Abstract
Mutations in several subgenomic regions of hepatitis C virus (HCV) have been implicated in influencing the response to interferon (IFN) therapy. Sequences within HCV NS5A (PKR binding domain [PKRBD], IFN sensitivity-determining region [ISDR], and variable region 3 [V3]) were analyzed for the pretreatment serum samples of 60 HCV genotype 1-infected patients treated with pegylated IFN plus ribavirin (1b, n = 47; 1a, n = 13) but with different treatment outcomes, those with sustained virologic responses (SVR; n = 36) or nonresponders (NR; n = 24). Additionally, the sequence of the PKR/eIF-2α phosphorylation homology domain (E2-PePHD) region was determined for 23 patients (11 SVR and 12 NR). The presence of >4 mutations in the PKRBD region was associated with SVR (P = 0.001) and early virologic responses (EVR; 12 weeks) (P = 0.037) but not rapid virologic responses (4 weeks). In the ISDR, the difference was almost statistically significant (68% of SVR patients with mutations versus 45% without mutations; P = 0.07). The V3 region had a very high genetic variability, but this was not related to SVR. Finally, the E2-PePHD (n = 23) region was well conserved. The presence of >4 mutations in the PKRBD region (odds ratio [OR] = 9.9; P = 0.006) and an age of ≤40 years (OR = 3.2; P = 0.056) were selected in a multivariate analysis as predictive factors of SVR. NS5A sequences from serum samples taken after 1 month of treatment and posttreatment were examined for 3 SVR and 15 NR patients to select treatment-resistant viral subpopulations, and it was found that in the V3 and flanking regions, the mutations increased significantly in posttreatment sera (P = 0.05). The genetic variability in the PKRBD (>4 mutations) is a predictive factor of SVR and EVR in HCV genotype 1 patients treated with pegylated IFN and ribavirin.
The hepatitis C virus (HCV) is one of the most frequent causes of chronic viral hepatitis, liver cirrhosis, and hepatocellular carcinoma. Current therapy, a combination of pegylated-alpha interferon (peg-IFN-α) and ribavirin (RBV), achieves a response rate between 48% and 88% (29). The factors that have been shown to predict the response to therapy include HCV genotypes other than 1, low viral load, rapid virologic response (RVR; 4 weeks), and early virologic response (EVR; 12 weeks) (3). It is believed that both host and viral factors, including several viral genomic regions, are essential for an effective response to IFN therapy (9).
The clinical importance of amino acid mutations within the functional regions of the HCV proteins in correlation with chronic hepatitis C (CHC) has been questioned. NS5A has been studied as a possible mediator of IFN-α resistance (17). Information, derived mainly from the subgenomic replicon system, suggests that NS5A is involved in genomic RNA replication (21, 23). Prior to this, Enomoto et al. (8) suggested that the genetic heterogeneity of a specific domain of the NS5A region, termed the IFN sensitivity-determining region (ISDR), was related closely to the response in Japanese patients with HCV genotype 1b, so that patients with at least four mutations within the ISDR achieved a sustained virologic response (SVR) to IFN-α monotherapy (8, 27, 40). However, the reported results in Europe and the United States concerning the correlation of the substitutions within the ISDR to the treatment outcome are conflicting (13, 26, 41). It is well known that the ISDR is necessary but not sufficient for the interaction between NS5A and the PKR enzyme (which is very important for the activation of IFN), with an additional 26 amino acids distal to the ISDR being required (12). This region is termed the PKR binding domain (PKRBD). This region-mediate disruption of PKR dimerization resulted in the repression of PKR function and the inhibition of PKR-mediated eIF-2α phosphorylation. The introduction of multiple mutations within the PKR-binding region, including those within the ISDR, abrogated the ability of NS5A to bind to PKR. Recent studies have found that mutations within the PKRBD of HCV type 1 are associated with a long-term sustained response to IFN-α and IFN-α/RBV therapy (2, 24, 33).
Recently, clinical studies have proposed that the number of amino acid variations within variable region 3 (V3) may be associated with the treatment outcome (7, 26, 30, 33).
A PKR/eIF-2α phosphorylation homology domain (PePHD) within the E2 protein has been found to interact with PKR and inhibit PKR in vitro, suggesting a possible mechanism of HCV to evade the antiviral effects of IFN (35). Mutations in this region are believed to influence the response to IFN therapy (16, 31, 32); however, the results of different studies are conflicting because this region is highly conserved (2, 5, 13, 18, 38).
It has been speculated that mutations in the functional regions of the HCV proteins may be correlated with the response to IFN therapy, but few studies have been made of combinate peg-IFN-α and RBV therapy, and what results are available are contradictory, possibly due to geographical factors. Furthermore, few studies exist that include all regions involving IFN activity, and little has been published on correlating the predictive factors of the response to the treatment with the genetic variability of these genome regions. We also examined the sequences not only in SVR and NR patients but also in patients showing RVR. In this paper, we report the results of an extensive analysis of the pretreatment amino acid substitutions in the E2-PePHD, NS5A-PKRBD, NS5A-ISDR, and NS5A-V3 and flanking regions and their correlation with factors to predict responses to peg-IFN-α plus RBV therapy in patients with genotype 1. We also analyzed changes in the sequence motif during therapy for 18 patients.
MATERIALS AND METHODS
In this prospective study, 60 patients infected with HCV genotype 1 were treated with peg-IFN-α-2a (180 μg/week) and RBV (1,000 to 1,200 mg/day) for 48 weeks. The diagnosis of CHC was based on raised aminotransferase levels for at least 6 months, biopsy-proven CHC, and the permanent detection of serum HCV-RNA. Individual data were collected on baseline clinical, biochemical, virological, and histological parameters. Finally, HCV-RNA analysis was performed at 4, 12, 24, 48, and 72 weeks. The histologic study was carried out in accordance with Scheuer's grading of necroinflammatory activity, with slight modifications (4). The patients showed no evidence of hepatitis B virus-infected, alcoholic, autoimmune, or drug-induced liver disease.
Patients who obtained normalization of alanine aminotransferase levels and HCV-RNA clearance for more than 6 months after therapy were considered as presenting SVR (n = 36; 60%). At week 12, 52 (87%) patients presented EVR (HCV-RNA negative or had a ≥2 log10 decrease in viral load). Patients without EVR (non-EVR) were considered nonresponders (NR; n = 8). Also, patients who became HCV-RNA negative during a course of therapy but then relapsed and redeveloped HCV-RNA after stopping treatment (n = 16; 27%) were considered NR (n = 24). Finally, in 40 patients we studied who had RVR at 4 weeks, 21 (53%) patients were HCV-RNA negative. The sequences of the NS5A-PKRBD (codons 2209 to 2274, including the ISDR [codons 2209 to 2248]), NS5A-V3 (codons 2356 to 2379), and flanking regions for the dominant variant of HCV were determined for the pretreatment sera of 60 patients. These sequences were also analyzed for the treatment and posttreatment serum samples of 18 patients in order to identify changes in the sequence motif during therapy. In addition, the sequences of the E2-PePHD region (codons 659 to 670) and 5′ flanking region (codons 629 to 658) were determined for 23 patients with HCV genotype 1b.
Informed consent to participate was obtained from each patient. The study protocol conformed to the ethical guidelines of the 1975 Helsinki Declaration, and prior approval for the study was obtained from the ethics committee.
Virologic assays.
The virus genotype was determined by reverse hybridization (Inno-LIPA II HCV; Innogenetics, S.A., Ghent, Belgium). HCV-RNA was determined for sera with the Amplicor HCV kit. HCV-RNA serum levels were measured by the Cobas Amplicor monitor HCV test version 2.0.
Amplification and sequencing of NS5A and E2-PePHD.
HCV-RNA was extracted from 200 μl of the pretreatment serum samples by means of the High Pure Viral RNA kit (Roche Diagnostics GmbH, Penzberg, Germany). Isolated HCV-RNA was reverse transcribed by using avian myeloblastosis virus (Reflectase; Activ Motif) and 20 U of RNasin (Promega) at 42°C for 1 h in the presence of antisense primer (1 μM) (Table 1). For the nested PCR, we used Brilliant Sybr green QPCR master mix (Stratagene) in an Mx3000 thermocycler (Stratagene). The first PCR mixture consisted of 12.5 μl of the master mix, a sense and an antisense primer (0.25 μM) (Table 1), 0.3 μl of ROX (reference fluorescent dye), and 4 μl of cDNA; 1 μl of the first PCR mixture was used for the second PCR, and diethyl pyrocarbonate H2O was added to obtain a final volume of 25 μl. To analyze the NS5A region and the PePHD region, two PCRs were conducted, the first consisting of 1 cycle at 95°C for 10 min; 33 cycles at 95°C for 30 s (30 cycles for PePHD), 60°C for 30 s (58°C for PePHD), and 72°C for 65 s (50 s for PePHD); and 1 cycle at 72°C for 8 min. The second PCR was conducted in the same way as the first PCR, except for an annealing phase at 58°C for 50 s for elongation. The samples were subjected to a cycle of denaturation to visualize the dissociation curve. The size, purity, and approximate yield of the DNA obtained were verified by direct observation of 2% agarose gels. For sequencing, we used the Terminator Big Dye 3.1 (Aplay BioSystems) within a 3100 genetic analyzer (Aplay BioSystems). Between 10 ng and 40 ng of purified DNA (NucleoSpin extract; Macherey-Nagel) and 6.4 pM of sequence primer (Table 1) were added to obtain a 10-μl final volume. Analysis of the chromatograms was by means of Chromas Lite software (http://technelysium.com.au).
TABLE 1.
Oligonucleotide primers used in the present study
| Region and application | Direction | Primer | Sequence | Positiona |
|---|---|---|---|---|
| NS5A-1b | ||||
| RT and first PCR | Antisense | 1a1b-ISV3-AS | 5′-ACGCCTTCGCCTTCATCTCC-3′ | 7812-7793 |
| First PCR | Sense | 1a1b-ISV3-S | 5′-CCCATCAACGCATACACCACG-3′ | 6510-6531 |
| Second PCR and sequencing | Antisense | 2a1b-ISV3-AS | 5′-GGACATTGAGCAGCAKACGAC-3′ | 7595-7575 |
| Sense | 2a1b-ISV3-S | 5′-GACCCCTCYCAYATYACAGCAG-3′ | 6858-6879 | |
| NS5A-1a | ||||
| RT and first PCR | Antisense | 1a1a-ISV3-AS | 5′-GACATTGAGCAGCACACGAC-3′ | 7609-7590 |
| First PCR | Sense | 1a1a-ISV3-S | 5′-CCATTAAYGCCTACACCACG-3′ | 6523-6542 |
| Second PCR and sequencing | Antisense | 2a1a-ISV3-AS | 5′-TACTGACCGTYGACCATGAC-3′ | 7569-7560 |
| Sense | 2a1a-ISV3-S | 5′-ATCCCTCCCATATAACAGCAG-3′ | 6871-6891 | |
| PePHD-1b | ||||
| RT and first PCR | Antisense | 1a1b-PePHD-AS | 5′-CGCAGAAGAACACRAGRAAGGAG-3′ | 2627-2649 |
| First PCR | Sense | 1a1b-PePHD-S | 5′-CCATACAGRCTYTGGCACTACC-3′ | 2163-2184 |
| Second PCR | Antisense | 2a1b-PePHD-AS | 5′-ACRGACGCYGCATTGAG-3′ | 2605-2589 |
| Sense | 2a1b-PePHD-S | 5′-AAGGTYAGGATGTATGTGGG-3′ | 2211-2230 |
Nucleotide positions are according to prototypes HCV-J (genotype 1b) and HCV-1 (genotype 1a).
Sequence analysis.
NS5A and PePHD were aligned to the following reference sequences: HCV-J (GenBank accession number D90208) for genotype 1b (19) and HCV-1 (GenBank accession number M62321) for genotype 1a (6). The most common amino acid at each position of this study was taken as the consensus sequence. The factors associated with the mutations in each of the regions were studied, taking into account the median of the number of mutations in each region and distributing, in this way, the population of each region into two groups of similar size (ISDR, with and without mutations; PKRBD, > or ≤4 mutations; V3, > or ≤5 mutations). This allowed us to determine major statistical significances.
Phylogenetic analysis.
The phylogenetic tree of the NS5A region (codons 2191 to 2405) of the 60 sequences of all patients was constructed by the Clustal W method (36a). The sequences in FASTA format were pasted into the submission form (http://www.ebi.ac.uk/clustalw/), and the output obtained was represented by a phylogenetic tree.
Statistical analysis.
The quantitative variables are expressed as means ± standard deviations. Comparisons between groups were made by the χ2 or Fisher exact test for categorical variables and the Student t test for quantitative variables. Multivariate analysis was done by discriminant analysis by the stepwise forward selection method. A P value of <0.05 was considered statistically significant.
RESULTS
Baseline characteristics of patients and different responses to peg-IFN plus RBV therapy.
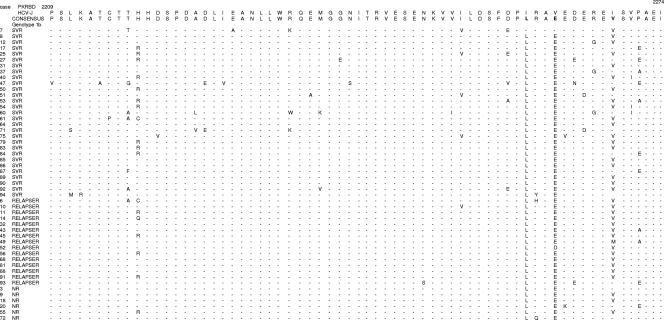
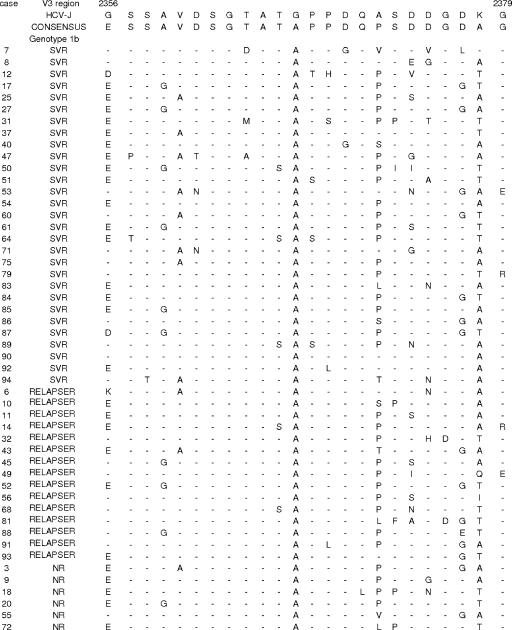
The general characteristics of the patients prior to therapy with different responses to combined therapy (SVR [SVR, n = 36; NR, n = 24], EVR [EVR, n = 52; non-EVR, n = 8], and RVR [RVR, n = 21; non-RVR, n = 19]) are summarized in Table 2. The sequences were aligned with the reference strands (HCV-J for genotype 1b and HCV-1 for genotype 1a) (Fig. 1, 2, and 3). We did not find any predictive factors of RVR (4 weeks). With respect to EVR, 19 patients (100%) who presented >4 mutations in the PKRBD region had EVR (P = 0.037). The only factors associated with SVR were an age of ≤40 years and the presence of >4 mutations in the PKRBD region. In the ISDR, the difference was almost statistically significant. Nevertheless, when the average mutations in the ISDR were determined, there was a statistically significant difference between SVR and NR (1.41 ± 1.46 mutations versus 0.58 ± 0.65 mutations, respectively; P = 0.005). In the logistic regression analysis, >4 mutations in the PKRBD and ages of ≤40 years were independent factors of SVR (Table 2); this does not preclude the possibility that both variables correlate to biological factor affecting the viability or fitness of the virus in the organism and therefore to the viral load. If the sequences were aligned with the consensus sequence, only the PKRBD region would be related to SVR (79.2% of the NR had ≤1 mutation and 58.3% of the SVR patients had >1 mutation; P = 0.004) (Fig. 1 and 3A). The V3 region had a very high genetic variability (Fig. 2 and 3B) but was not related to SVR (Table 2). The sequence of the E2-PePHD was determined for 23 patients with genotype 1b (19 EVR, 4 non-EVR, 11 SVR, and 12 NR). This region was well conserved. Two patients with one mutation had EVR, and later, they were viral RNA positive. Twenty-one patients had the wild type, which is why we did not continue sequencing this region. In the 5′ flanking region of E2-PePHD, 9 of 23 patients (5 SVR and 4 NR) had >6 mutations, but this fact was not statistically significant.
TABLE 2.
Baseline characteristics of patients and predictive factors of different types of responses to peg-IFN plus RBV therapya
| Patients | Age (yr)
|
Sex
|
Epidemiologyb
|
Genotype
|
Viral load
|
Activity gradec | Fibrosis stagec | PKRBD mutations
|
ISDR mutations
|
V3 mutations
|
PePHD mutations (n = 23)
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤40 | >40 | Male | Female | Parenteral | Non- parenteral | ALT (IU/liter) | AST (IU/liter) | GGT (IU/liter) | 1a | 1b | ≤600,000 | >600,000 | ≤4 | >4 | Without | ≥1 | ≤5 | >5 | Without | 1 | |||
| All (n = 60) | 30 (50) | 30 (50) | 34 (57) | 26 (43) | 46 (84) | 9 (16) | 117 ± 92 | 68 ± 67 | 74 ± 63 | 13 (22) | 47 (78) | 12 (20) | 48 (80) | 4 ± 2.2 | 2.1 ± 1.1 | 41 (68) | 19 (32) | 22 (37) | 38 (63) | 39 (65) | 21 (35) | 21 (91) | 2 (9) |
| RVR (n = 21) | 13 (62) | 10 (42) | 16 (59) | 7 (39) | 18 (58) | 3 (33) | 136 ± 115 | 86 ± 87 | 63 ± 82 | 8 (67) | 15 (45) | 4 (40) | 19 (54) | 3.89 ± 2.68 | 2.00 ± 1.19 | 15 (50) | 8 (53) | 8 (47) | 15 (54) | 12 (41) | 11 (69) | 9 (45) | 1 (50) |
| Non-RVR (n = 19) | 8 (38) | 14 (49) | 11 (41) | 11 (61) | 13 (42) | 6 (67) | 106 ± 67 | 60 ± 28 | 69 ± 40 | 4 (33) | 18 (55) | 6 (60) | 16 (46) | 4.63 ± 1.98 | 2.44 ± 1.29 | 15 (50) | 7 (47) | 9 (53) | 13 (46) | 17 (59) | 5 (31) | 11 (55) | 1 (50) |
| EVR (n = 52) | 27 (90) | 25 (83) | 31 (91) | 21 (81) | 42 (91) | 6 (67) | 113 ± 71 | 69 ± 41 | 59 ± 52 | 10 (77) | 42 (89) | 11 (92) | 41 (85) | 4.05 ± 2.3 | 2.07 ± 1.1 | 33 (80) | 19 (100)f | 17 (77) | 35 (92) | 33 (85) | 19 (90) | 17 (81) | 2 (100) |
| Non-EVR (n = 8) | 3 (10) | 5 (17) | 3 (9) | 5 (19) | 4 (9) | 3 (33) | 146 ± 183 | 107 ± 137 | 123 ± 116 | 3 (23) | 5 (11) | 1 (8) | 7 (15) | 3.67 ± 1.8 | 2.29 ± 0.44 | 8 (20) | 0 (0) | 5 (23) | 3 (8) | 6 (15) | 2 (10) | 4 (19) | 0 (0) |
| SVR (n = 36) | 22 (73)d,e | 14 (47) | 22 (65) | 14 (54) | 29 (63) | 4 (44) | 116 ± 98 | 72 ± 42 | 55 ± 41 | 9 (69) | 27 (57) | 9 (75) | 27 (56) | 4.3 ± 2.2 | 2 ± 1.1 | 19 (46) | 17 (89)g,h | 10 (45) | 26 (68) | 21 (54) | 15 (71) | 11 (52) | 0 (0) |
| NR (n = 24) | 8 (27) | 16 (53) | 12 (35) | 12 (46) | 17 (37) | 5 (56) | 173 ± 112 | 53 ± 17 | 52 ± 35 | 4 (31) | 20 (43) | 3 (25) | 21 (44) | 4 ± 2.4 | 2.1 ± 0.7 | 22 (54) | 2 (11) | 12 (55) | 12 (32) | 18 (46) | 6 (29) | 10 (48) | 2 (100) |
Data are expressed as means ± standard deviations or as numbers of patients with percentages in parentheses. Comparisons between groups were made by the χ2 or Fisher exact test for the categorical variables and the Student t test for the quantitative variables. A P value of <0.05 was considered statistically significant. The variables included in the multivariate analysis were age, epidemiology, viral load, PKRBD mutations, and ISDR mutations.
ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, gamma glutamyltransferase.
Scheuer's grading of necroinflammatory activity, with slight modifications. Activity grade is periportal inflammation (score, 0 to 4) plus intralobular degeneration and focal necrosis (score, 0 to 4); the total score for activity could range from 0 to 8. Fibrosis stages are no fibrosis (0), periportal fibrous expansion without septum formation (1), periportal fibrous expansion with some septum formation (2), periportal fibrous expansion with septum formation (3), and cirrhosis (4).
Statistically significant difference between the values for SVR and NR patients (P = 0.03).
Logistic regression analysis of the values for SVR patients yielded an odds ratio of 3.2, a 95% confidence interval of 1 to 10.3, and a P value of 0.056.
Statistically significant difference between the values for EVR and non-EVR patients (P = 0.037).
Statistically significant difference between the values for SVR and NR patients (P = 0.001).
Logistic regression analysis of the values for SVR patients yielded an odds ratio of 9.9, a 95% confidence interval of 1.9 to 50.3, and a P value of 0.006.
FIG. 1.
Major PKRBD sequences in genotype 1b-infected patients. Each sequence was compared to the HCV-J 1b prototype sequence and consensus sequence. The positions of the first and last amino acids of the PKRBD in the HCV polyprotein are indicated over the HCV-J sequence. Each patient is designated by a number and response to treatment.
FIG. 2.
Major V3 sequences in genotype 1b-infected patients. Each sequence was compared to the HCV-J 1b prototype sequence and consensus sequence. The positions of the first and last amino acids of the V3 domain in the HCV polyprotein are indicated over the HCV-J sequence. Each patient is designated by a number and response to treatment.
FIG. 3.
Major PKRBD sequences (A) and V3 sequences (B) in genotype 1a-infected patients. Each sequence was compared to the HCV-1 1a prototype sequence and consensus sequence. The positions of the first and last amino acids of the PKRBD domain and V3 domain in the HCV polyprotein are indicated over the HCV-1 sequence. Each patient is designated by a number and response to treatment.
When we compared patients with SVR (n = 36) and relapsers (n = 16), the predictive factors of SVR were ages of <40 years (P = 0.04) and >4 mutations in the PKRBD (P = 0.001) as before; also, the differences in viral load were almost statistically significant (P = 0.07).
Analysis of factors associated with mutations in the NS5A-PKRBD.
All patients presented amino acid substitutions in the PKRBD region when the sequences were aligned with the reference strands (Fig. 1 and 3A). Ninety-two percent of the women (P < 0.0001) and 79% of the patients with viral loads of >600,000 IU/ml (P = 0.006) were related with ≤4 mutations (Table 3). On examining the consensus sequence and considering the number of mutations, the women had less variability (77% of women with ≤1 mutation versus 41% of men with ≤1 mutation; P = 0.006). Moreover, the patients with low viral loads had more mutations (75% of patients with viral loads of ≤600,000 IU/ml with >1 mutation versus 25% of patients with viral loads of ≤600,000IU/ml with ≤1 mutation; P = 0.016).
TABLE 3.
Factors associated with mutations in NS5A-PKRBD, NS5A-ISDR, and NS5A-V3a
| Region and mutations | Age (yr)
|
Sex
|
Genotype
|
Viral load (IU/ml)
|
||||
|---|---|---|---|---|---|---|---|---|
| ≤40 (n = 30) | >40 (n = 30) | Male (n = 34) | Female (n = 26) | 1a (n = 13) | 1b (n = 47) | ≤600,000 (n = 12) | >600,000 (n = 48) | |
| PKRBD | ||||||||
| ≤4 | 19 (63) | 22 (73) | 17 (50) | 24 (92)d | 9 (69) | 32 (68) | 3 (25) | 38 (79) |
| ≤4 | 11 (37) | 8 (27) | 17 (50) | 2 (8) | 4 (31) | 15 (32) | 9 (75)g | 10 (21) |
| ISDR | ||||||||
| Without | 7 (23) | 15 (50) | 9 (27) | 13 (50) | 2 (15) | 20 (43) | 1 (8) | 21 (44) |
| With | 23 (77)b | 15 (50) | 25 (73)c | 13 (50) | 11 (85)f | 27 (57) | 11 (92)h | 27 (56) |
| V3 | ||||||||
| ≤5 | 19 (63) | 20 (67) | 17 (50) | 22 (85)e | 10 (77) | 29 (62) | 8 (67) | 31 (65) |
| ≤5 | 11 (37) | 10 (35) | 17 (50) | 4 (15) | 3 (23) | 18 (38) | 4 (33) | 17 (35) |
Data are expressed as numbers of patients with percentages in parentheses. Comparisons between groups were made by the χ2 or Fisher exact test for the categorical variables. A P value of <0.05 was considered statistically significant.
Statistically significant difference (P = 0.03).
Statistically significant difference (P = 0.05).
Statistically significant difference (P = 0.0001).
Statistically significant difference (P = 0.0001).
Statistically significant difference (P = 0.06).
Statistically significant difference (P = 0.006).
Statistically significant difference (P = 0.021).
Analysis of factors associated with mutations in NS5A-ISDR and the flanking region.
In the ISDR, 38 patients (63%) had genetic variability and 22 (37%) had a prototype ISDR sequence. When the sequences were aligned with the reference strands, the patients aged ≤40 years, the men, and those with viral loads of ≤600,000 IU/ml presented high genetic variability (Table 3). With respect to the consensus sequence, 65% (22/34) of the men presented mutations, whereas 62% (16/26) of the women did not (P = 0.039), and the patients with low viral loads had more mutations in this region (92% with mutations versus 8% without mutations; P = 0.003).
In the 5′ flanking region (codons 2191 to 2208), 83% (50/60) of the patients did not present mutations, and statistically significant differences were found only with genotypes (genotype 1a, 31% of patients without mutations and 69% with mutations; genotype 1b, 98% of patients without mutations and 2% with mutations; P < 0.0001).
Analysis of factors associated with mutations in NS5A-V3 and the flanking region.
In the V3 region, when the sequences were aligned with the reference strands, the only factor associated with the number of mutations was gender (Table 3). When the sequences were aligned with the consensus sequence, only the genotype was statistically significant (for genotype 1a, 92% of patients presented >3 mutations (median); for genotype 1b, 64% of patients presented <3 mutations; P < 0.0001) (Fig. 2 and 3B).
The flanking regions, preV3 (codons 2275 to 2355) and postV3 (codons 2380 to 2405), were notable for their lack of homology between genotypes 1a and 1b. As happened with the V3 region consensus, the patients with genotype 1a had a greater number of mutations in both regions than the patients with genotype 1b (preV3 region, P = 0.013; postV3 region, P = 0.025) (data not shown). We have not found a correlation between the number of mutations in these regions (preV3 and postV3) and the response to the treatment (data not shown).
Evolutive study.
We analyzed the genetic variability of the different regions of NS5A before, during, and after therapy to determine the selection of treatment-resistant viral subpopulations in 18 patients (3 SVR, 15 NR) (Table 4). Because of a rapid decrease in the viral loads in responders, sequence data were available for three patients undergoing therapy, who were sampled during the first month of treatment, but no consistent change was detected in the PKRBD, V3, or flanking regions. Serum samples from 15 NR patients from the first month of treatment and posttreatment were sequenced, and statistically significant differences were found between the number of mutations from the first month of treatment and the number of posttreatment mutations in the V3 and flanking regions (P = 0.05) (Table 4).
TABLE 4.
Treatment and posttreatment sequence changesa
| Region and treatment | SVR (n = 3) | NR (n = 15) |
|---|---|---|
| PKRBD region | ||
| Baseline | 4.67 ± 0.57 | 3.8 ± 0.86 |
| First-mo treatment | 4.67 ± 0.57 | 4 ± 1 |
| Posttreatment | 4.38 ± 1.68 | |
| V3 and flanking region | ||
| Baseline | 11.33 ± 2.3 | 11.67 ± 1.67b |
| First-mo treatment | 11.67 ± 2.0 | 10.83 ± 2.25c |
| Posttreatment | 13.29 ± 0.56 |
Data are expressed as means ± standard deviations. Comparisons between groups were made by the Student t test for the quantitative variables. A P value of <0.05 was considered statistically significant.
Posttreatment versus basal value differences were statistically significant (P = 0.06).
Posttreatment versus first-month treatment value differences were statistically significant (P = 0.05).
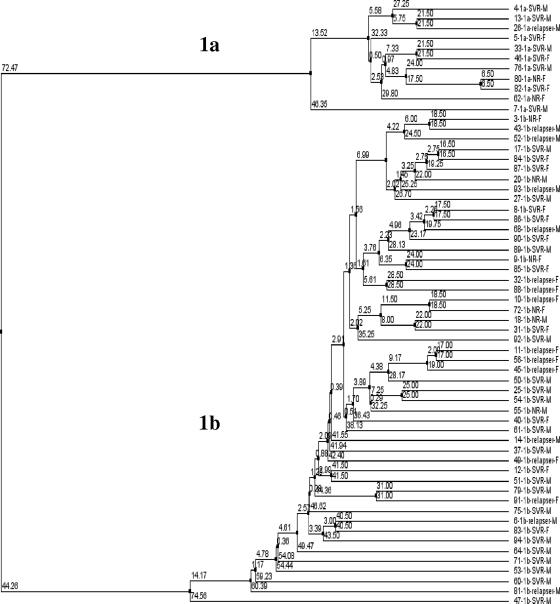
Phylogenetic analysis.
The NS5A region has been used to make the phylogenetic tree (codons 2191 to 2405) using 60 sequences from all of the patients. Patients are listed by patient number, genotype, treatment response, and gender (Fig. 4). No clustering occurred by treatment response or gender. However, the patients were clustered by genotype, as was to be expected.
FIG. 4.
Phylogenetic tree of the NS5A region in 60 patients. Genotype (1b and 1a), treatment response, and sex (M, male; F, female) are stated for all patients.
DISCUSSION
In the present study, we set out to determine whether the numbers of mutations in the E2-PePHD, NS5A-PKRBD, NS5A-ISDR, and NS5A-V3 regions isolated from the pretreatment serum samples of chronically infected patients with genotype 1 treated with peg-IFN-α-2a and RBV were correlated with the RVR and final outcome. The majority of the previous studies were done with a notably lower number of patients than the number of patients used in this study (n = 60) and using other treatments like IFN monotherapy or standard IFN and ribavirin. In addition, we sought to correlate the predictive factors of virologic response, such as age, gender, genotype, viral load, and EVR, with the genetic variability of the virus in these regions.
We know that other regions exist that have been less widely investigated, such as the polymorphisms in the RNA viral 5′ region (IRES). This region has been studied in relation to the response to treatment with IFN. Nevertheless, in several studies, no significant difference between the polymorphisms in this region and the response to IFN was found (34, 36).
As we found mutations in relation to HCV-J and HCV-1 that occurred repeatedly in all our patients, we decided to perform a consensus sequence with all the patients participating in this study. These mutations have been demonstrated in other reference sequences (37). This suggests that it is reasonable to use the consensus sequences to assess the substitutions in NS5A, but when the sequences were aligned with the consensus strand, we obtained results similar to those with the reference strand.
It has been suggested that the NS5A protein plays a role in HCV-RNA replication. It has also been demonstrated that the number of mutations in the ISDR regulates the replication of replicons in vitro (21). It is likely that specific amino acid variations within the NS5A regions that modulate HCV-RNA replication are important for the antiviral response to IFN-α.
The inhibition of the antiviral activity of IFN by NS5A has been shown in vitro in NS5A/PKR interaction via an NS5A-specific PKRBD (10, 12) and in mammalian cells using encephalomyocarditis virus and vesicular stomatitis virus (11). However, an inconsistent association between this in vitro interaction and the clinical response has also been reported (28). In this work, multivariate analysis revealed that the presence of >4 mutations in the PKRBD was only an independent factor associated with SVR and EVR to combination therapy. Previous papers have also reported a positive correlation between the number of mutations and SVR in this region (2, 24, 33). However, there are only a few studies in which such relationships have not been found (20, 22); these studies either investigated only genotype 1a patients (22) or performed the statistical analysis with the remaining portion of the PKRBD located downstream from the ISDR (26 amino acids) (20). A high level of significance for this correlation was observed when the analysis was restricted to nonconserved amino acid variations (33); this was found, too, with genotype 3 patients (15). Our data back the idea that the PKRBD and not the ISDR alone is a functional domain of NS5A and may produce resistance or sensitivity to IFN therapy.
It has been reported that a subset of genotype 1 patients who had RVR (undetectable serum HCV-RNA levels of <29 IU/ml) at 4 weeks of treatment achieved SVR (89%) with only 24 weeks of peg-IFN and ribavirin treatment and that data at 12 weeks are good for predicting NR (42). It is easier to routinely determine HCV-RNA and viral loads rather than sequence HCV, but the objective of this work was to study the importance of genetic variability in relation to viral response. This type of study (analysis of genomic viral sequences) can have many future repercussions with progressive cost reduction and easy use of sequencing technologies. Therefore, we think that in a not very distant future, the genetic sequence might apply in the clinical routine, facilitating a more personalized follow-up of patients. Nevertheless, we did not find any predictive factors for RVR.
In the present study, women presented less variability than men in the PKRBD. Likewise, patients with high viral loads presented fewer mutations in the PKRBD region than those with low viral loads. The fact that 100% of the patients who have >4 mutations in the PKRBD region presented EVR is important.
The ISDR is useful as a predictive marker of the response to IFN therapy for patients with a Japanese-specific subtype (HCV-J) (8, 25, 40), but similar correlations were not observed in studies conducted in Europe and the United States (13, 26, 41). Nevertheless, a meta-analysis of 655 Japanese and 525 European patients that focused on geographical differences (27) concluded that there was a significant positive correlation between the number of ISDR mutations and the SVR rate, irrespective of geographical region, although the likelihood of SVR with each additional mutation within the ISDR was considerably more pronounced for the Japanese patients than for the European patients. In our study, the existence or absence of mutations in the ISDR was not correlated with SVR (Table 2). Nevertheless, the patients with more mutations were correlated with a greater response to treatment (1.41 ± 1.46 mutations for SVR patients versus 0.58 ± 0.65 mutations for NR patients; P = 0.005). Enomoto et al. (8) found a significant positive correlation between the number of ISDR mutations and the SVR rate, when considering three groups of patients: wild type (0 mutations), intermediate type (1 to 3 mutations), and mutant type (>4 mutations). In our cases, only 7% of the patients were mutant type, whereas 57% were intermediate type and 37% wild type, as has been found in other European studies. Interestingly, we found that predictive factors of SVR, such as ages of ≤40 years and viral loads of ≤600,000 IU/ml, were correlated significantly with a higher number of mutations in the ISDR.
Studies of the V3 domain have revealed that responsiveness to IFN/RBV therapy is correlated with mutations (7, 26, 30, 33). In the present study, no correlation was found between the number of mutations in the V3 and flanking regions and treatment response. The number of mutations in the V3 region was only correlated with gender, with women presenting fewer mutations. We have not found a correlation between the mutations in the flanks and V3 region and the possible cooperation with the PKRBD to influence the response to treatment. We believe that this can be possible, but we think that a major number of sequences are needed to be able to demonstrate these facts.
In the present study, women presented less variability in the PKRBD and V3 regions. We do not have a clear explanation for this result. Nevertheless, we found a paper (39) for which it was found that females and males have different numbers of mutations in their pretreatment sequences. However, there are very few cases, and they have not been studied. At first, we thought that the variability could be due to a more aggressive immune response in men than women. Another possibility is that men generate more radical oxygen species in cells, causing more viral mutations. In any case, we believe that it is an interesting finding that needs more study.
The present study shows that PePHD was highly conserved, and thus, only 2 NR patients had one mutation, this being corroborated by other studies. There are no significant correlations between the substitutions in PePHD and the response to IFN therapy (2, 5, 13, 18, 32, 38). Nevertheless, the flanking region of PePHD (the N-terminal variable region in the C-terminal part of E2) correlates with both the response to IFN monotherapy and viral load (38). We did not find these correlations in the flanking region. Attempts have also been made to correlate substitutions in PePHD with hepatocellular carcinoma (1, 14). Further studies on the correlation of substitutions of PePHD with the clinical aspects of chronic HCV infection are needed.
In the evolutive study (Table 4), we observed for the NR patients that the PKRBD region did not present significant changes so that the dominant quasispecies of the pretreatment were persistent during treatment. We think that IFN will be an important selective agent for the present quasispecies and that these could change the composition of the viral population. Nevertheless, most of our patients from the evolutionary study (15 of 18) were NR, from which we found some viral populations that finally resisted the pressure of IFN. In the V3 and flanking regions, we found a statistically significant increase in mutations between the first month of treatment and 6 months posttreatment. It is possible that important modifications in the number of mutations or of changes in concrete positions in the V3 region could modify the structure of certain functional domains as the PKRBD and ISDR. Nevertheless, we consider that this region alone does not play an important role in the treatment outcome.
We think that specific mutations, not just the total number of mutations, can be sufficient to impede the PKRBD's interaction with PKR. Because of this, some authors (21) have proposed different amino acids (2209, 2216 and 2227) whose substitutions might be associated with a high rate of response. In our study, only one patient with an SVR presented substitutions in amino acids 2209 and 2216. Likewise, we believe that the substitution of such amino acids as proline, which breaks the alpha helix in the protein structure, means a drastic change in the structure and function of the protein. Nevertheless, in our study, we found only one change of this type (proline), and it corresponded to a patient with a response. In the same way, we think that a change in an amino acid's group (hydrophobic, polar, acidic, and basic) presupposes an alteration of function and structure. We have analyzed changes in the groups of amino acids (data not shown) which are very similar to those that we show in this paper. These results have not been given in this paper because we think that the identification of important positions and/or concrete substitutions of amino acids that alter function requires the analysis of more patients or sequences to obtain trustworthy conclusions.
In summary, our results concerning the ISDR are generally in line with those reported in European studies. It is clear that a large cohort, divided into wild-type, intermediate-type, and mutant-type ISDR groups, needs to be analyzed to draw conclusions similar to those of Enomoto et al. (8). From our study of HCV genotype 1-infected patients with a strong statistical analysis method and with a big sample of the population, we established that the presence of >4 mutations in the PKRBD region of NS5A protein from pretreatment serum samples is correlated with the virologic response to peg-IFN/RBV therapy, and therefore, PKRBD sequences might be used as a prognostic guide for treating HCV-1-infected patients.
Acknowledgments
This work was supported in part by Instituto de Salud Carlos III (Fondo de Investigaciones Sanitarias) grant PI03/1018, and Ciberehd is funded by the Instituto de Salud Carlos III.
Footnotes
Published ahead of print on 30 April 2008.
REFERENCES
- 1.Bagaglio, S., M. S. De Mitri, S. Lodrini, C. Paties, R. Cassini, G. Bianchi, M. Bernardi, A. Lazzarin, and G. Morsica. 2005. Mutations in the E2-PePHD region of hepatitis C virus type 1b in patients with hepatocellular carcinoma. J. Viral Hepat. 12243-250. [DOI] [PubMed] [Google Scholar]
- 2.Berg, T., A. Mas Marques, M. Höhne, et al. 2000. Mutations in the E2-PePHD and NS5A region of hepatitis C virus type 1 and the dynamics of hepatitis C viremia decline during interferon alfa treatment. Hepatology 321386-1395. [DOI] [PubMed] [Google Scholar]
- 3.Berg, T., C. Sarrazin, E. Herrmann, et al. 2003. Prediction of treatment outcome in patients with chronic hepatitis C: significance of baseline parameters and viral dynamics during therapy. Hepatology 37600-609. [DOI] [PubMed] [Google Scholar]
- 4.Caballero, T., A. Pérez-Milena, M. Masseroli, F. O'Valle, F. J. Salmeron, R. M. G. Del Moral, and G. Sanchez-Salgado. 2001. Liver fibrosis assessment with semiquantitative indexes and image analysis quantification in sustained-responder and non-responder interferon-treated patients with chronic hepatitis C. J. Hepatol. 34740-747. [DOI] [PubMed] [Google Scholar]
- 5.Chayama, K., F. Suzuki, A. Tsubota, M. Kobayashi, Y. Arase, S. Saitoh, Y. Suzuki, N. Murashima, K. Ikeda, N. Takahashi, M. Kinoshita, and H. Kumada. 2000. Association of amino acid sequence in the PKR-eIF2 phosphorylation homology domain and response to interferon therapy. Hepatology 321138-1144. [DOI] [PubMed] [Google Scholar]
- 6.Choo, Q.-L., K. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, A. Medina-Selby, P. J. Barr, A. Weiner, D. W. Bradley, G. Kuo, and M. Houghton. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 882451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duverlie, G., H. Khorsi, S. Castelain, O. Jaillon, J. Izopet, F. Lunel, F. Eb, F. Penin, and C. Wychowski. 1998. Sequence analysis of the NS5A protein of European hepatitis C virus 1b isolates and relation to interferon sensitivity. J. Gen. Virol. 791373-1381. [DOI] [PubMed] [Google Scholar]
- 8.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, Y. Ogura, N. Izumi, F. Marumo, and C. Sato. 1996. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N. Engl. J. Med. 33477-81. [DOI] [PubMed] [Google Scholar]
- 9.Feld, J. J., and J. H. Hoofnaglr. 2005. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436967-972. [DOI] [PubMed] [Google Scholar]
- 10.Gale, M., Jr., C. M. Blakely, B. Kwieciszewski, S. L. Tan, M. Dossett, N. M. Tang, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 185208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gale, M., Jr., B. Kwieciszewski, M. Dossett, H. Nakao, and M. G. Katze. 1999. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J. Virol. 736506-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gale, M. J., Jr., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230217-227. [DOI] [PubMed] [Google Scholar]
- 13.Gerotto, M., F. Dal Pero, P. Pontisso, et al. 2000. Two PKR inhibitor HCV proteins correlate with early but not sustained response to interferon. Gastroenterology 1191649-1655. [DOI] [PubMed] [Google Scholar]
- 14.Gimenez-Barcons, M., S. Franco, Y. Suarez, X. Forns, S. Ampurdanes, F. Puig-Basagoiti, A. Sanchez-Fueyo, J. M. Barrera, J. M. Llovet, J. Bruix, J. M. Sanchez-Tapias, J. Rodes, and J. C. Saiz. 2001. High amino acid variability within the NS5A of hepatitis C virus (HCV) is associated with hepatocellular carcinoma in patients with HCV-1b-related cirrhosis. Hepatology 34158-167. [DOI] [PubMed] [Google Scholar]
- 15.Goyal, A., W. P. Hoofmann, E. Hermann, S. Traver, S. S. Hissar, N. Arora, H. E. Blum, S. Zeuzem, C. Sarrazin, and S. K. Sarin. 2007. The hepatitis C virus NS5A protein and response to interferon alpha: mutational analyses in patients with chronic HCV genotype 3a infection from India. Med. Microbiol. Immunol. 19611-21. [DOI] [PubMed] [Google Scholar]
- 16.Gupta, R., M. Subramani, M. N. Khaja, C. Madhavi, S. Roy, C. M. Habibullah, and S. Das. 2006. Analysis of mutations within the 5′ untranslated region, interferon sensitivity region, and PePHD region as a function of response to interferon therapy in hepatitis C virus-infected patients in India. J. Clin. Microbiol. 44709-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann, W. P., S. Zeuzem, and C. Sarrazin. 2005. Hepatitis C virus-related resistance mechanisms to IFN-α based antiviral therapy. J. Clin. Virol. 3286-91. [DOI] [PubMed] [Google Scholar]
- 18.Hung, C. H., C. M. Lee, S. N. Lu, J. F. Lee, J. H. Wang, H. D. Tung, T. M. Chen, T. H. Hu, W. J. Chen, and C. S. Changchien. 2003. Mutations in the NS5A and E2-PePHD region of hepatitis C virus type 1b and correlation with the response to combination therapy with interferon and ribavirin. J. Viral Hepat. 1087-94. [DOI] [PubMed] [Google Scholar]
- 19.Kato, N., M. Hijikata, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 879524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kmieciak, D., L. Kruszyna, P. Migdalski, M. Lacinski, J. Juszczyk, and W. H. Trzeciak. 2006. Mutations within protein kinase R-binding domain of NS5A protein of hepatitis C virus (HCV) and specificity of HCV antibodies in pretreatment sera of HCV-chronically infected patients and their effect on the result of treatment. Jpn. J. Infect. Dis. 5992-99. [PubMed] [Google Scholar]
- 21.Kohashi, T., S. Maekawa, N. Sakamoto, M. Kurosaki, H. Watanabe, Y. Tanabe, C.-H. Chen, N. Kanazawa, M. Nakagawa, S. Kakinuma, T. Yamashiro, Y. Itsui, T. Koyama, N. Enomoto, and M. Watanabe. 2006. Site-specific mutation of the interferon sensitivity-determining region (ISDR) modulates hepatitis C virus replication. J. Viral Hepat. 13582-590. [DOI] [PubMed] [Google Scholar]
- 22.Layden-Almer, J. E., C. Kuiken, R. M. Ribeiro, K. J. Kunstman, A. S. Perelson, T. J. Layden, and S. M. Wolinsky. 2005. Hepatitis C virus genotype 1a NS5A pretreatment sequence variation and viral kinetics in African American and white patients. J. Infect. Dis. 1921078-1087. [DOI] [PubMed] [Google Scholar]
- 23.Lohmann, V., F. Korner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285110-113. [DOI] [PubMed] [Google Scholar]
- 24.Macquillan, G. C., X. Niu, D. Speers, S. Englihs, G. Garas, G. B. Harnett, W. D. Reed, J. E. Allan, and G. P. Jeffrey. 2004. Does sequencing the PKRBD of hepatitis C virus NS5A predict therapeutic response to combination therapy in an Australian population? J. Gastroenterol. Hepatol. 19551-557. [DOI] [PubMed] [Google Scholar]
- 25.Murayama, M., Y. Katano, I. Nakano, M. Ishigami, K. Hayashi, and T. Honda. 2007. A mutation in the interferon sensitivity-determining region is associated with responsiveness to interferon-ribavirin combination therapy in chronic hepatitis patients infected with a Japan-specific subtype of hepatitis C virus genotype 1b. J. Med. Virol. 7935-40. [DOI] [PubMed] [Google Scholar]
- 26.Murphy, M. D., H. R. Rosen, G. I. Marousek, et al. 2002. Analysis of sequence configurations of the ISDR, PKR-binding domain, and V3 region as predictors of response to induction interferon-alpha and ribavirin therapy in chronic hepatitis C infection. Dig. Dis. Sci. 471195-1205. [DOI] [PubMed] [Google Scholar]
- 27.Pascu, M., P. Martus, M. Höhne, B. Wiedenmann, U. Hopf, E. Schreier, and T. Berg. 2004. Sustained virological response in hepatitis C virus type 1b infected patients is predicted by the number of mutations within the NS5A-ISDR: a meta-analysis focused on geographical differences. Gut 531345-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paterson, M., C. D. Laxton, H. C. Thomas, A. M. Ackrill, and G. R. Foster. 1999. Hepatitis C virus NS5A protein inhibits interferon antiviral activity, but the effects do not correlate with clinical response. Gastroenterology 1171187-1197. [DOI] [PubMed] [Google Scholar]
- 29.Poynard, T., M. F. Yuen, V. Ratziu, and C. L. Lai. 2003. Viral hepatitis C. Lancet 3622095-2100. [DOI] [PubMed] [Google Scholar]
- 30.Puig-Basagoiti, F., X. Forns, I. Furcic, S. Ampurdanes, M. Gimenez-Barcons, S. Franco, et al. 2005. Dynamics of hepatitis C virus NS5A quasispecies during interferon and ribavirin therapy in responder and non-responder patients with genotype 1b chronic hepatitis C. J. Gen. Virol. 861067-1075. [DOI] [PubMed] [Google Scholar]
- 31.Saito, T., T. Ito, H. Ishiko, M. Yonaha, K. Morikawa, A. Miyokawa, and K. Mitamura. 2003. Sequence analysis of PePHD within HCV E2 region and correlation with resistance of interferon therapy in Japanese patients infected with HCV genotypes 2a and 2b. Am. J. Gastroenterol. 981377-1383. [DOI] [PubMed] [Google Scholar]
- 32.Sarrazin, C., M. Bruckner, E. Herrmann, B. Ruster, K. Bruch, W. K. Roth, and S. Zeuzem. 2001. Quasispecies heterogeneity of the carboxy-terminal part of the E2 gene including the PePHD and sensitivity of hepatitis C virus 1b isolates to antiviral therapy. Virology 289150-163. [DOI] [PubMed] [Google Scholar]
- 33.Sarrazin, C., E. Herrmann, K. Bruch, and S. Zeuzem. 2002. Hepatitis C virus nonstructural 5A protein and interferon resistance: a new model for testing the reliability of mutational analyses. J. Virol. 7611079-11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soler, M., M. Pellerin, C. E. Malnou, D. Dhumeaux, K. M. Kean, and J.-M. Pawlotsky. 2002. Quasispecies heterogeneity and constraints on the evolution of the 5′ noncoding region of hepatitis C virus (HCV): relationship with HCV resistance to interferon-alpha therapy. Virology 298160-173. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285107-110. [DOI] [PubMed] [Google Scholar]
- 36.Thelu, M.-A., E. Drouet, M.-N. Hilleret, and J.-P. Zarski. 2004. Lack of clinical significance of variability in the internal ribosome entry site of hepatitis C virus. J. Med. Virol. 72396-405. [DOI] [PubMed] [Google Scholar]
- 36a.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomson, M., M. Nascimbeni, S. Gonzales, K. K. Murthy, B. Rehermann, and T. J. Liang. 2001. Emergence of a distinct pattern of viral mutations in chimpanzees infected with a homogeneous inoculum of hepatitis C virus. Gastroenterology 1211226-1233. [DOI] [PubMed] [Google Scholar]
- 38.Ukai, K., M. Ishigami, K. Yoshioka, N. Kawabe, Y. Katano, K. Hayashi, T. Honda, M. Yano, and H. Goto. 2006. Mutations in carboxy-terminal part of E2 including PKR/eIF2alpha phosphorylation homology domain and interferon sensitivity determining region of nonstructural 5A of hepatitis C virus 1b: their correlation with response to interferon monotherapy and viral load. World J. Gastroenterol. 123722-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veillon, P., C. Payan, H. Le Guillou-Guillemette, C. Gaudy, and F. Lunel. 2007. Quasispecies evolution in NS5A region of hepatitis C virus genotype 1b during interferon or combined interferon-ribavirin therapy. World J. Gastroenterol. 131195-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe, K., K. Yoshioka, M. Yano, M. Ishigami, K. Ukai, H. Ito, F. Miyata, T. Mizutani, and H. Goto. 2005. Mutations in the nonstructural region 5B of hepatitis C virus genotype 1b: their relation to viral load, response to interferon, and the nonstructural region 5A. J. Med. Virol. 75504-512. [DOI] [PubMed] [Google Scholar]
- 41.Zeuzem, S., J. H. Lee, and W. K. Roth. 1997. Mutations in the NS5A gene of European hepatitis C virus isolates and response to IFN alfa. Hepatology 25740-744. [DOI] [PubMed] [Google Scholar]
- 42.Zeuzem, S., M. Buti, P. Ferenci, et al. 2006. Efficacy of 24 weeks treatment with peginteferon alfa-2b plus ribavirin in patients with chronic hepatitis C with genotype 1 and low pretreatment viremia. J. Hepatol. 4497-103. [DOI] [PubMed] [Google Scholar]