Abstract
The human parainfluenza virus type 2 (hPIV2) V protein plays important roles in inhibiting the host interferon response and promoting virus growth, but its role in hPIV2 replication and transcription is not clear. A green fluorescent protein (GFP)-expressing a negative-sense minigenomic construct of hPIV2 has been established by standard technology, with helper plasmids expressing the nucleocapsid protein (NP), phosphoprotein (P), and large RNA polymerase (L) protein, to examine the role of V protein. We found that the simultaneous expression of wild-type V protein in the minigenome system inhibited GFP expression, at least in part, by inhibiting minigenome replication. In contrast, expression of C terminally truncated or mutant hPIV2 V proteins had no effect. Moreover, the V protein of simian virus 41, the rubulavirus most closely related virus to hPIV2, also inhibited GFP expression, whereas that of PIV5, a more distantly related rubulavirus, did not. Using these other rubulavirus V proteins, as well as various mutant hPIV2 V proteins, we found that the ability of V protein to inhibit GFP expression correlated with its ability to bind to L protein via its C-terminal V protein-specific region, but there was no correlation with NP binding. A possible role for this inhibition of genome replication in promoting viral fitness is discussed.
Human parainfluenza virus type 2 (hPIV2) is a member of the Rubulavirus genus of the family Paramyxoviridae. This family includes many well-known human and animal pathogens, such as Sendai virus (SeV), hPIV types 1 to 4, simian virus 41 (SV41), parainfluenza virus type 5 (PIV5; formerly known as SV5), mumps virus, Newcastle disease virus, measles virus (MeV), and respiratory syncytial virus, as well as important emerging viruses such as Hendra and Nipah viruses. The negative-stranded RNA genome of hPIV2 is 15,654 nucleotides long and encodes seven viral proteins from six genes (30). The nucleocapsid protein (NP), phosphoprotein (P), and large RNA polymerase (L) protein are important for transcription and replication of the viral RNA genome. All viruses of the Paramyxoviridae (with the notable exception of hPIV1) contain an mRNA-editing site at which G residues are inserted into the P gene mRNA in a programmed manner during its synthesis. In respiroviruses and morbilliviruses, the P mRNA is a faithful copy of the genome RNA, and the V mRNA results from the insertion of one additional pseudotemplated G nucleotide. In only rubulaviruses, it is the V mRNA that is a faithful transcript of the V/P gene, whereas the P mRNA is synthesized through a cotranscriptional insertion of two pseudotemplated G residues. Thus, the N-terminal 164 amino acids (aa) of the V and P proteins are common, while their C termini are unique (43). Since insertion of the G residues in hPIV2 occurs ca. 50% of the time, roughly equal amounts of V and P mRNAs are produced. The C termini of the V proteins contain seven invariant cysteines that bind two atoms of zinc and is ca. 50% identical in sequence among all paramyxoviruses (30, 47). The structure of the PIV5 V protein has recently been reported (31).
The hPIV2 V protein appears to be multifunctional. As summarized in Table 1 and Fig. 6A, the V protein has two NP-binding sites: the N-terminal 47 aa on the P/V common region (42, 61) and the C-terminal 50 aa on the V-specific region (35). It also has a V-oligomerization domain on the C-terminal 28 aa of the V-specific region (35) and shows a diffuse nuclear and cytoplasmic distribution in infected cells. In contrast, the P protein has two independent NP-binding sites, aa 1 to 47 and aa 357 to 395, and a P-multimerization domain, aa 211 to 248. P protein is organized in numerous granules with the NP protein in the cytoplasm of infected cells. P protein granule formation is due to the binding between residues 357 to 395 on the C-terminal domain of P protein and residues 295 to 400 of the NP, presumably of assembled nucleocapsids (40, 41, 42). It is presumed that the P protein forms a complex with both unassembled NP (soluble NP, NP0) and assembled NP (NP in helical nucleocapsids, NPNC), but that the V protein forms a complex only with NP0, similar to SeV and PIV5 V proteins (21, 48).
TABLE 1.
Summary of the various properties of the hPIV2 V protein mutants
| Mutation | IFN signaling block | Virus growth in Vero cells | Interaction of the:
|
V protein oligomerization | |
|---|---|---|---|---|---|
| N terminus of V protein with NP (aa 401 to 493) | C terminus of V protein with NP (aa 1 to 82) | ||||
| wt | + | ++ | +++ | +++ | + |
| F143S | - | ++ | +++ | +++ | + |
| W178H/W182E | - | + | +++ | - | - |
| C193/197A | - | + | +++ | ++ | + |
| C209/211/214A | - | + | +++ | + | - |
| C218/221A | - | + | +++ | + | - |
FIG. 6.
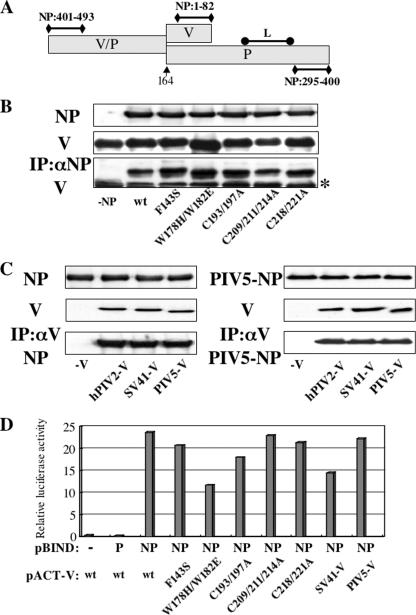
Analysis of interactions between NP and V proteins by immunoprecipitation. (A) Schematic diagram of P/V regions required for binding to NP and L proteins previously identified. The numbers show amino acid residues on the NP protein. The arrow marks the editing site. (B) BSR T7/5 cells were transfected with plasmids expressing hPIV2 NP and mutant hPIV2 V proteins. At 48 hpt, the cell extracts were either analyzed directly by Western blot analysis (anti-NP; upper panel, anti-V; middle panel) or immunoprecipitated with anti-NP before Western blot analysis (anti-V; lower panel). The asterisk on the right indicates the immunoglobulin light chain. (C) BSR T7/5 cells were transfected with plasmids expressing hPIV2 (left panel) or PIV5 (right panel) NP and rubulavirus V proteins. At 48 hpt, the cell extracts were either analyzed directly by Western blot analysis (anti-NP, upper panel; anti-V and PIV5-V, middle panel) or immunoprecipitated with anti-V or PIV5-V before Western blot analysis (anti-NP, lower panel). (D) Mammalian two-hybrid analysis for NP and various V proteins. COS cells were cotransfected with pBIND and pACT plamids together with luciferase reporter plasmid. After 48 h, the cells were harvested and assayed by dual-luciferase reporter assay system. The pACT-V wt and mutant constructs refer to the hPIV2 V proteins; those of the other virus V proteins are specified.
Many paramyxoviruses have evolved specific proteins that inhibit the interferon (IFN)-induced antiviral responses through direct inhibition of cellular STAT proteins. The V proteins of MeV (a morbillivirus) and the Nipah and Hendra viruses (henipaviruses) inhibit IFN signaling by preventing STAT1 and STAT2 nuclear accumulation (44, 49, 50, 55). The V proteins of most rubulaviruses, such as PIV5, SV41, and mumps virus, as well as an avulavirus, Newcastle disease virus, block IFN signaling by targeting STAT1 for degradation (2, 10, 11, 22, 29, 38, 39, 46, 59, 60, 63, 64), whereas the V protein of hPIV2 targets STAT2 for degradation (38, 39, 45). The essential residues of hPIV2 V protein needed to block IFN signaling are summarized in Table 1. SeV and hPIV3 (respiroviruses) also block IFN signaling, and this anti-IFN ability has been shown to be a property of their C proteins (12, 13, 14, 15, 16, 24, 27, 33, 56). As well as blocking IFN signaling, the paramyxovirus V proteins also limit the production of IFN-β by binding to the cellular RNA helicase mda-5 (1, 6).
Recombinant morbilliviruses (51), respiroviruses (9, 23), and an avulavirus (22, 46) that cannot express their V and W proteins have been recovered, and all of these viruses grow similarly to their respective parent viruses, at least in some cell lines such as Vero cells. In the case of the rubulaviruses, PIV5 that lacked the V protein C-terminal specific domain (rPIV5VΔC) was recovered. rPIV5VΔC induces apoptosis in many cells types but grows similarly to rPIV5 in Vero cells (18). V and W-minus hPIV2, in contrast, is highly debilitated, and its growth is very limited even in Vero cells. Moreover, the virus yields of rPIV2VΔC and rPIV2s carrying mutations in the C-terminal V protein-specific domain are 2 to 3 orders of magnitude lower than that of wild-type (wt) hPIV2, even in Vero cells (25, 38). The hPIV2 V protein is thus clearly important for promoting virus growth, independent of the anti-IFN activity.
In the cases of SeV and hPIV3, viral RNA synthesis is downregulated by the C proteins, which bind to the L polymerase subunit (5, 8, 17, 20, 52). In studies using recombinant Rinderpest virus (RPV), a member of genus Morbillivirus, the absence of the V protein has little effect on the replication rate but does lead to increased synthesis of genome and antigenome RNAs. RPV that does not express the C protein, on the other hand, is clearly impaired for growth in cell culture, and its mRNA transcription is reduced (3). The RPV V and C proteins were found to interact with the L protein (54). Recently, the negative modulatory activity of V proteins encoded by PIV5 and MeV has been reproduced in transient minireplicon expression systems (32, 62). However, the mechanisms of the V protein inhibition of these minigenome systems are not clear. Since the hPIV2 V protein shares the N-terminal 164 aa with the P protein, which is essential for viral RNA transcription and replication, it is thought that V may also play a role in viral RNA transcription and replication.
In the present study we investigated the role of the V protein in hPIV2 replication, using a minigenome system free of vaccinia virus. We show here that the hPIV2 V protein inhibits genome replication. Using mutant hPIV2 V proteins and other rubulavirus V proteins, we found that the C terminus of the V protein was essential for this inhibition and for interaction with the L protein but not for interaction with the NP protein. These data suggest that the inhibitory effect of the hPIV2 V protein is the result of L protein binding and not that of NP.
MATERIALS AND METHODS
Cells and antibodies.
COS cells were grown in Eagle's minimal essential medium supplemented with 10% fetal calf serum. BSR T7/5 (4) cells were cultured in Eagle's minimal essential medium supplemented with 10% fetal calf serum and 1 mg of G418 (Geneticin; Gibco)/ml.
Monoclonal antibodies (MAbs) against hPIV2 P/V protein (315-1), hPIV2 P protein (335A), hPIV2 V protein (53V), hPIV2 NP protein (306-1), and hPIV2 L protein (L70-6) were as described previously (36, 40, 41). The MAb against hPIV2 P/V protein 315-1 has cross-reactivity with SV41 P/V protein, and MAbs against hPIV2 P protein 335A and NP protein 306-1 have cross-reactivities with PIV5 proteins (57). The MAb against hPIV2 L protein, 8-2-1, was obtained by immunizing mice with 1,004 to 1,285 aa of the L protein recombinantly expressed in Escherichia coli and is cross-reactive with PIV5 L protein (unpublished data). Anti-V5 antibody and antibody to green fluorescent protein (GFP; sc-8334) were purchased from Invitrogen or Santa Cruz Biotechnology (Santa Cruz, CA).
Construction of expression plasmids.
Various V genes, hPIV2 NP gene, P gene, and L gene cloned into pTM1, which contains a T7 promoter and an encephalomyocarditis virus internal ribosome entry site (B. Moss, National Institutes of Health), were as described previously (38). PIV5 P, NP, L, and V genes were amplified by PCR and subcloned into pTM1 vector. Plasmid pPIV2-GFP was constructed by using standard molecular biology techniques. Various V genes and NP or P gene of hPIV2 were amplified by PCR and subcloned into pBIND or pACT vector (Promega) for mammalian two-hybrid assay. All of these constructs were confirmed by DNA sequencing.
Transient-expression analysis.
Analysis of transient minigenome-encoded gene expression was performed in BSR T7/5 cells cultured in six-well plates. Plasmids pPIV2-GFP, pTM1-P, NP, L, and/or V were transfected into the cells by using FuGENE 6 (Roche) according to the manufacturer's instructions. The amounts of plasmids per well were as follows: pPIV2-GFP, 1 μg; pTM1-NP (nucleoprotein), 0.75 μg; pTM1-P (phosphoprotein), 0.4 μg; and pTM1-L (RNA-dependent RNA polymerase), 0.75 μg with or without pTM1-V at various amounts. pTM1 vector was used to normalize the amount of DNA in each sample. After 2 days, the transfected cells were lysed in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.6% NP-40, 4 mM phenylmethylsulfonyl fluoride). The cell extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and analyzed by a Western blot technique with appropriate antibodies as described previously (34).
RT-PCR of viral genome.
Three days after transfection, cells were lysed with radioimmunoprecipitation assay buffer and immunoprecipitated with anti-NP antibody (306-1), followed by ISOGEN (Nippongene) to purify NP-binding viral RNA. Purified RNAs in different transfection combinations were dissolved in 30 μl of H2O. A total of 2 μl of each sample was used for one-step reverse transcription-PCR (RT-PCR; Qiagen) using the oligonucleotide pair hPIV2 trailer (5′-ACCAAGGGGAAAATCAATATG-3′) and GFP (5′-GACAACCACTACCTGAGCACCCAGTCCGCC-3′), which anneal to antigenomic sense viral RNA and GFP gene, respectively. A total of 2 μl of each RNA sample was used for PCR for 30 cycles as controls for possible contamination of plasmid DNA.
Immunoprecipitation analysis.
BSR T7/5 cells in six-well plates were transfected with 2 μg of pTM1-V or mutants, 2 μg of pTM1-NP or pTM1-L, and 7 μl of FuGENE 6 according to the manufacturer's instructions. At 42 h posttransfection (hpt), cells were lysed in lysis buffer. The supernatants obtained by centrifugation were incubated with MAbs and protein A-Sepharose for 6 h as described previously (42). Polypeptides were analyzed by a Western blotting technique. Cell lysates were also subjected directly to Western blotting with MAbs to confirm expression of the proteins.
Mammalian two-hybrid assay.
A CheckMate mammalian two-hybrid system (Promega) was used for the mammalian two-hybrid assay, and experiments were performed according to the manufacturer's protocol. Plasmids for this assay were prepared as described above. COS cells were transfected with the indicated pBIND and pACT plasmids, together with the pG5luc reporter plasmid. At 48 hpt, the cells were harvested and assayed by the dual-luciferase reporter assay system (Promega).
RESULTS
A minigenome system free of vaccinia virus.
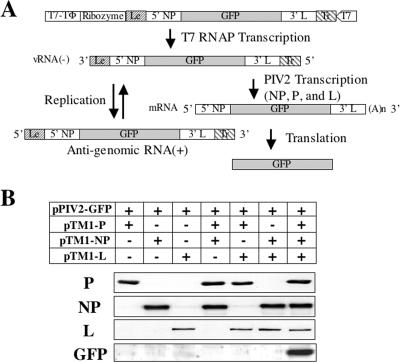
To study the role of the hPIV2 V protein in viral transcription and replication, we established a minigenome system that is free of vaccinia virus. We constructed a mini-genome plasmid (pPIV2-GFP), containing the hPIV2 leader (Le), trailer (Tr), and a reporter gene (GFP) under the control of a T7 RNAP promoter (Fig. 1A). Similar ratios of NP, P, and L expression plasmids that were determined to rescue rPIV2 previously were used in the minigenome system. Transcription from the T7 promoter results in a negative-strand minigenome. In the presence of NP, P, and L support plasmids, this template is assembled and transcribed into the reporter gene mRNA, resulting in the expression of GFP, as well as being used as a template for replication. When plasmids encoding NP, P, and L plus pPIV2-GFP were transfected into BSR T7/5 cells, GFP was detected in cells transfected with all of the plasmids but not in cells lacking any one of the plasmids (Fig. 1B).
FIG. 1.
Establishment of a minigenome system free of vaccinia virus. (A) Schematic diagram of the minigenome system. Plasmid pPIV2-GFP contains an hPIV2 minigenome flanked at one end by a bacteriophage T7 RNA polymerase (T7 RNAP) promoter (T7) and at the other end by a hepatitis delta virus ribozyme (Ribozyme) and T7 transcriptional terminator (T7-TΦ). T7 RNA transcripts can be synthesized under the control of the T7 promoter to generate viral negative-sense PIV2 RNA. pPIV2-GFP contains three extra G residues after the T7 RNAP promoter and prior to the PIV2 trailer sequence (Tr) in order to increase T7 RNAP transcription efficiency. The extra leader sequence (Le) is generated by cleavage with hepatitis delta virus ribozyme. The plasmids pTM1-NP, pTM1-P, and pTM1-L were used to express NP, P, and L proteins in BSR T7/5 cells, a cell line that constitutively expresses T7 RNAP. GFP gene expression can be generated from transcription of primary T7 transcript and vRNA sense genome through viral RNA replication. 5′ NP, 5′ sequence of NP gene; 3′ L, 3′ sequence of L gene. (B) GFP expression from the minigenome system. Plasmids encoding NP, P, L, and pPIV2-GFP at various combinations were transfected into BSR T7/5 cells. At 48 hpt, cells were assayed by Western blotting with anti-NP, P, L, and GFP antibodies.
Repression of GFP expression by the V protein.
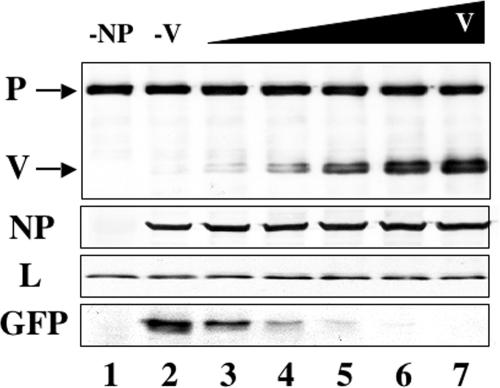
To examine the role of V in hPIV2 RNA replication and transcription, cells were cotransfected with increasing amounts of pTM1-V and constant amounts of pPIV2-GFP plus plasmids encoding NP, P, and L proteins. As shown in Fig. 2, increasing amounts of pTM1-V resulted in increased amounts of the V protein expressed in the cells. Comparison of GFP in the positive control (lane 2) with those in lanes 3 to 7 revealed that V protein repressed GFP expression in a dose-dependent manner.
FIG. 2.
Repression of GFP expression from the minigenome system by V. All transfections include equal amounts of total DNA including plasmids pPIV2-GFP (1 μg), pTM1-NP (0.75 μg), pTM1-P (0.4 μg), and pTM1-L (0.75 μg). Lane 1 contained a negative control derived from cells transfected without pTM1-NP. Increasing amounts of V-expressing plasmid pTM1-V (0, 0.05, 0.1, 0.5, 1, and 2 μg) were used (lanes 2 to 7). The total mass of transfected DNA was held constant by including the appropriate amount of pTM1 vector lacking an insert. After 48 h, the cells were assayed by Western blotting with anti-P/V, NP, L, and GFP antibodies.
V inhibits genome replication in the minigenome system.
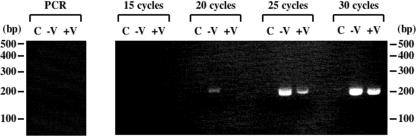
To investigate the mechanism of this inhibition of GFP expression, the effects of V protein on the amount of genomes in the minigenome system were examined. As genome RNAs are assembled with NP as nucleocapsids, these nucleocapsids were purified by immunoprecipitation with anti-NP MAb. The genome RNAs were then reverse transcribed, and PCRs with increasing numbers of cycles were carried out, which quantify the amount of DNA more accurately. As shown in Fig. 3 (right panel), smaller amounts of PCR products were observed from the transfected cells with V protein than without V protein at all stages when PCR product was detectable. These results suggest that V protein inhibits genome replication, either by directly inhibiting the replication itself or by inhibiting genome assembly that is required for replication. These results do not exclude the possibility that V protein inhibits transcription, as well as replication.
FIG. 3.
Effect of V on hPIV2 RNA replication. At 72 hpt, BSR T7/5 cells were lysed and immunoprecipitated with anti-NP MAb. NP-encapsidated RNAs were purified, and RT-PCR was carried out as described in Materials and Methods. The expected size of the PCR product is about 200 bp. Ethidium bromide staining of products from the RT-PCRs is shown. In the left panel, purified RNAs were used directly for PCR without the RT reaction. In the right panel, products from staged RT-PCRs are shown. Lanes: C, control cells transfected with pTM1-P, pTM1-NP, and pTM1-L without pPIV2-GFP; −V, cells transfected with pTM1-P, pTM1-NP, pTM1-L, and pPIV2-GFP; +V, cells transfected with pTM1-P, pTM1-NP, pTM1-L, and pPIV2-GFP plus pTM1-V. The migration of 100-bp DNA size makers is indicated adjacent to the blot.
Repression of GFP expression mediated by mutant hPIV2 proteins.
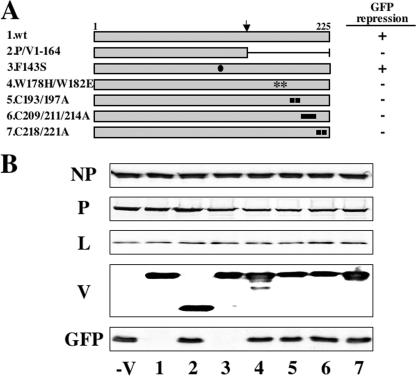
We previously identified hPIV2 V protein residues essential to induce STAT degradation and block IFN signaling, by their mutation to Ala or other residues, as summarized in Table 1. These residues included the seven conserved cysteines (193, 197, 209, 211, 214, 218, and 221 Cys), three tryptophans (174, 182, and 192 Trp), Phe207 in the C-terminal V-unique domain, and Phe143 in the P/V common domain (38). Interestingly, all of these residues except for Phe143 were also required to promote virus growth independent of their anti-IFN effects. We have used these mutants and a C-terminally truncated V protein (Fig. 4A, lane 2) to identify the residues of V protein important for inhibiting minigenome GFP expression. As shown in Fig. 4B, the C-terminal truncated and mutant V proteins in the V-specific domain had all lost the ability to repress GPF expression. In contrast, the F143S mutant repressed GFP expression like wt V protein (Fig. 4B, lanes 1 and 3). Thus, the ability to inhibit GFP expression requires the C-terminal V-unique region and correlates more strongly with the ability to promote virus growth rather than to counter IFN action.
FIG. 4.
Repression of GFP expression from the minigenome system by mutant hPIV2 V proteins. (A) Schematic diagram of the V proteins. The closed circle indicates the position of the mutation residue of Phe143. The asterisks indicate the positions of the mutation residues of Trp-motif. The closed squares indicate the positions of the mutation residues of the Cys motif. The arrow above marks the editing site. (B) Effects of mutant V proteins on the GFP expression in the minigenome system. BSR T7/5 cells were transfected with plasmids pPIV2-GFP (1 μg), pTM1-NP (0.75 μg), pTM1-P (0.4 μg), and pTM1-L (0.75 μg) plus various pTM1-V (1 μg). After 48 h, the cells were assayed by Western blotting with anti-NP, P/V, L, and GFP antibodies. The numbers on the bottom of the figure correspond to each V protein described in panel A.
hPIV2 GFP minigenome expression by combinations of rubulavirus NP, P, and L proteins and repression mediated by different rubulavirus V proteins.
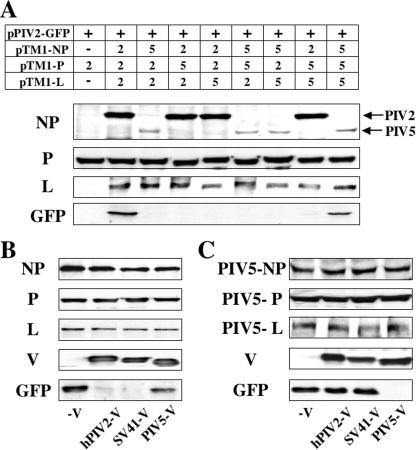
hPIV2, SV41, and PIV5 are all rubulaviruses, and their proteins have highly homology. The NP proteins of hPIV2 and the more distantly related PIV5 are 57.0% identical at the amino acid level, the P proteins are 39.8% identical, and the L proteins are 63.9% identical (19, 43, 58). In order to assess which combinations of the NP, P, and L proteins were able to cooperate in the minigenome system of hPIV2, all possible combinations of the NP, P, and L proteins derived from hPIV2 and PIV5 were tested (Fig. 5A). We found that a homogeneous set of the PIV5 plasmids was also able to drive GFP expression from the hPIV2 minigenome, but all heterogeneous combinations were inactive.
FIG. 5.
GFP expression from the minigenome system by combinations of rubulavirus NP, P, L, and V proteins. (A) Effect of heterogeneous sets containing NP, P, and L plasmids derived from hPIV2 and PIV5. BSR T7/5 cells were transfected with pPIV2-GFP and all possible combinations of hPIV2 and PIV5 NP, P, and L expression plasmids. −, No plasmid was included as a negative control. Repression of GFP expression from minigenome system of homologous sets of hPIV2 (B) or PIV5 (C) by rubulavirus V proteins. After 48 h, the cells were assayed by Western blotting with anti-NP, P/V, PIV5-V, L, and GFP antibodies.
Next, the SV41 and PIV5 V proteins were tested to determine whether the repressive activity was conserved by other rubulaviruses. The hPIV2 V protein shows 69.3 and 41.1% homology with those of SV41 and PIV5, respectively (26, 28). As shown in Fig. 5B, lane 3, the SV41 V protein also repressed the minigenome GFP expression, whereas the PIV5 V was inactive (Fig. 5B, lane 4). In an analogous fashion for GFP expression by a homologous set of PIV5 support plasmids, the PIV5 V protein repressed GFP expression, but the hPIV2 and SV41 V proteins were inactive (Fig. 5C). These results suggest that the different V proteins inhibit minigenome GFP expression by interacting with their NP, P, or L proteins.
Interactions of various V and NP proteins.
The SeV V protein interacts with its NP protein to regulate genome RNA replication (21). It was postulated that the formation of V-NP complex sequestered sufficient NP to limit SeV encapsidation and replication. The hPIV2 V protein interacts with NP via two binding sites: one located at the N-terminal part of the protein (aa 1 to 47) and the other located in the C-terminal 50 aa (Fig. 6A and Table 1) (35, 42, 61). To examine the interaction between the hPIV2 V and NP proteins, NP and various hPIV2 V proteins were coexpressed in BSR T7/5 cells, and the cell lysates were immunoprecipitated with anti-NP (see Materials and Methods). As shown in Fig. 6B, the wt and all mutant V proteins were coprecipitated, indicating that the N-terminal binding site of V was sufficient for this interaction. The interactions between hPIV2, SV41, or PIV5 V and the hPIV2 or PIV5 NP proteins were also examined by coimmunoprecipitation (Fig. 6C). Both NP proteins of hPIV2 and PIV5 bind to all of the V proteins. Thus, the repressive activity of the V protein does not correlate with its ability to form a complex with NP.
We further tested the V-NP interactions by mammalian two-hybrid analysis (Fig. 6D). COS cells were cotransfected with GAL4-fused NP (pBIND), a series of VP16-fused V protein mutants (pACT) and a firefly luciferase reporter plasmid. At 48 hpt, luciferase activities of cell lysates were measured. The bar graph of luciferase activity of Fig. 6D shows that the NP protein interacted with all of the V proteins. The activities between NP and hPIV2-V/W178H/W182E or SV41-V protein were lower than those between NP and other V proteins, that is, ca. 50 or 60% reduced compared to that of hPIV2-V, respectively. However, these activities also did not correlate with the ability of repression by V proteins. Taken together, these data suggest that the V-NP complex formation and minigenome repression are unrelated.
We have previously shown that the V protein cannot directly bind to P by coimmunoprecipitation (35). As shown in Fig. 6D, this V-P interaction is also not detected by mammalian two-hybrid analysis.
Interactions of various V and L proteins.
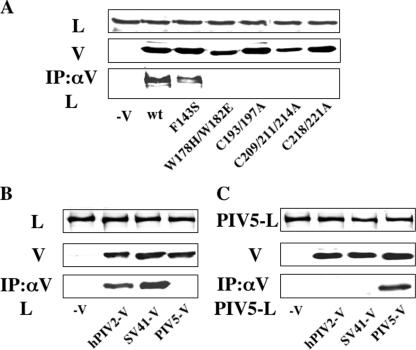
As indicated in Fig. 6A, hPIV2 P protein interacts with L protein on the P-specific region (aa 278 to 353) (36). Although the P/V common domain is not essential for this interaction with L protein, whether the V protein interacts with L protein is not known. To examine the interaction between the V and L proteins, hPIV2 L protein and various mutants of hPIV2 V protein were expressed in BSR T7/5 cells, and the cell lysates were immunoprecipitated by anti-V MAb. Since our previous study showed that V protein binds to viral RNA (37), we carried out immunoprecipitation after treating the cell extracts with 0.25 μg of RNase A (Roche)/μl. As shown in Fig. 7A, the wt and F143S V proteins were coprecipitated, but all of the other V proteins that have mutations on the V-specific region were not. We also examined the interactions between the hPIV2 L protein and the various rubulaviruses V proteins (Fig. 7B). We found that the SV41 V protein also binds to the hPIV2 L protein, whereas PIV5 V protein does not. In an analogous fashion, the PIV5 V protein binds to the PIV5 L protein, but hPIV2 and SV41 V proteins do not (Fig. 7C). There is thus a perfect correlation between the bindings of various V proteins to L proteins and their ability to inhibit minigenome GFP expression.
FIG. 7.
Analysis of interactions between L and V proteins by immunoprecipitation. (A) BSR T7/5 cells were transfected with plasmids expressing hPIV2 L and mutant hPIV2 V proteins. At 48 hpt, the cell extracts were either analyzed directly by Western blot analysis (anti-L, upper panel; anti-V, middle panel) or immunoprecipitated with anti-V before Western blot analysis (anti-L, lower panel). (B and C) BSR T7/5 cells were transfected with plasmids expressing hPIV2 (B) or PIV5 (C) L and rubulavirus V proteins. At 48 hpt, the cell extracts were either analyzed directly by Western blot analysis (anti-L, upper panel; anti-V and PIV5-V, middle panel) or immunoprecipitated with anti-V (B) or PIV5-V (C) before Western blot analysis (anti-L, lower panel).
DISCUSSION
Previous studies showed that the conserved carboxyl terminus of the hPIV2 V protein plays important roles in preventing IFN signaling (and the establishment of an antiviral state) and in promoting virus growth independent of its anti-IFN effects (e.g., in Vero cells) as summarized in Table 1 (35, 38). The mechanism by which the first of these roles is carried out is well described (by inducing STAT protein degradation), but little is known about how hPIV2 V promotes virus growth. We report here another role for this rubulavirus V protein, namely, as a negative regulator of genome replication. To date, the roles of the SeV, MeV, CDV, PIV5, and hPIV2 V proteins in viral RNA synthesis have been examined by using a similar transient-expression approach (32, 62) or by examining defective interfering particle genome replication (7, 20). In all cases, these V proteins were found to act as negative regulators. However, only in the case of SeV was the mechanism of this inhibition previously examined. The SeV V protein appears to inhibit genome replication by binding to unassembled N protein (N°), thereby preventing the formation of the P-N° complex that is required for assembling the nascent genome chain during genome replication (21). The hPIV2 V protein, in contrast, appears to inhibit genome replication in a very different manner. Using a panel of hPIV2 V proteins that have lost the ability to block IFN signaling, as summarized in Table 1, and the V proteins of the closely related SV41 or the more distantly related PIV5, we have found no correlation between their ability to bind to NP and to inhibit minigenome reporter gene expression (Fig. 6). In contrast, we found a perfect correlation between their ability to bind to L protein and to inhibit the reporter gene expression (Fig. 7). In this respect, the hPIV2 V protein appears to act similarly to the SeV C protein, which also binds to its L protein to inhibit genome replication (20).
Sun et al. (53) recently reported that PIV5 V protein interacts with Akt, a serine/threonine kinase, and Akt plays a critical role in PIV5 replication. We have examined the interaction of the hPIV2 V and Akt. Except for W178H/W182E, whose mutations are very close to the putative Akt target site at Ser179, all of the other mutant V proteins bind to Akt similar to the wt hPIV2 V protein (data not shown). These data suggest that hPIV2 V inhibits the expression of the reporter gene through its interaction with L protein rather than its inhibition of Akt.
The paramyxovirus C and V genes are found as overlapping open reading frames of the P gene when they are present. C and V are sometimes referred to as accessory genes, since some viruses do not express one or the other. Except for hPIV1, all paramyxoviruses express a V protein, whereas rubulaviruses do not express C proteins. Rubulaviruses are also unique within this subfamily since their V proteins are translated from the unedited P gene mRNA. Moreover, in contrast to respirovirus (SeV) and morbillivirus (MeV) V proteins, rubulavirus V proteins are not excluded from virus particles. Perhaps more importantly, whereas respiroviruses (SeV) and morbilliviruses (MeV) that cannot express V proteins grow well in cell culture (at least in Vero cells), PIV2 which cannot express V protein grows very poorly in cell culture, even in Vero cells. Moreover, PIV5 which cannot express V protein has not as yet been recovered despite several attempts (Bob Lamb, unpublished data). In this respect, V-minus rubulaviruses resemble C-minus SeV, which are also difficult to prepare and which grow very poorly even in Vero cells. Given all of these unique properties of rubulavirus V proteins, one might have expected that they would exert a positive effect on viral RNA synthesis, but this is clearly not the case.
Leaving aside for the moment the essential but poorly described property of these accessory proteins in promoting virus growth, the paramyxovirus C and V proteins appear to have two main properties: to counteract the host innate immune response and to inhibit virus replication. At first glance, these two properties appear to act in a contradictory fashion. On the one hand, these accessory proteins directly interact with key players of the innate immune response by which cells establish an antiviral state, thus suppressing this cellular response and promoting virus replication. On the other hand, they interact directly with the virus replication machinery (the NP and L proteins) to inhibit virus replication. Although these two effects appear to be work in opposite directions, we assume that both effects contribute to viral fitness since these proteins also promote virus growth. It is possible that these two apparently contradictory effects are both part of the balancing act that viruses use to delay triggering apoptosis and maintain cells in a state that supports virus replication. A virus that inhibits the innate immune response and does not limit virus replication will presumably trigger apoptosis prematurely and thus not produce much viable progeny. Similarly, a virus that directly inhibits it own replication but does not inhibit the innate immune response will simply not get very far in producing progeny. Only viruses that have learned to balance their inhibition of the host cell antiviral state versus their inhibition of their own replication will have survived in the presence of the innate immune response that ultimately leads to programmed cell death. Presumably, this balancing act can be best accomplished when the same protein carries out both functions. This may be one explanation for why the paramyxovirus accessory proteins that counteract the innate immune response also directly inhibit genome replication.
Acknowledgments
This study was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Culture, Sports, and Technology of Japan.
Footnotes
Published ahead of print on 16 April 2008.
REFERENCES
- 1.Andregeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-β promoter. Proc. Natl. Acad. Sci. USA 10117264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrejeva, J., E. Poole, D. F. Young, S. Goodbourn, and R. E. Randall. 2002. The p127 subunit (DDB1) of the UV-DNA damage repair binding protein is essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5. J. Virol. 7611379-11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron, M., and T. Barrett. 2000. Rinderpest viruses lacking the C and V proteins show specific defects in growth and transcription of viral RNAs. J. Virol. 742603-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadd, T., D. Garcin, C. Tapparel, M. Ito, M. Homma, L. Poux, J. Curran, and D. Kolakofsky. 1996. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J. Virol. 705067-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Childs, K., N. Stock, C. Ross, J. Andrejeva, L. Hilton, M. Skinner, R. Randall, and S. Goodbourn. 2007. mda-5, but not RIG-1, is a common target for paramyxovirus V proteins. Virology 359190-200. [DOI] [PubMed] [Google Scholar]
- 7.Curran, J., R. Boeck, and D. Kolakofsky. 1991. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA syntheses by shuffling modules via mRNA editing. EMBO J. 103079-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curran, J., J.-B. Marq, and D. Kolakofsky. 1992. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology 189647-656. [DOI] [PubMed] [Google Scholar]
- 9.Delenda, C., S. Hausmann, D. Garcin, and D. Kolalofsky. 1997. Normal cellular replication of Sendai virus without the trans-frame, nonstructural V protein. Virology 22855-62. [DOI] [PubMed] [Google Scholar]
- 10.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J. Virol. 733125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 for protease-mediated degradation. J. Virol. 739928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcin, D., J. Curran, and D. Kolakofsky. 2000. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol. 748823-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 736559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcin, D., J.-B. Marq, L. Strahle, P. Le Mercier, and D. Kolakofsky. 2002. All four Sendai virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology 295256-265. [DOI] [PubMed] [Google Scholar]
- 15.Gotoh, B., K. Takeuchi, T. Komatsu, and J. Yokoo. 2003. The STAT2 activation process is a crucial target of Sendai virus C protein for the blockade of alpha interferon signaling. J. Virol. 773360-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gotoh, B., K. Takeuchi, T. Komatsu, J. Yokoo, Y. Kimura, A. Kurotani, A. Kato, and Y. Nagai. 1999. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-alpha/beta-mediated responses. FEBS Lett. 459205-210. [DOI] [PubMed] [Google Scholar]
- 17.Grogan, C. C., and S. A. Moyer. 2001. Sendai virus wild-type and mutant C proteins show a direct correlation between L polymerase binding and inhibition of viral RNA synthesis. Virology 28896-108. [DOI] [PubMed] [Google Scholar]
- 18.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodburn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-β induction and interferon signaling. Virology 30315-32. [DOI] [PubMed] [Google Scholar]
- 19.Higuchi, Y., Y. Miyahara, M. Kowano, M. Tsurudome, H. Matsumura, S. Kusagawa, H. Komada, M. Nishio, and Y. Ito. 1992. Sequence analysis of the large (L) protein of simian virus 5. J. Gen. Virol. 731005-1010. [DOI] [PubMed] [Google Scholar]
- 20.Horikami, S. M., R. E. Hector, S. Smallwood, and S. A. Moyer. 1997. Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology 235261-270. [DOI] [PubMed] [Google Scholar]
- 21.Horikami, S. M., S. Smallwood, and S. A. Moyer. 1996. The Sendai virus V protein interacts with the NP protein to regulate viral genome RNA replication. Virology 222383-390. [DOI] [PubMed] [Google Scholar]
- 22.Huang, Z., S. Krishnamurthy, A. Panda, and S. K. Samal. 2003. Newcastle disease virus V protein is associated with viral pathogenesis and functions as an alpha interferon antagonist. J. Virol. 778676-8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato, A., K. Kiyotani, Y. Sakai, T. Yoshida, and Y. Nagai. 1997. The paramuxovirus Sendai virus V protein encodes a luxury function required for virus pathogenesis. EMBO J. 16578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato, A., Y. Ohnishi, M. Kohase, S. Saito, M. Tashiro, and Y. Nagai. 2001. Y2 the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting virus RNA synthesis. J. Virol. 753802-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawano, M., M. Kito, Y. Kozuka, H. Komada, N. Noda, K. Nanba, M. Tsurudome, M. Ito, M. Nishio, and Y. Ito. 2001. Recovery of infectious human parainfluenza type 2 virus from cDNA clones and properties of the defective virus without V-specific cysteine-rich domain. Virology 28499-112. [DOI] [PubMed] [Google Scholar]
- 26.Kawano, M., M. Tsurudome, N. Oki, M. Nishio, H. Komada, H. Matsumura, S. Kusagawa, H. Ohta, and Y. Ito. 1993. Sequence determination of the P gene of simian virus 41: presence of irregular deletions near the RNA-editing sites of paramyxoviruses. J. Gen. Virol. 74911-916. [DOI] [PubMed] [Google Scholar]
- 27.Komatsu, T., K. Takeuchi, Y. Yokoo, and B. Gotoh. 2000. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J. Virol. 742477-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondo, K., H. Bando, M. Tsurudome, M. Kawano, M. Nishio, and Y. Ito. 1990. Sequence analysis of the phosphoprotein (P) genes of human parainfluenza type 4A and 4B viruses and RNA editing at transcript of the P genes: the number of G residues added is imprecise. Virology 178321-326. [DOI] [PubMed] [Google Scholar]
- 29.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C-terminal CYS-RICH region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT 1-alpha. Biochem. Biophys. Res. Commun. 283255-259. [DOI] [PubMed] [Google Scholar]
- 30.Lamb, R. A., and G. D. Parks. 2007. Paramyxoviridae: the viruses and their replication, p. 1449-1490. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott/Williams & Wilkins, Philadelphia, PA.
- 31.Li, T., X. Chan, K. C. Garbutt, P. Zhou, and N. Zheng. 2006. Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell 124105-117. [DOI] [PubMed] [Google Scholar]
- 32.Lin, Y., F. Horvath, J. A. Aligo, R. Wilson, and B. He. 2005. The role of simian virus 5 V protein on viral RNA synthesis. Virology 338270-280. [DOI] [PubMed] [Google Scholar]
- 33.Malur, A. G., S. Chattopadhyay, R. K. Maitra, and A. K. Banerjee. 2005. Inhibition of STAT1 phosphorylation by human parainfluenza virus type 3 C protein. J. Virol. 797877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishio, M., D. Garcin, V. Simonet, and D. Kolakofsky. 2002. The carboxyl segment of the mumps virus V protein associates with Stat proteins in vitro via a tryptophan-rich motif. Virology 30092-99. [DOI] [PubMed] [Google Scholar]
- 35.Nishio, M., M. Tsurudome, H. Ishihara, M. Ito, and Y. Ito. 2007. The conserved carboxyl terminus of human parainfluenza virus type 2 V protein plays an important role in virus growth. Virology 37285-98. [DOI] [PubMed] [Google Scholar]
- 36.Nishio, M., M. Tsurudome, M. Ito, and Y. Ito. 2000. Mapping of domains on the human parainfluenza type 2 virus P and NP proteins that are involved in the interaction with the L protein. Virology 273241-247. [DOI] [PubMed] [Google Scholar]
- 37.Nishio, M., M. Tsurudome, M. Ito, and Y. Ito. 2006. Identification of RNA-binding regions on the P and V proteins of human parainfluenza virus type 2. Med. Microbiol. Immunol. 19529-36. [DOI] [PubMed] [Google Scholar]
- 38.Nishio, M., M. Tsurudome, M. Ito, D. Garcin, D. Kolakofsky, and Y. Ito. 2005. Identification of paramyxovirus V protein residues essential for STAT protein degradation and promotion of virus replication. J. Virol. 798591-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, H. Komada, and Y. Ito. 2001. High resistance of human parainfluenza type 2 virus protein-expressing cells to the antiviral and anti-cell proliferative activities of alpha/beta interferons: cysteine-rich V-specific domain is required for high resistance to the interferons. J. Virol. 759165-9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, S. Kusagawa, H. Komada, and Y. Ito. 1999. Mapping of domains on the human parainfluenza virus type 2 nucleocapsid protein (NP) required for NP-phosphoprotein or NP-NP interaction. J. Gen. Virol. 802017-2022. [DOI] [PubMed] [Google Scholar]
- 41.Nishio, M., M. Tsurudome, M. Ito, N. Watanabe, M. Kawano, H. Komada, and Y. Ito. 1997. Human parainfluenza virus type 2 phosphoprotein: mapping of monoclonal antibody epitopes and location of the multimerization domain. J. Gen. Virol. 781303-1308. [DOI] [PubMed] [Google Scholar]
- 42.Nishio, M., M. Tsurudome, M. Kawano, N. Watanabe, S. Ohgimoto, M. Ito, H. Komada, and Y. Ito. 1996. Interaction between nucleocapsid protein (NP) and phosphoprotein (P) of human parainfluenza virus type 2: one of the two NP binding sites on P is essential for granule formation. J. Gen. Virol. 772457-2463. [DOI] [PubMed] [Google Scholar]
- 43.Ohgimoto, S., H. Bando, M. Kawano, K. Okamoto, K. Kondo, M. Tsurudome, M. Nishio, and Y. Ito. 1990. Sequence analysis of P gene of human parainfluenza type 2 virus: P and cysteine-rich proteins are translated by two mRNAs that differ by two nontemplated G residues. Virology 177116-123. [DOI] [PubMed] [Google Scholar]
- 44.Palosaari, H., J.-P. Parisien, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 777635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parisien, J.-P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283230-239. [DOI] [PubMed] [Google Scholar]
- 46.Park, M. S., A. Garcia-Sastre, J. F. Cros, C. F. Basler, and P. Palese. 2003. Newcastle disease virus V protein is a determinant of host range restriction. J. Virol. 779522-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paterson, R. G., G. P. Leser, M. A. Shqughnessy, and R. A. Lamb. 1995. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 208121-131. [DOI] [PubMed] [Google Scholar]
- 48.Randall, R. E., and A. Bermingham. 1996. NP:P and NP:V interactions of the paramyxovirus simian virus 5 examined using a novel protein:protein capture assay. Virology 224121-129. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez, J. J., J.-P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 7611476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez, J. J., L. F. Wang, and C. M. Horvath. 2003. Hendra virus V protein inhibits interferon signaling by preventing STAT1 and STAT2 nuclear accumulation. J. Virol. 7711842-11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider, H., K. Kaelin, and M. A. Billeter. 1997. Recombinant measles viruses defective for RNA editing and V protein synthesis are viable in cultured cells. Virology 227314-322. [DOI] [PubMed] [Google Scholar]
- 52.Smallwood, S., and S. A. Moyer. 2004. The L polymerase protein of parainfluenza virus 3 forms an oligomer and can interact with the heterologous Sendai virus L, P, and C proteins. Virology 318439-450. [DOI] [PubMed] [Google Scholar]
- 53.Sun, M., S. M. Fuentes, K. Timani, D. Sun, C. Murphy, Y. Lin, A. August, M. N. Teng, and B. He. 2008. Akt plays a critical role in replication of nonsegmented negative-stranded RNA viruses. J. Virol. 82105-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sweetman, D. A., A. Miskin, and M. D. Baron. 2001. Rinderpest virus C and V proteins interact with the major (L) component of the viral polymerase. Virology 281193-204. [DOI] [PubMed] [Google Scholar]
- 55.Takeuchi, K., S. I. Kadota, M. Takeda, N. Miyajima, and K. Nagata. 2003. Measles virus V protein blocks interferon (IFN)-alpha/beta but not IFN-gamma signaling by inhibiting STAT1 and STAT2 phosphorylation. FEBS Lett. 545177-182. [DOI] [PubMed] [Google Scholar]
- 56.Takeuchi, K., T. Komatsu, J. Yokoo, A. Kato, T. Shioda, Y. Nagai, and B. Gotoh. 2001. Sendai virus C protein physically associates with Stat1. Genes Cells 6545-557. [DOI] [PubMed] [Google Scholar]
- 57.Tsurudome, M., M. Nishio, H. Komada, H. Bando, and Y. Ito. 1989. Extensive antigenic diversity among human parainfluenza type 2 virus isolates and immunological relationships among paramyxoviruses revealed by monoclonal antibodies. Virology 17138-48. [DOI] [PubMed] [Google Scholar]
- 58.Tsurudome, M., N. Oki, Y. Higuchi, K. Miyahara, M. Yoshimoto, N. Mutsuga, S. Kitada, M. Ogawa, Y. Miyahara, K. Okamoto, M. Kawano, H. Komada, H. Matsumura, S. Kusagawa, M. Nishio, and Y. Ito. 1991. Molecular relationships between human parainfluenza virus type 2, and simian viruses 41 and 5: determination of nucleoprotein gene sequences of simian viruses 41 and 5. J. Gen. Virol. 722289-2292. [DOI] [PubMed] [Google Scholar]
- 59.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304160-166. [DOI] [PubMed] [Google Scholar]
- 60.Ulane, C. M., J. J. Rodriguez, J.-P. Parisien, and C. M. Horvath. 2003. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J. Virol. 776385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe, N., M. Kawano, M. Tsurudome, M. Nishio, M. Ito, S. Ohgimoto, S. Suga, H. Komada, and Y. Ito. 1996. Binding of the V proteins to the nucleocapsid proteins of human parainfluenza type 2 virus. Med. Microbiol. Immunol. 18589-94. [DOI] [PubMed] [Google Scholar]
- 62.Witko, S. E., C. Lotash, M. S. Sidhu, S. A. Udem, and C. L. Parks. 2006. Inhibition of measles virus minireplicon-encoded reporter gene expression by V protein. Virology 248107-119. [DOI] [PubMed] [Google Scholar]
- 63.Yokosawa, N., S. Yokota, T. Kubota, and N. Fujii. 2002. C-terminal region of STAT-1α is not necessary for its ubiquitination and degradation caused by mumps virus V protein. J. Virol. 7612683-12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269383-390. [DOI] [PubMed] [Google Scholar]