] [Google Scholar]
- 10.Fargette, D., G. Konate, C. Fauquet, E. Muller, M. Peterschmitt, and J. M. Thresh. 2006. Molecular ecology and emergence of tropical plant viruses. Ann. Rev. Phytopathol. 44235-260. [DOI] [PubMed] [Google Scholar]
- 11.Fauquet, C. M., M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball. 2005. Virus taxonomy: Eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, Inc., New York, NY.
- 12.Folliot, M., S. Galzi, N. Laboureau, M.-L. Caruana, P.-Y. Teycheney, and F.-X. Côte. 2005. Risk assessment of spreading Banana Streak Virus (BSV) through in vitro culture. XIIIth International Congress of Virology, San Francisco, CA.
- 13.Gaut, B. S., S. I. Wright, C. Rizzon, J. Dvorak, and L. K. Anderson. 2007. Recombination: an underappreciated factor in the evolution of plant genomes. Nat. Rev. Genet. 877-84. [DOI] [PubMed] [Google Scholar]
- 14.Gawel, N. J., and R. L. Jarret. 1991. A modified CTAB DNA extraction procedure for Musa and Ipomea. Plant Mol. Biol. Reporter 9262-266. [Google Scholar]
- 15.Geering, A. D. W., N. E. Olszewski, G. Dahal, J. E. Thomas, and B. E. L. Lockhart. 2001. Analysis of the distribution and structure of integrated Banana streak virus DNA in a range of Musa cultivars. Mol. Plant Pathol. 2207-213. [DOI] [PubMed] [Google Scholar]
- 16.Geering, A. D. W., N. E. Olszewski, G. Harper, B. E. L. Lockhart, R. Hull, and J. E. Thomas. 2005. Banana contains a diverse array of endogenous badnaviruses. J. Gen. Virol. 86511-520. [DOI] [PubMed] [Google Scholar]
- 17.Gregor, W., M. F. Mette, C. Staginnus, M. A. Matzke, and A. J. M. Matzke. 2004. A distinct endogenous pararetrovirus family in Nicotiana tomentosiformis, a diploid progenitor of polyploid tobacco. Plant Physiol. 1341191-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 4195-98. [Google Scholar]
- 19.Hansen, C., and J. S. Heslop-Harrison. 2004. Sequences and phylogenies of plant pararetroviruses, viruses, and transposable elements. Adv. Bot. Res. Adv. Plant Pathol. 41165-193. [Google Scholar]
- 20.Harper, G., R. Hull, B. Lockhart, and N. Olszewski. 2002. Viral sequences integrated into plant genomes. Annu. Rev. Phytopathol. 40119-136. [DOI] [PubMed] [Google Scholar]
- 21.Harper, G., J. O. Osuji, J. S. P. Heslop-Harrison, and R. Hull. 1999. Integration of Banana streak badnavirus into the Musa genome: molecular and cytogenetic evidence. Virology 255207-213. [DOI] [PubMed] [Google Scholar]
- 22.Hull, R. 1999. Classification of reverse transcribing elements: a discussion document. Arch. Virol. 144209-214. [DOI] [PubMed] [Google Scholar]
- 23.Hull, R., and S. N. Covey. 1995. Retroelements: propagation and adaptation. Virus Genes 11105-118. [DOI] [PubMed] [Google Scholar]
- 24.Hull, R., G. Harper, and B. Lockhart. 2000. Viral sequences integrated into plant genomes. Trends Plant Sci. 5362-365. [DOI] [PubMed] [Google Scholar]
- 25.Iskra Caruana, M. L., F. Lheureux, J. C. Noa-Carrazana, P. Piffanelli, F. Carreel, C. Jenny, N. Laboureau, and B. E. L. Lockhart. 2003. Unstable balance of relation between pararetrovirus and its host plant: the BSV-EPRV banana pathosystem, p. 8. EMBO Workshop: Genomic Approaches in Plant Virology, Keszthely, Hungary.
- 26.Jakowitsch, J., M. F. Mette, J. van der Winden, M. A. Matzke, and A. J. M. Matzke. 1999. Integrated pararetroviral sequences define a unique class of dispersed repetitive DNA in plants. Proc. Natl. Acad. Sci. USA 9613241-13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunii, M., M. Kanda, H. Nagano, I. Uyeda, Y. Kishima, and Y. Sano. 2004. Reconstruction of putative DNA virus from endogenous rice tungro bacilliform virus-like sequences in the rice genome: implications for integration and evolution. BMC Genomics 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurtz, S., and C. Schleiermacher. 1999. REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics 15426-427. [DOI] [PubMed] [Google Scholar]
- 29.Le Provost, G., M. L. Iskra-Caruana, I. Acina, and P. Y. Teycheney. 2006. Improved detection of episomal banana streak viruses by multiplex immunocapture PCR. J. Virol. Methods 1377-13. [DOI] [PubMed] [Google Scholar]
- 30.Lheureux, F., F. Carreel, C. Jenny, B. Lockhart, and M. Iskra-Caruana. 2003. Identification of genetic markers linked to banana streak disease expression in inter-specific Musa hybrids. TAG Theor. Appl. Genet. 106594-598. [DOI] [PubMed] [Google Scholar]
- 31.Lheureux, F., N. Laboureau, E. Muller, B. E. Lockhart, and M. L. Iskra-Caruana. 2007. Molecular characterization of banana streak acuminata Vietnam virus isolated from Musa acuminata siamea (banana cultivar). Arch. Virol. 1521409-1416. [DOI] [PubMed] [Google Scholar]
- 32.Lheureux, F. 2002. Etude des mécanismes génétiques impliqués dans l'expression des séquences EPRVs pathogènes des bananiers au cours de croisements génétiques interspécifiques. Ph.D. thesis. Université Sciences et Techniques du Languedoc, Montpellier, France.
- 33.Lockhart, B., and D. Jones. 2000. Banana streak, p. 263-274. In D. R. Jones (ed.), Diseases of banana, abaca, and enset. CAB International, Wallingford, United Kingdom.
- 34.Lockhart, B. E., J. Menke, G. Dahal, and N. E. Olszewski. 2000. Characterization and genomic analysis of tobacco vein clearing virus, a plant pararetrovirus that is transmitted vertically and related to sequences integrated in the host genome. J. Gen. Virol. 811579-1585. [DOI] [PubMed] [Google Scholar]
- 35.Lukashin, A. V., and M. Borodovsky. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 261107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maori, E., E. Tanne, and I. Sela. 2007. Reciprocal sequence exchange between non-retroviruses and hosts leading to the appearance of new host phenotypes. Virology 362342-349. [DOI] [PubMed] [Google Scholar]
- 37.Mette, M. F., T. Kanno, W. Aufsatz, J. Jakowitsch, J. van der Winden, M. A. Matzke, and A. J. M. Matzke. 2002. Endogenous viral sequences and their potential contribution to heritable virus resistance in plants. EMBO J. 21461-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ndowora, T., G. Dahal, D. LaFleur, G. Harper, R. Hull, N. E. Olszewski, and B. Lockhart. 1999. Evidence that badnavirus infection in Musa can originate from integrated pararetroviral sequences. Virology 255214-220. [DOI] [PubMed] [Google Scholar]
- 39.Noreen, F., R. Akbergenov, T. Hohn, and K. R. Richert-Poggeler. 2007. Distinct expression of endogenous petunia vein clearing virus and the DNA transposon dTph1 in two Petunia hybrida lines is correlated with differences in histone modification and siRNA production. Plant J. 50219-229. [DOI] [PubMed] [Google Scholar]
- 40.Puchta, H. 2005. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 561-14. [DOI] [PubMed] [Google Scholar]
- 41.Richert-Poggeler, K. R., F. Noreen, T. Schwarzacher, G. Harper, and T. Hohn. 2003. Induction of infectious petunia vein clearing (pararetro) virus from endogenous provirus in petunia. EMBO J. 224836-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richert-Poggeler, K. R., and R. J. Shepherd. 1997. Petunia vein-clearing virus: a plant pararetrovirus with the core sequences for an integrase function. Virology 236137-146. [DOI] [PubMed] [Google Scholar]
- 43.Safar, J., J. C. Noa-Carrazana, J. Vrana, J. Bartos, O. Alkhimova, X. Sabau, H. Simkova, F. Lheureux, M. L. Caruana, J. Dolezel, and P. Piffanelli. 2004. Creation of a BAC resource to study the structure and evolution of the banana (Musa balbisiana) genome. Genome 471182-1191. [DOI] [PubMed] [Google Scholar]
- 44.Salamov, A. A., and V. V. Solovyev. 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10516-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Schuermann, D., J. Molinier, O. Fritsch, and B. Hohn. 2005. The dual nature of homologous recombination in plants. Trends Genet. 21172-181. [DOI] [PubMed] [Google Scholar]
- 47.Sironi, M., G. Menozzi, G. P. Comi, M. Cereda, R. Cagliani, N. Bresolin, and U. Pozzoli. 2006. Gene function and expression level influence the insertion/fixation dynamics of distinct transposon families in mammalian introns. Genome Biol. 7R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slotkin, R. K., and R. Martienssen. 2007. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8272-285. [DOI] [PubMed] [Google Scholar]
- 49.Soderlund, C., S. Humphray, A. Dunham, and L. French. 2000. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 101772-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Solovyev, V., A. Zharkikh, and N. Kolchanov. 1985. Context analysis of polynucleotide sequences: methods of detecting non-random repeats. I. Direct repeats in genes of β, β′, σ subunits of Escherichia coli RNA-polymerase. Mol. Biol. 19524-536. (In Russian.) [PubMed] [Google Scholar]
- 51.Sonnhammer, E. L., and R. Durbin. 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167GC1-GC10. [DOI] [PubMed] [Google Scholar]
- 52.Staginnus, C., W. Gregor, M. F. Mette, C. H. Teo, E. G. Borroto-Fernandez, M. L. Machado, M. Matzke, and T. Schwarzacher. 2007. Endogenous pararetroviral sequences in tomato (Solanum lycopersicum) and related species. BMC Plant Biol. 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staginnus, C., and K. R. Richert-Poggeler. 2006. Endogenous pararetroviruses: two-faced travelers in the plant genome. Trends Plant Sci. 11485-491. [DOI] [PubMed] [Google Scholar]
- 54.Sulston, J., F. Mallett, R. Durbin, and T. Horsnell. 1989. Image analysis of restriction enzyme fingerprint autoradiograms. Comput. Appl. Biosci. 5101-106. [DOI] [PubMed] [Google Scholar]
- 55.Temin, H. M. 1993. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc. Natl. Acad. Sci. USA 906900-6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vaillant, I., I. Schubert, S. Tourmente, and O. Mathieu. 2006. MOM1 mediates DNA-methylation-independent silencing of repetitive sequences in Arabidopsis. EMBO Rep. 71273-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vilarinhos, A. D., P. Piffanelli, P. Lagoda, S. Thibivilliers, X. Sabau, F. Carreel, and A. D'Hont. 2003. Construction and characterization of a bacterial artificial chromosome library of banana (Musa acuminata Colla). Theor. Appl. Genet. 1061102-1106. [DOI] [PubMed] [Google Scholar]
- 59.Wang, G. D., P. F. Tian, Z. K. Cheng, G. Wu, J. M. Jiang, D. B. Li, Q. Li, and Z. H. He. 2003. Genomic characterization of Rim2/Hipa elements reveals a CACTA-like transposon superfamily with unique features in the rice genome. Mol. Genet. Genomics 270234-242. [DOI] [PubMed] [Google Scholar]