Abstract
The Ebola virus nucleoprotein (NP) is an essential component of the nucleocapsid, required for filovirus particle formation and replication. Together with virion protein 35 (VP35) and VP24, this gene product gives rise to the filamentous nucleocapsid within transfected cells. Ebola virus NP migrates aberrantly, with an apparent molecular mass of 115 kDa, although it is predicted to encode an ∼85-kDa protein. In this report, we show that two domains of this protein determine this aberrant migration and that this region mediates its incorporation into virions. These regions, amino acids 439 to 492 and amino acids 589 to 739, alter the mobility of Ebola virus NP by sodium dodecyl sulfate-polyacrylamide gel electrophoresis by 5 and 15 kDa, respectively, and confer similar effects on a heterologous protein, LacZ, in a position-independent fashion. Furthermore, when coexpressed with VP40, VP35, and VP24, this region mediated incorporation of NP into released viruslike particles. When fused to chimeric paramyxovirus NPs derived from measles or respiratory syncytial virus, this domain directed these proteins into the viruslike particle. The COOH-terminal NP domain comprises a conserved highly acidic region of NP with predicted disorder, distinguishing Ebola virus NPs from paramyxovirus NPs. The acidic character of this domain is likely responsible for its aberrant biochemical properties. These findings demonstrate that this region is essential for the assembly of the filamentous nucleocapsids that give rise to filoviruses.
Ebola virus is a member of the family Filoviridae, most closely related to the paramyxoviruses among the negative-stranded RNA viruses. Filoviruses are characterized by their enveloped, nonsegmented, and filamentous forms, and these viruses replicate rapidly in monocytes/macrophages, fibroblasts, and endothelial cells during acute infection (7, 10, 24). Ebola virions are characterized by their longitudinal fibers, with forms ∼970 nm in length displaying optimal infectivity (9). They otherwise show a uniform diameter. In previous studies, three viral proteins—Ebola virus nucleoprotein (NP), virion protein 35 (VP35), and VP24—were shown to be necessary and sufficient for the formation of nucleocapsid-containing viruslike structures in transfected cells (13). Although expression of VP40 gives rise to filamentous vesicles when expressed in cells, it does not by itself allow formation of nucleocapsid-containing viruslike particles (2, 11, 15, 19, 26). Transfection of these genes gave rise to nucleocapsids confirmed by transmission electron microscopy, and they appear to be necessary and sufficient for the formation of these particles within cells. In a previous study, NP was found to interact with glycan-binding proteins (lectins) (13), and it was suggested that this posttranslational modification may account for its aberrant migration. A subsequent investigation provided further insight into the structure-function relationship within NP, including the role of the first 450 amino acids in NP-NP interactions and viral assembly (31). These investigators also found biochemical evidence supporting possible posttranslational modification, although its precise nature was not defined. They further showed that the next 150 amino acids (aa) were involved in nucleocapsid formation, but the precise domains responsible for its aberrant migration, and its ability to direct NP or other proteins into virus particles was not examined. To address these unresolved questions, we have localized regions of the NP that account for its aberrant migration and evaluated its requirement for nucleocapsid incorporation into virions by cotransfection with complementary viral genes. We found that the same domain responsible for the aberrant migration of NP is also required for its incorporation into virions, and this region confers this property on heterologous proteins, suggesting that this filovirus-unique region of NP contributes to both the assembly and the morphology of this distinct class of viruses.
MATERIALS AND METHODS
Plasmids.
Plasmids containing NP cDNA (provided by A. Sanchez [23]) were subcloned into the mammalian expression vector pcDNA3 or CMV/R (29) as previously described (13). Truncation and domain swapping mutants were created by mutagenesis PCR (Stratagene). For making the NP-truncated mutations, we used pcDNA3-NP as a template (data not shown). The PCR fragments were digested with BamHI and PmeI and cloned into the same sites of pcDNA3. Measles virus (aa 1 to 401) or respiratory syncytial virus (RSV; aa 1 to 387) NP and COOH-terminal Ebola virus NP (aa 384 to 739) were fused using overlapping PCR and cloned into a CMV/R vector. All mutations and fused constructs were confirmed by sequencing.
Transient transfection, immunoprecipitation, and Western blot analysis.
293T cells were maintained in Dulbecco modified Eagle medium (Invitrogen), supplemented with 10% fetal bovine serum. Plasmid DNAs were purified by using double cesium chloride sedimentation gradients. Approximately 5 × 105 293T cells were placed in six-well dishes 1 day before transfection. Then, 2 μg of each plasmid was used to transfect 293T cells by using the calcium phosphate method (4). Western blotting was performed 12 to 24 h after transfection as described previously (8). Polyclonal antibodies against Ebola virus NP were produced as described in Sullivan et al. (25). Mouse anti-VP24 and anti-VP35 antiserum were also described previously (13). Rabbit anti-β-galactosidase antibody and anti-measles NPs were purchased from Abcam (Cambridge, MA). The anti-Flag monoclonal antibody was from Sigma (St. Louis, MO). The anti-RSV antibody was obtained from Maine Biotechnologies (Portland, ME).
Glycomic analysis of NP.
Reductive elimination and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) searches for putative O-glycans were carried out as described for O-glycosylated proteins (32). Briefly, lyophilized NP (50 μg) was incubated in an aqueous solution of 0.05 M NaOH-1 M NaBH4 at 45°C overnight and desalted through a Dowex 50W-X8(H) column. Excess borates were removed by coevaporation with 10% (vol/vol) acetic acid in methanol under a stream of nitrogen, and the sample was then permethylated, purified on a Sep-Pak cartridge, and analyzed by MALDI-TOF. Mass spectrometry (MS) data were acquired on a Voyager-DE sSTR mass spectrometer (PerSeptive Biosystems) in the reflectron mode with delayed extraction. Permethylated samples were dissolved in 10 μl of 80% (vol/vol) methanol in water, and 1 μl of dissolved sample was premixed with 1 μl of matrix (2,5-dihydroxybenzoic acid) before being loaded onto the MALDI plate.
Proteomic analysis of NP.
NP was digested with cyanogen bromide and trypsin according to our established procedures (5). Pepsin digests were carried out by incubating with pepsin (EC 3.4.23.1; Sigma) at a 50:1 ratio (wt/wt) in 5% formic acid for 4 h at 37°C. The digestion was terminated by lyophilization. Digests were analyzed by nanoscale liquid chromatography-electrospray-MS (nano-LC-ES-MS) and tandem MS (MS/MS) using a reversed-phase nano-HPLC system (Dionex, Sunnyvale, CA) connected to a quadrupole TOF mass spectrometer (Q-STAR Pulsar I; MDS Sciex) as previously described (18).
Biochemical interaction of NP with other Ebola virus proteins.
A total of 2 μg of each expression plasmid (Ebola-NP, chimera measles-Ebola NP, or RSV-Ebola NP) was cotransfected with VP24, VP35, and/or VP40 expression vectors (13) in a 10-cm dish of 293T cells by using the calcium phosphate method (Promega). The wild-type measles NP, RSV NP, and backbone plasmid were used as a control. At 48 h after transfection, the supernatants were clarified by centrifugation at 1,500 rpm at 4°C for 10 min. The cells were washed twice with cold phosphate-buffered saline (PBS) and then lysed with radioimmunoprecipitation assay buffer (150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris-HCl [pH 8.0]) on ice for 30 min. Cell lysates were clarified by centrifuging at 15,000 rpm for 10 min at 4°C. Protein expression was detected by using Western blotting analysis. Protein-protein interactions were assessed by immunoprecipitation using indicated antiserum or antibodies, followed by Western blotting analysis.
Primary structure analysis.
The sequence for Ebola virus NP (Swiss-Prot entry P18272) was analyzed by using the following disorder prediction programs via their respective web servers: PONDR VL-XT (http://www.pondr.com) (16), RONN (http://www.strubi.ox.ac.uk/RONN) (33), DISOPRED2 (http://bioinf.cs.ucl.ac.uk/disopred/disopred.html) (30), and FoldIndex (http://bip.weizmann.ac.il/fldbin/findex) (21). The secondary structure was predicted with PSIPRED (17) by using the DISOPRED2 web server.
MALDI-TOF MS analysis of the COOH-terminal domain of NP (aa 439 to 739).
The COOH terminal fragment of NP was expressed in Escherichia coli and purified by using affinity chromatography. A few microliters of the protein solution (optical density at 220 nm of 0.2) was diluted 10 times in pure water. Then, 1 μl of this solution was mixed with 1 μl of sinapinic acid (10 mg/ml in 0.1% (vol/vol) trifluoroacetic acid-acetonitrile (70:30 [vol/vol]). MS data were acquired in linear mode and in positive ion mode by using a MALDI-TOF Voyager DE-STR instrument (Applied Biosystems, Warrington, United Kingdom). A minimum of 500 shots per spectrum were accumulated. A careful calibration was performed prior and after each spectrum acquisition using bovine serum albumin (BSA) as the standard (catalog no. 2-2158-00; Applied Biosystems). The BSA doubly charged ion has a theoretical mass/charge (m/z) ratio of 33,216. The m/z values obtained in seven different spectra were used to calculate the average m/z values and the corresponding standard deviations. The same approach was applied to the BSA standard. The calculated average of the m/z value of the doubly charged ion of BSA was 33,218 ± 15.
Electron microscopy.
Approximately 3 × 106 293T cells were placed in a 10-cm dish the day before transfection. Then, 2 μg of each plasmid was used to transfect 293T cells using the calcium phosphate method (Promega). The backbone plasmid was used as filler DNA to maintain the same amount of total DNA used. At 48 h after transfection, the cells were lifted from plates by resuspending them with PBS and then pelleted in a 15-ml conical tube by centrifugation at 1,000 rpm for 5 min. The supernatant was removed, and 2 ml of fixing solution was added (3% glutaraldehyde and 3% formaldehyde in 0.1 M sodium cacodylate buffer [pH 7.3]; Tousimis Research Corp., Rockville, MD). The specimens were mixed gently and analyzed in the EM facility at National Cancer Institute-Frederick.
RESULTS
Mapping of the COOH-terminal domains that determine aberrant NP migration.
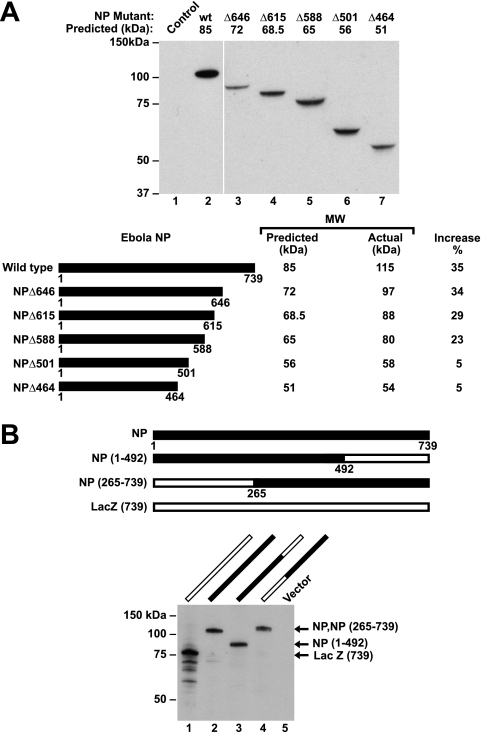
To identify the sequences responsible for the aberrant migration of Ebola virus NP, COOH-terminal deletion mutants were prepared. As progressive truncations were made, the molecular size progressively diminished, and several proteins show a decrease in size beyond the predicted molecular mass of the deletion. For example, a deletion at aa 646 led to reduction of size in excess of 6%, which was subsequently reduced further by deletion to aa 588 (Fig. 1A, lanes 2 versus 3 and 5), but both proteins still migrated with an unexpectedly high apparent molecular weight by SDS-polyacrylamide gel electrophoresis (PAGE). Deletion to residue 501 eliminated much of the aberrant increase in molecular weight (Fig. 1A, lane 7). These data suggested that protein sequence downstream of this amino acid residue contributed in large measure, although not entirely, to the increased apparent molecular mass of this protein.
FIG. 1.
COOH-terminal deletion mapping of NP that determines its aberrant molecular mass, and chimeras of Ebola virus NP and LacZ confirm that COOH-terminal sequences are responsible for increased apparent molecular mass, which can be observed on a heterologous protein. (A) Western blot analysis of NP in cell lysates from 293T cells transfected with NP mutant plasmids reveals that sequences downstream of aa 501 are responsible for increased size (upper panel). The predicted and observed molecular masses of truncated and deleted Ebola virus NP protein are indicated, along with the percent increase (lower panel). NP mutants were separated by 4 to 12% gradient SDS-PAGE, transferred to nitrocellulose filters, and analyzed by immunoblotting with mouse anti-NP as described in Materials and Methods. (B) The upper panel shows a schematic representation of Ebola virus NP deletion mutants, in which the NH2-terminal 265 and COOH-terminal 245 aa of NP were replaced with LacZ. The lower panel shows the results of a Western blot analysis of the indicated chimeric NP and LacZ proteins. Extracts were prepared from transfected 293T cells, separated by 7.5% gradient SDS-PAGE, transferred to nitrocellulose filters, and analyzed by immunoblotting with mouse anti-NP and rabbit anti-β-galactosidase. LacZ has been terminated at this amino acid residue to make it identical in length to NP. Lane numbers refer to expression from the indicated plasmids shown in each panel.
Preparation and analysis of chimeric forms of Ebola virus NP and LacZ.
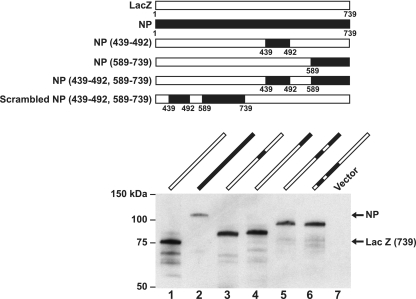
To further define the regions responsible for this effect, chimeric proteins were prepared in which selected COOH- and NH2-terminal domains of NP and LacZ were exchanged. The aberrant migration of NP was markedly reduced by the inclusion of amino acid residues from LacZ at position 492 to the COOH terminus of the molecule (Fig. 1B, lanes 2 versus 3). In contrast, the retention of the COOH terminus of NP resulted in an increased molecular size (Fig. 1B, lane 4), confirming that the COOH-terminal region of NP determined its aberrantly high molecular mass. To define the regions more precisely, additional chimeric proteins were expressed and analyzed by SDS-PAGE (data not shown). This analysis implicated two subdomains within the COOH-terminal domain, one between aa 439 and 510 and another beyond aa 564 (data not shown). The regions were characterized further with chimeric proteins containing regions from aa 439 to 492 and/or aa 589 to 739, inserted into LacZ (Fig. 2, upper panel). These subdomains were placed at different positions within LacZ. Two different molecular mass forms of NP were detected, depending on whether the proximal or distal subdomain of NP was included (Fig. 2, lanes 3 and 4). Inclusion of the proximal aa 439 to 492 region significantly reduced the mobility of the chimeric protein independent of the distal subdomain (Fig. 2, lower panel, lane 1 versus lane 3). Inclusion of the COOH-terminal subdomain from aa 589 to 739 also caused a change in the apparent molecular mass independent of the proximal subdomain (Fig. 2, lower panel, lanes 1 versus lane 4). Finally, the inclusion of both segments together further increased the size compared to the native LacZ protein (Fig. 2, lanes 1 versus lane 5). Notably, the aberrant molecular mass was observed regardless of the position of these two fragments within the heterologous protein sequence, LacZ (Fig. 2, lower panel, lanes 1 versus lane 5 or 6).
FIG. 2.
Two NP domains, aa 439 to 92 and aa 589 to 739, cause aberrant NP migration independent of position within LacZ. Further definition of the COOH-terminal sequences of NP required for aberrant migration on SDS-PAGE with representation of chimeric proteins (upper panel) and analysis of NP/LacZ proteins by Western blotting with mouse anti-NP and rabbit anti-LacZ (lower panel). The lane numbers refer to the plasmids shown schematically in the upper panel.
Analysis of serine and threonine residues within the subdomains responsible for altered mobility.
Previous studies had indicated that NP was able to interact with several lectins, including Datura stramonium agglutinin, which binds specifically to galactose β(1-4)-glucosamine in complex or in hybrid type glycans; Galantus nivalis agglutinin, which recognizes terminal mannose-linked α(1-3)-, α(1-6)-, or α(1-2)-mannose found in N-glycans; and Maackia amurensis agglutinin or Sanbucus nigra agglutinin, which react specifically with α(2-3)-linked or α(2-6)-linked sialic acids, respectively (13). Because such binding can occur through ill-defined, noncarbohydrate determinants, these potential posttranslational modifications were evaluated in two ways. First, glycomic and proteomic experiments were carried out on purified NP protein to search for putative glycans. In the glycomic experiments, NP was subjected to reductive elimination using conditions that are known to efficiently release O-glycans from glycoproteins (32). The products of reductive elimination were permethylated and analyzed by MALDI-TOF MS. This methodology is capable of detecting picomolar levels of glycans. No signals corresponding to O-glycans were observed in the MALDI spectra (data not shown). In contrast, control experiments on human erythrocyte glycophorin yielded spectra containing signals at the expected masses for sialylated O-glycans. These experiments, which cast doubt on the presence of sialylated O-glycans in NP, were corroborated by an exhaustive proteomics study of NP. A variety of proteolytic digests were examined by nano-LC-ES-MS and MS/MS. These included single digestions with cyanogen bromide and pepsin, as well as sequential treatment with cyanogen bromide and trypsin (data not shown). Of the entire amino acid sequence, 739 residues were mapped and shown to lack carbohydrate modifications. The residues in the COOH-terminal portion of NP (residue 401 onward) were confirmed by the proteomics analyses to be nonglycosylated (data not shown). Two stretches of this part of the protein could not be detected, probably because of the absence of proteolytic cleavage sites. They include aa 439 to 470 and aa 561 to 594; both of these segments are rich in Asp and/or Glu and are therefore likely to ionize poorly in proteomics experiments.
To complement the proteomics experiments, specific serine or threonine mutations were made, and migration of these mutants was analyzed by Western blotting. In addition, because proline residues can also alter conformation and mobility, selected proline mutations were made. Within subdomain 1, mutation of all serines and threonines had no effect on mobility, nor did prolines alter migration (data not shown). Similarly, mutations of a variety of serines, threonines, and prolines within the distal subdomain (data not shown) of the protein had no effect on protein mobility. In addition, NP migrates similarly on SDS-PAGE gels after treatment with 6 M urea or boiling in SDS with 2-mercaptoethanol and 6 M urea, where it is monomeric. Furthermore, the COOH-terminal fragment reported here has been expressed alone and shows similar aberrant migration in mammalian cells or in bacteria. MALDI profiling of the E. coli expressed COOH-terminal subdomain (aa 439 to 739) showed that the true molecular mass was consistent with the polypeptide sequence (Fig. 3). Full-length protein expressed in bacteria also has similar migration properties. Finally, the localization of the COOH-terminal subdomains within the filovirus family and in related paramyxoviruses was analyzed. Although some parts of subdomain 1 were retained within a conserved region of the NP protein, to a large extent, the downstream sequence was found within the region unique to filoviruses (Fig. 4).
FIG. 3.
Expanded region of the MALDI-TOF MS spectrum of the COOH-terminal NP domain (aa 439 to 739) acquired in linear positive ion mode. The m/z values of 36,036 and 35,838 are consistent with the expected molecular weight of the polypeptide with or without the N-terminal two amino acids, respectively (theoretical 36,051 Da and also 35,849 Da). The “+/−” numbers correspond to the calculated standard deviations (n = 7).
FIG. 4.
Amino acid sequence similarity among negative-strand RNA viruses and localization of domains 1 and 2 to a region unique to filoviruses.
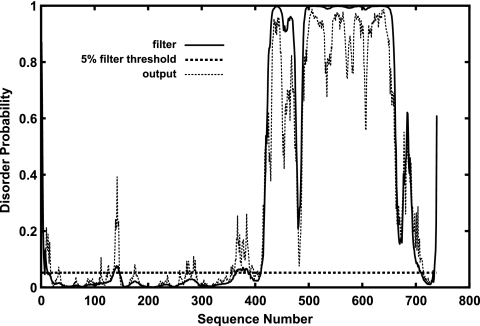
In an effort to further characterize NP, we analyzed the amino acid sequence by using several computer algorithms to predict the secondary structure and intrinsic structural disorder. All four disorder prediction programs we used PONDR (16), RONN (33), DISOPRED2 (30), and FoldIndex (21) predicted that the majority of the COOH-terminal region of NP is structurally disordered, especially residues 424 to 651 (Fig. 5). Consistent with this result, the secondary structure prediction program PSIPRED (17) predicts that residues 399 to 676 in the COOH-terminal half of NP have neither β-sheet nor helical structures, whereas NH2-terminal residues 1 to 398 are predicted to be largely helical, and residues 677 to 739 are predicted to have some β-sheet and helical structure.
FIG. 5.
The disorder probability by residue of Ebola virus NP predicted by DISOPRED (30). Disorder probability (0-1) is plotted on the y axis against the protein sequence (x axis). The dotted horizontal line represents the order or disorder threshold for a false-positive rate of 5%.
Ability of chimeric paramyxovirus NPs fused to COOH-terminal Ebola virus NP to interact with VP35 and VP24 in cells and viruslike particles.
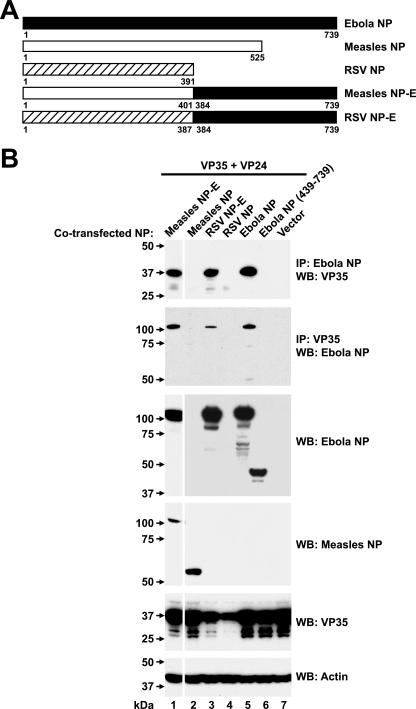
The role of the COOH-terminal sequences of Ebola virus NP in viral assembly was further investigated by preparing chimeric fusion proteins in which the NPs of related paramyxoviruses, measles virus and RSV, were fused at a region homologous to Ebola virus NP. Various chimeric fusion proteins were prepared in the region that was homologous between these proteins, at aa 401 and 387, respectively (Fig. 6A). The ability of these proteins, or the COOH-terminal fragment of Ebola virus NP (aa 439 to 739) alone, to associate with VP35 was examined after cotransfection into 293 cells with VP24 and VP35 expression vectors. Similar to previous studies that showed an association between Ebola virus NP and VP35 in the presence of VP24 (13), wild-type Ebola virus NP associated readily with VP35 (Fig. 6B, lane 5, upper two panels). In contrast, COOH-terminal Ebola virus NP (aa 439 to 738) was unable to interact. However, when this region was appended to either measles NP or RSV NP, the fusion protein, in contrast to the parental NP, was able to bind VP35 by immunoprecipitation and Western blotting (Fig. 6B, lanes 1 and 3 versus lanes 2 and 4). The expression of wild-type RSV NP and chimeric RSV NP with Ebola virus NP COOH-terminal was also confirmed by Western blotting with antisera to RSV (data not shown).
FIG. 6.
Chimeric measles virus or RSV NP fused to COOH-terminal NP interact biochemically with VP35 and VP24 within transfected cells. (A) The indicated measles virus NP or RSV NP fusion proteins were prepared by substituting homologous regions of their NH2-terminal regions to the COOH-terminal domain of Ebola virus NP to generate the indicated expression vectors. (B) The ability of measles virus, RSV, and Ebola virus NPs and chimeras to interact with VP35 was determined by immunoprecipitation of cell extracts with anti-Ebola virus NP (upper panel) or anti-VP35 (second panel), followed by Western blotting for VP35 or NP, respectively, after cotransfection into 293 cells of plasmid expression vectors encoding VP35, VP24, and chimeric measles virus/Ebola virus (lane 1), measles virus NP (lane 2), RSV/Ebola virus NP (lane 3), RSV NP (lane 4), Ebola virus NP (lane 5), COOH-terminal Ebola virus NP (aa 439 to 739) (lane 6), or a negative control (lane 7). Western blot analysis confirmed expression of the Ebola virus NP, measles virus NP, and VP35 antigens in cell extracts relative to a β-actin standard (third to sixth panels, respectively).
To determine whether this interaction occurred in viruslike particles released from cells, Ebola virus NP and measles virus NP-E chimeric proteins were evaluated in comparison to Ebola virus NP (aa 439 to 739) or measles virus NP alone in cells cotransfected with VP24 and VP35 or with VP24, VP35, and VP40. Previous studies have shown that expression of VP40 promotes release of vesicles with filamentous morphology from transfected cells (2, 11, 15, 19, 26). Consistent with these studies, cotransfection of these NPs with VP24 and VP35 failed to release particles into the culture media (Fig. 7A, lanes 1 to 4). In contrast, when cotransfected with VP40, wild-type NP, measles virus NP-E chimeric protein, and COOH-terminal NP (aa 439 to 739) but not measles virus, NP was readily detected in the supernatant associated with VP35 (Fig. 7A, lanes 5 to 8 versus lane 9). The COOH-terminal region of NP was therefore necessary for incorporation of NP into viruslike particles and also conferred this effect on a heterologous, but related nuclear capsid protein lacking this structure.
FIG. 7.
NP chimeras but not measles virus or RSV NP are incorporated into viruslike particles released from cells cotransfected with VP40, VP35, and VP24. (A) Detection of Ebola NP, COOH-terminal Ebola virus NP, and measles virus NP-E, but not measles virus NP in cell supernatants was not observed when these expression vectors were cotransfected with VP24 and VP35 (lanes 1 to 4) but was seen in the presence of cotransfected VP40 (lanes 5 to 8 versus lane 9). The indicated expression plasmids are described in Fig. 6A. The presence of VP35 and VP40 was confirmed by Western blotting with antisera to VP35 (middle panel) or a flag antibody specific for flag-tagged VP40 (lower panel). (B) Biochemical evidence for release of viruslike particles in cell supernatants. Cells were cotransfected with expression vectors encoding GP, NP (Ebola virus NP, NP 439-739, NP 1-450, measles virus NP-E, or measles virus NP, lanes 1 to 5, respectively), VP40, VP35, and VP24, virions purified by sucrose density sedimentation, and analyzed by immunoprecipitation with control sera, NP, or GP, followed by Western blotting (lanes 6 to 10) for Ebola virus NP (upper panel) or VP35 (lower panel). Precipitation with GP reveals associated virion proteins, a finding consistent with the formation of viruslike particles.
To confirm that these proteins interacted biochemically in viruslike particles, virions were purified by using sucrose density gradient sedimentation. Ebola virus GP was included, in addition to the plasmids described above. After purification, particles were immunoprecipitated with anti-GP monoclonal antibodies and analyzed by Western blotting for the presence of the viral NP (Fig. 7B, lanes 1 to 5 and lanes 6 to 10). Consistent with the biochemical analysis in cell culture supernatants and in transfected cells, only Ebola virus NP and measles virus NP-E, in contrast to the Ebola virus 1-450 or measles virus NPs, were found in the viral particle immunoprecipitated with GP (Fig. 7B, lanes 1 and 4). There is an apparent discrepancy between the interaction of VP35 and the carboxy-terminal region of NP in cell extracts (Fig. 6) and its incorporation into virions (Fig. 7), possibly because its inclusion into the particle is facilitated through interactions mediated by additional viral proteins, such as VP40.
Analysis of nucleocapsid formation in viruslike particles by electron microscopy.
To confirm the presence of the nucleocapsid in viruslike particles, electron microscopy was performed on 293L cells cotransfected with VP24, VP35, VP40, and either Ebola virus NP or Ebola virus NP(1-450). Consistent with the biochemical analysis, an electron-dense core with a radial structure, similar to those reported previously (13, 31) showed the presence of a viral nucleocapsid. This density was seen using full-length Ebola virus NP but was not observed in the mutant lacking the COOH-terminal region of Ebola virus NP [Fig. 8A, upper panel, Ebola NP, versus lower panel, Ebola (1-450)]. Although this finding is consistent with the presence of a nucleocapsid in the former but not the latter case, it cannot be considered definitive in the absence of immunogold staining for NP. Similarly, when the measles NP-E expression vector was cotransfected with these plasmids, an electron-dense core was observed within these viruslike particles that was not observed with cotransfection by measles virus NP (Fig. 8B, upper panel, measles NP-E, versus lower panel, measles NP).
FIG. 8.
Electron microscopic confirmation of virus particle formation and requirement of COOH-terminal NP for incorporation of capsid into virions. (A) Electron microscopic imaging of 293L cells cotransfected with expression vectors encoding Ebola virus NP (upper) or Ebola virus NP 1-450 (lower) with VP40, VP35, and VP24. Arrows indicate the presence of capsid proteins in cross-sections seen with full-length NP but not NP 1-450. (B) Images of 293L cells cotransfected with expression vectors encoding measles virus NP-E (upper) or measles virus NP (lower) with VP40, VP35, and VP24. Arrows indicate the presence of capsid proteins in cross-sections seen with measles virus NP-E chimera but not measles virus NP.
DISCUSSION
In this study, we have analyzed the determinants of the NP that determine its aberrant migration. We show that the region responsible for this effect also mediates incorporation of the nucleocapsid into the viral particle, and this property can be conferred to a homologous NP from the measles virus that is not normally incorporated into filoviruses. Specifically, two domains within this COOH terminal domain, one between aa 439 to 492 and another at aa 589 to 739, are responsible for its unusual biochemical properties. We suggest that its unexpected migration on SDS-PAGE gels is likely due to the high acidic content of the two domains. Informatic analysis of the protein suggests such behavior. In addition, extensive analysis of the COOH-terminal domain shows no evidence of posttranslational modification, including evaluation of all peptides by MALDI-TOF MS.
Previous studies have shown that NP migrates aberrantly by SDS-PAGE (3, 13, 31), but the biochemical basis of this effect is not well understood. A previous report of phosphorylation of Marburg virus NP may suggest Ebola virus NP is also phosphorylated and account for this size change (3). However, this protein is not substantially phosphorylated, and such phosphorylation is unlikely to cause substantial changes in the migration of this protein. The possibility of posttranslational carbohydrate modification has been raised in two independent studies (13, 31). The finding that [3H]glucosamine could be incorporated into the protein further supported this possibility. However, lectin binding has been observed in the absence of carbohydrate modification, and in vitro labeling may be nonspecific.
Glycomic and proteomic analysis failed to identify distinct posttranslational modification sites. Instead, we find that there are two discontinuous subdomains that give rise to this aberrant migration, which are notable for their highly charged acidic character. Point mutations of a variety of amino acids in these regions suggest that neither serines nor threonines nor prolines within these affected regions were responsible for aberrant migration. In addition to posttranslational modifications, acidic domains have also been shown to cause aberrant migration of proteins. For example, in an early observation of this phenomenon, a 52-kDa component of the U1 small nuclear ribonucleoprotein was found to migrate on PAGE at 70 kDa (22). Its aberrant electrophoretic migration was attributed to a carboxy-terminal charged domain. Similarly, the anomalous electrophoretic behaviors of the human papillomavirus type 16 E7 protein and the DNA repair protein XPA were found to be due to the high content of acidic amino acid residues (1, 14). Within the two subdomains of NP that cause aberrant migration, such acidic regions comprise ca. 30% of the sequence, well beyond the remainder of this protein, ca. 10%, or of other proteins in general. In addition, this region of the protein is predicted to show significant disorder (Fig. 5). Many proteins are now known to require regions of disorder for function, particularly when molecular recognition is used (27, 28). Indeed, recent bioinformatics studies have revealed a small but significant correlation between intrinsic structural disorder and participation in multiple protein interactions (12, 20). Such disordered regions generally have a higher percentage of polar and charged residues, as well as Gly, Ala, and Pro. Despite their lack of rigidly defined structure, disordered regions of proteins involved in macromolecular interactions can have numerous advantages over structured regions such as more rapid association rates, lower entropy losses upon binding, greater topological flexibility, the potential to use shorter amino acid sequences for molecular recognition, the flexibility to recognize multiple partners depending on the microenvironment, and the ability to provide entropic exclusion of other macromolecules in spacer regions. Furthermore, many, although not all, disordered regions are induced to fold into distinct structures upon interaction with target molecules (6). The prediction of a large intrinsically disordered region in the COOH-terminal end of an NP with high acidic content is thus consistent with NP's role as a critical component of a large macromolecular assembly involving VP35 and VP24. These results suggest that the COOH-terminal region of NP may play a direct role in the assembly of the nucleocapsid.
The filoviruses are characterized by their filamentous nucleocapsids, a feature not observed with uniformity in their most closely related phylogenetic class of viruses, the paramyxoviruses. Interestingly, the region responsible for this aberrant migration is lacking in the paramyxoviruses. When the COOH-terminal region is fused to either measles or RSV NP, it associated in nucleocapsids with VP35 and was incorporated into viruslike particles by biochemical analysis. This finding suggests that these Ebola virus subdomains are required for incorporation of NP into the virion. Taken together, these data indicate that the subdomains of the Ebola virus NP responsible for its aberrant biochemistry contribute to NP localization in the virus and may help to determine the unique characteristics and morphology of this class of viruses.
Acknowledgments
We dedicate this study to the memory of Yue Huang, who passed away unexpectedly and tragically in 2006. At the time, he was a faculty member at the Pasteur Institute in Shanghai, China.
We thank Ati Tislerics for assistance with manuscript preparation, Brenda Hartman and Michael Cichanowski for figure preparation, and members of the Nabel lab for helpful discussions and advice.
This research was supported by the Intramural Research Program of the National Institutes of Health, Vaccine Research Center, National Institute of Allergy and Infectious Disease. The Imperial College work was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) and the Wellcome Trust. A.D. was a BBSRC Professorial Fellow.
Footnotes
Published ahead of print on 16 April 2008.
REFERENCES
- 1.Armstrong, D. J., and A. Roman. 1993. The anomalous electrophoretic behavior of the human papillomavirus type 16 E7 protein is due to the high content of acidic amino acid residues. Biochem. Biophys. Res. Commun. 1921380-1387. [DOI] [PubMed] [Google Scholar]
- 2.Bavari, S., C. M. Bosio, E. Wiegand, G. Ruthel, A. B. Will, T. W. Geisbert, M. Hevey, C. Schmaljohn, A. Schmaljohn, and M. J. Aman. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, S., S. Huppertz, H.-D. Klenk, and H. Feldmann. 1994. The nucleoprotein of Marburg virus is phosphorylated. J. Gen. Virol. 75809-818. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 72745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dell, A., H. R. Morris, R. L. Easton, M. Panico, M. Patankar, S. Oehniger, R. Koistinen, H. Koistinen, M. Seppala, and G. F. Clark. 1995. Structural analysis of the oligosaccharides derived from glycodelin, a human glycoprotein with potent immunosuppressive and contraceptive activities. J. Biol. Chem. 27024116-24126. [DOI] [PubMed] [Google Scholar]
- 6.Dyson, H. J., and P. E. Wright. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell. Biol. 6197-208. [DOI] [PubMed] [Google Scholar]
- 7.Feldmann, H., H. Bugany, F. Mahner, H. D. Klenk, D. Drenckhahn, and H. J. Schnittler. 1996. Filovirus-induced endothelial leakage triggered by infected monocytes/macrophages. J. Virol. 702208-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402889-894. [DOI] [PubMed] [Google Scholar]
- 9.Geisbert, T. W., and P. B. Jahrling. 1995. Differentiation in filoviruses by electron microscopy. Virus Res. 39129-150. [DOI] [PubMed] [Google Scholar]
- 10.Geisbert, T. W., P. B. Jahrling, M. A. Hanes, and P. M. Zack. 1992. Association of Ebola-related Reston virus particles and antigen with tissue lesions of monkeys imported to the United States. J. Comp. Pathol. 106137-152. [DOI] [PubMed] [Google Scholar]
- 11.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 9713871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haynes, C., C. J. Oldfield, F. Ji, N. Klitgord, M. E. Cusick, P. Radivojac, V. N. Uversky, M. Vidal, and L. M. Iakoucheva. 2006. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, Y., L. Xu, Y. Sun, and G. J. Nabel. 2002. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol. Cell 10307-316. [DOI] [PubMed] [Google Scholar]
- 14.Iakoucheva, L. M., A. L. Kimzey, C. D. Masselon, R. D. Smith, A. K. Dunker, and E. J. Ackerman. 2001. Aberrant mobility phenomena of the DNA repair protein XPA. Protein Sci. 101353-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jasenosky, L. D., G. Neumann, I. Lukashevich, and Y. Kawaoka. 2001. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 755205-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, X., P. Romero, M. Rani, A. K. Dunker, and Z. Obradovic. 1999. Predicting protein disorder for N-, C-, and internal regions. Genome Inform. Ser. Workshop Genome Inform. 1030-40. [PubMed] [Google Scholar]
- 17.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16404-405. [DOI] [PubMed] [Google Scholar]
- 18.Morris, H. R., S. Chalabi, M. Panico, M. Sutton-Smith, G. F. Clark, D. Goldberg, and A. Dell. 2007. Glycoproteomics: past, present, and future. Int. J. Mass Spectrom. 25916-31. [Google Scholar]
- 19.Noda, T., H. Sagara, E. Suzuki, A. Takada, H. Kida, and Y. Kawaoka. 2002. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 764855-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patil, A., and H. Nakamura. 2006. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 5802041-2045. [DOI] [PubMed] [Google Scholar]
- 21.Prilusky, J., C. E. Felder, T. Zeev-Ben-Mordehai, E. H. Rydberg, O. Man, J. S. Beckmann, I. Silman, and J. L. Sussman. 2005. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 213435-3438. [DOI] [PubMed] [Google Scholar]
- 22.Query, C. C., R. C. Bentley, and J. D. Keene. 1989. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell 5789-101. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez, A., M. P. Kiley, B. P. Holloway, and D. D. Auperin. 1993. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 29215-240. [DOI] [PubMed] [Google Scholar]
- 24.Schnittler, H. J., F. Mahner, D. Drenckhahn, H. D. Klenk, and H. Feldmann. 1993. Replication of Marburg virus in human endothelial cells: a possible mechanism for the development of viral hemorrhagic disease. J. Clin. Investig. 911301-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan, N. J., A. Sanchez, P. E. Rollin, Z.-Y. Yang, and G. J. Nabel. 2000. Development of a preventive vaccine for Ebola virus infection in primates. Nature 408605-609. [DOI] [PubMed] [Google Scholar]
- 26.Timmins, J., S. Scianimanico, G. Schoehn, and W. Weissenhorn. 2001. Vesicular release of Ebola virus matrix protein VP40. Virology 2831-6. [DOI] [PubMed] [Google Scholar]
- 27.Tompa, P. 2005. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 5793346-3354. [DOI] [PubMed] [Google Scholar]
- 28.Tompa, P., and M. Fuxreiter. 2008. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 332-8. [DOI] [PubMed] [Google Scholar]
- 29.Tooze, J., S. Tooze, and G. Warren. 1988. Site of Addition of N-acetyl-galactosamine to the E1 glycoprotein of mouse hepatitis virus-A59. J. Cell Biology. 1061475-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward, J. J., J. S. Sodhi, L. J. McGuffin, B. F. Buxton, and D. T. Jones. 2004. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337635-645. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe, S., T. Noda, and Y. Kawaoka. 2006. Functional mapping of the nucleoprotein of Ebola virus. J. Virol. 803743-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong, N. K., R. L. Easton, M. Panico, M. Sutton-Smith, J. C. Morrison, F. A. Lattanzio, H. R. Morris, G. F. Clark, A. Dell, and M. S. Patankar. 2003. Characterization of the oligosaccharides associated with the human ovarian tumor marker CA125. J. Biol. Chem. 27828619-28634. [DOI] [PubMed] [Google Scholar]
- 33.Yang, Z. R., R. Thomson, P. McNeil, and R. M. Esnouf. 2005. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 213369-3376. [DOI] [PubMed] [Google Scholar]