Abstract
Herpes simplex virus type 1 glycoprotein K (gK) and the UL20 protein (UL20p) are coordinately transported to the trans-Golgi network (TGN) and cell surfaces and are required for cytoplasmic virion envelopment at the TGN. In addition, cell surface expression of gK and UL20p is required for virus-induced cell fusion. Previously, confocal microscopy colocalization and intracellular transport experiments strongly suggested direct protein-protein interactions between gK and UL20p. Direct protein-protein interactions between gK and UL20p were demonstrated through reciprocal coimmunoprecipitation experiments, as well as with glutathione S-transferase (GST) pull-down experiments. A fusion protein consisting of the amino-terminal 66 amino acids of UL20p fused in-frame with GST was expressed in Escherichia coli and purified via glutathione column chromatography. Precipitation of GST-UL20p from mixtures of GST-UL20p fusion protein with cellular extracts containing gK specifically coprecipitated gK but not other viral glycoproteins. The purified UL20p-GST fusion protein reacted with all gK-associated protein species. It was concluded that the amino terminus of UL20p, most likely, interacted with gK domain III, which is predicted to lie intracellularly. UL20p-gK domain-specific interactions must serve important functions in the coordinate transport of UL20p and gK to the TGN, because retention of UL20p in the endoplasmic reticulum (ER) via the addition of an ER retention signal at the carboxyl terminus of UL20p forced the ER retention of gK and drastically inhibited intracellular virion envelopment and virus-induced cell fusion.
Herpes simplex viruses (HSVs) specify at least 11 virally encoded glycoproteins, as well as several nonglycosylated membrane-associated proteins, which serve important functions in virion infectivity and spread. Virus spread occurs either via direct egress of enveloped virions or via virus-induced cell fusion of adjacent cellular membranes. Mutations that cause extensive virus-induced cell-to-cell fusion have been mapped to at least four regions of the viral genome: the UL20 gene (2, 28, 31), the UL24 gene (25, 42), the UL27 gene (encoding glycoprotein B [gB]) (6, 37), and the UL53 gene (coding for gK) (4, 9, 23, 38, 39, 41). The UL20 and UL53 (gK) genes encode multipass transmembrane proteins of 222 and 338 amino acids, respectively, and are conserved in all alphaherpesviruses (9, 29, 40). Both proteins have multiple sites where posttranslational modification can occur; however, only gK is posttranslationally modified by N-linked carbohydrate addition (9, 23, 40). The specific membrane topologies of both gK and the UL20 protein (UL20p) have been predicted and experimentally confirmed via the use and detection of epitope tags within predicted intracellular and extracellular domains (12, 14, 31). Syncytial mutations in gK map predominantly in extracellular domains of gK, and particularly within the amino-terminal portion of gK (domain I) (12), while syncytial mutations of UL20 are located within the amino terminus of UL20p, which has been shown to be located intracellularly (31).
Virus-induced cell fusion is thought to occur via a concerted action of glycoproteins gD, gB, and gH/gL. Accordingly, transient coexpression of gB, gD, and gH/gL causes cell-to-cell fusion (36, 44). However, this glycoprotein-mediated cell fusion phenomenon does not accurately model virus-induced cell fusion, since it does not require gB or gK containing syncytial mutations, nor is it dependent on other viral glycoproteins known to be important for virus-induced cell fusion (3, 8, 21). Specifically, wild-type gK expression inhibited cell fusion in the transient glycoprotein coexpression assay, while expression of gK carrying a syncytial mutation did not (1). Furthermore, gK and UL20p are absolutely required for virus-induced cell fusion (14, 33), and syncytial mutations within gK (4, 9, 23, 38, 39, 41) or UL20 (2, 28, 31) promote extensive virus-induced cell fusion. Together, these observations suggest that gK and UL20p directly or indirectly interact with gB and/or other viral glycoproteins involved in virus-induced cell fusion.
According to the most prevalent model for herpesvirus intracellular morphogenesis, initially capsids assemble within the nuclei and virions acquire an initial envelope by budding into the perinuclear spaces. Subsequently, these enveloped virions lose their envelope by fusion with the outer nuclear lamellae. Within the cytoplasm, tegument proteins associate with the viral nucleocapsid and final envelopment occurs by budding of cytoplasmic capsids into specific trans-Golgi network (TGN)-associated membranes (5, 19, 34, 45). Mature virions subsequently traffic to cell surfaces, presumably following the cellular secretory pathway (22, 34, 43). In addition to their significant roles in virus-induced cell fusion, gK and UL20p are required for cytoplasmic virion envelopment. Specifically, viruses with deletions in either the gK or UL20 gene were unable to translocate from the cytoplasm to extracellular spaces and accumulate enveloped virions within TGN-like cytoplasmic vesicles (2, 9, 13, 14, 18, 23, 24, 26, 31, 40). Current evidence suggests that the functions of gK and UL20p in cytoplasmic virion envelopment and virus-induced cell fusion are carried out by different domains of UL20p, inasmuch as specific UL20 mutations within the amino and carboxyl termini of UL20p allowed cotransport of gK and UL20p to cell surfaces, virus-induced cell fusion, and TGN colocalization while effectively inhibiting cytoplasmic virion envelopment (31, 32).
A variety of experimental evidence has strongly suggested that HSV type 1 (HSV-1) gK and UL20p functionally and physically interact. Specifically, transport of gK to cell surfaces and gK-mediated cell-to-cell fusion were abolished in UL20-null virus-infected cells (14). UL20p expression was found to be necessary for intracellular transport and cell surface expression of gK in transient-expression experiments (12). Importantly, detailed confocal colocalization experiments have shown a strong interdependence of gK and UL20p for intracellular transport, cell surface expression and localization at the TGN, virus-induced cell fusion, and cytoplasmic virion envelopment, suggesting physical interactions between gK and UL20p (11, 12, 14, 18, 31). Additional evidence for the formation of heterodimeric or multimeric structures of gK with UL20p was obtained with equine herpesvirus type 1-infected cells (20).
In this paper, we demonstrate for the first time direct physical interactions between HSV-1 gK and UL20p mediated, most likely, by interactions of the amino-terminal portion of UL20p with cytoplasmic gK domain III. In addition, we show that retention of UL20p in the endoplasmic reticulum (ER) compartment leads to retention of gK in the ER of virus-infected cells, suggesting that gK and UL20p can be intracellularly transported past the ER only after forming a heterodimeric or multimeric structure. Importantly, UL20p-mediated retention of gK in the ER leads to abrogation of virus-induced cell fusion and cytoplasmic virion envelopment, underlining the high importance of UL20p and gK in these two virus life cycle steps.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero) cells were obtained from ATCC (Manassas, VA). The UL20-complementing cell line G5 was a gift of P. Desai (Johns Hopkins Medical Center) and was maintained as previously described (10, 12, 13). The parental wild-type strain used in this study, HSV-1 (KOS), was originally obtained from P. A. Schaffer (Harvard Medical School). The UL53-gK protein C epitope-tagged virus gKprotC and the V5 epitope-containing virus gKV5DI were propagated in Vero cells as described previously (12, 17). The UL20-null virus Δ20/gKD1V5 specifies a V5 epitope tag within the amino terminus of gK and was described previously (14). The UL20-null viruses Δ20/gKsyn1 and Δ20/gBsyn3, which specify the gKsyn1 and gBsyn3 syncytial mutations, respectively, as well as an enhanced green fluorescent protein (EGFP) gene cassette in place of the UL20 gene were described previously (14). All UL20-null virus stocks were prepared on UL20-null-complementing G5 cells.
Plasmids.
Plasmids for the untagged UL20 gene, the 3× FLAG-tagged UL20 gene, the 3× FLAG-tagged UL20 gene with the ER retention (pUL20KKSL) and control ER retention (pUL20KKSLAL) motifs, and the gK with a protein C epitope in domain 3 (gKprtCD3) were as described previously (12, 14, 17). The HSV-1 gB/gD-coexpressing plasmid was kindly provided by A. Minson (44). The recombination plasmid p20F used for the generation of recombinants within the UL20 gene and the rescue of the UL20 deletion with a copy of UL20 specifying an epitope-tagged UL20p were described previously (14). The 3× FLAG-tagged UL20 was PCR amplified with primers that specified the UL20 gene with the ER retention or ER retention control motifs and BamHI restriction sites for cloning into the p20F recombination vector, as we described previously for the generation of UL20 recombinant viruses (14). Plasmid pF20 contains upstream and downstream HSV genomic flanking regions to facilitate homologous recombination with viral genomes (Fig. 1).
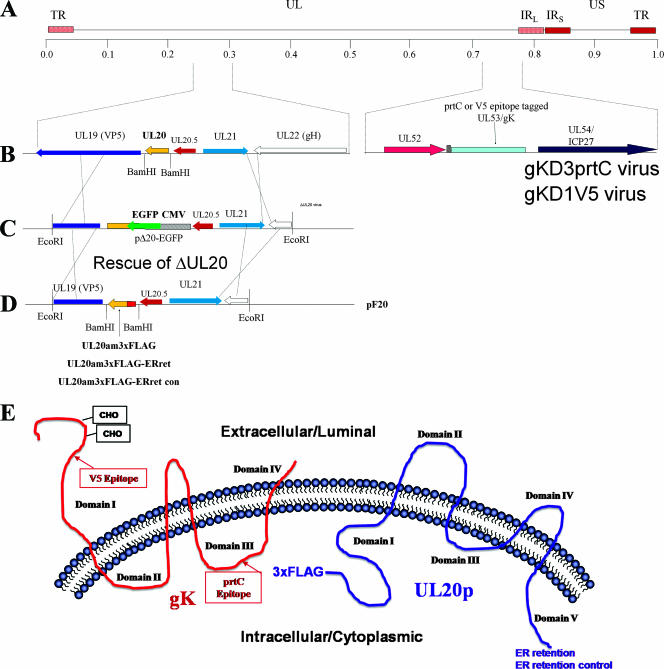
FIG. 1.
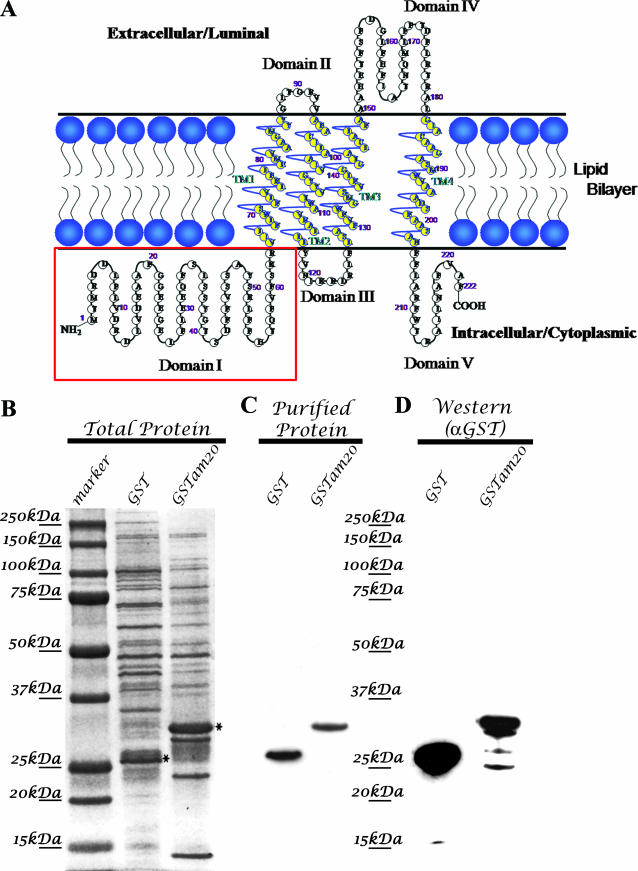
Schematic for generation of recombinant viruses that specify epitope-tagged UL20p and gK. (A) The top line represents the prototypic arrangement of the HSV-1 genome with the unique long (UL) and unique short (US) regions flanked by the terminal repeat (TR) and internal repeat (IR) regions. (B) An expanded genomic region between map units 0.25 and 0.3 containing the UL19, UL20, UL20.5, UL21, and UL22 genes (left) and the region between map units 0.7 and 0.8 containing the UL52, UL53, and UL54 open reading frames (right). (C) Diagram of the UL20-null virus, which contains an EGFP gene cassette within the UL20p open reading frame. (D) Homologous recombination plasmids that encode the indicated 3× FLAG epitope-tagged UL20 genes as well as flanking sequences for recombination were used to rescue the UL20 deletion and transfer the UL20p genes with their 3× FLAG epitope tags into the virus. (E) Schematic depicting the experimentally determined gK and UL20p membrane topologies, as well as the sites of insertion of each of the epitope tags and ER retention motifs.
Recombinant virus construction.
Individual plasmids that specified the UL20 genes with their respective specific epitope tags and ER retention motifs were transfected into Vero cells and subsequently infected with the parental UL20-null virus that specified the pertinent gK epitope tag (either prtC or V5). ER retention motif-containing viruses were subsequently isolated on the UL20-null-complementing G5 cells, whereas all other recombinant viruses were isolated on Vero cells. Viral plaque isolates were picked, plaque purified at least seven times, and tested by diagnostic PCR, DNA sequencing, and immunofluorescence using anti-FLAG for the presence of the UL203×FLAG, as we have described previously.
Plaque assays.
Plaque morphology was assessed as we have described previously (14). Briefly, Vero or G5 cells were infected at a multiplicity of infection (MOI) of 0.001 with the indicated virus and visualized by immunohistochemistry at 48 h postinfection (hpi), utilizing horseradish peroxidase-conjugated anti-HSV antibodies (DAKO) and Novared (VectorLabs) substrate development.
Transfections.
Subconfluent cells in six-well plates were transfected with the indicated plasmids utilizing the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's directions with a total of 5 μg of the indicated plasmids. Cells were processed for immunoprecipitation and Western analysis at 36 h posttransfection.
SDS-PAGE and Western immunoblots.
For visualization of virus-infected cell extracts, subconfluent Vero cell monolayers were infected with the indicated virus at an MOI of 5. At 36 hpi, cells were collected by low-speed centrifugation, washed with Tris-buffered saline (TBS), and lysed at room temperature for 15 min in mammalian protein extraction reagent supplemented with a cocktail of protease inhibitors (Invitrogen-Life Technologies, Carlsbad, CA). Insoluble cell debris was pelleted, and supernatants were suspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (0.1 M Tris, 5% SDS, 20% glycerol, 0.2% bromophenol blue, 10% β-mercaptoethanol). Samples were heated for 30 min at 50°C and electrophoretically separated in a 10 to 14.5% SDS-polyacrylamide gel. Following separation, the proteins were electrotransferred to nitrocellulose membranes, visualized with Ponceau S (0.1% Ponceau S in 3% trichloroacetic acid), and destained. Blots were blocked against nonspecific binding for 2 h using 10% skim milk in TBS supplemented with 0.135 M CaCl2 and 0.11 M MgCl2 (TBS-Ca/Mg). Blots were then incubated with the specified antibodies overnight at a 1:5,000 dilution in TBS-Ca/Mg and 0.1% Tween 20 (TBS-T), washed five times for 10 minutes each with TBS-T, incubated for 1 h with horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Pierce, Rockford, IL) at a 1:50,000 dilution in TBS-T, and washed five times for 15 min each with TBS-T. Blots were visualized by autoradiography using the Pierce SuperSignal chemiluminescent detection kit (Pierce, Rockford, IL) as per the manufacturer's instructions. All antibody dilutions and buffer washes were performed in TBS-T. For assessment of bacterially expressed glutathione S-transferase (GST)-UL20p and purified protein, aliquots of bacterial cell lysates or purified proteins were analyzed by SDS-PAGE followed by staining with Gel-Code Blue (Pierce Chemical), according to the manufacturer's directions.
Bacterial protein expression and purification of GST-UL20am.
BL21 Escherichia coli cells with either the GST vector alone or the GST-UL20p amino fusion (GST-UL20am) vector were grown in LB (Luria-Bertani) (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl, pH 7.0) medium with ampicillin (50 μg/ml) for approximately 2.5 h (30°C, 250 rpm). After the optical density at 600 nm of the culture approached 0.5, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM to induce expression, followed by 3 h of culturing at 30°C at a shaking rate of 250 rpm. After 2.5 h of expression, the bacterial cells where harvested by centrifugation (4°C, 7,700 × g, 10 min). The harvested cells were lysed by sonication in B-PER (Pierce Chemical) with 20 mg/liter lysozyme and protease inhibitor cocktail. Affinity purification, using glutathione-Sepharose 4B columns, was carried out as described by the manufacturer (Pierce Chemical). Protein concentrations were determined by the Bradford method relative to bovine serum albumin (BSA) protein standards.
Protein-protein interaction assays by coimmunoprecipitation and GST pull-down experiments.
For coimmunoprecipitation of interacting proteins within HSV-infected cells, Vero cells were infected with the dual-epitope-tagged gKD3prtC/UL20FLAG virus at an MOI of 5 and incubated at 37°C. At 24 hpi, cells were lysed for 15 min on ice in mammalian protein extraction reagent (Pierce Chemical) supplemented with complete protease inhibitor cocktail and 1% Triton X-100. After lysis, cell debris was pelleted from solution three times by high-speed centrifugation, and clarified supernatants were subsequently diluted at least threefold in TBS-Ca/Mg binding buffer (TBS-Ca/Mg supplemented with 0.25% Triton X-100 and 3% BSA). For each immunoprecipitation reaction, 1,000 μl of cellular extracts (approximately 1,000 μg of total protein) were incubated with 3 μl of the indicated primary antibody overnight at 4°C on a spinning nutating shaker. Fifty microliters of protein G-Sepharose bead slurry was added to each reaction mixture and allowed to incubate for an additional 2 h. Beads were collected by centrifugation and washed five times in TBS-Ca/Mg binding buffer and twice in TBS-Ca/Mg. Interacting proteins were eluted from the beads either by incubation in TBS without Ca and Mg (anti-FLAG and anti-prtC binding activity requires calcium) or with 0.1 M glycine HCl (pH 3.5) in TBS. For interactions in transfected cells, Vero cells in six-well plates were transiently transfected with the indicated plasmids in equimolar ratios and incubated at 37°C for 36 h prior to coimmunoprecipitation reactions as described for infected cells.
For GST pull-down experiments, cell lysates were processed and clarified as described above for immunoprecipitations, except that instead of adding antibodies, 150 μg of purified GST or the amino terminus of UL20 fused with GST was added to cell lysates. Following incubation, a glutathione-agarose slurry was added to each reaction mixture and incubated for 2 h a 4°C. The beads were collected by centrifugation at 2,500 × g in spin cups and extensively washed (at least five times for 5 min each) at 4°C with lysis buffer, followed by three washes with phosphate-buffered saline. Bound proteins were eluted by gentle rocking for 20 min at room temperature with 100 mM glutathione in phosphate-buffered saline and drop dialyzed against TBS-Ca/Mg prior to loading for SDS-PAGE.
Confocal microscopy.
Cell monolayers grown on coverslips in six-well plates were infected with the indicated virus at an MOI of 10. For cell surface biotinylation, prior to fixation cells were washed with TBS-Ca/Mg and incubated for 15 min at room temperature in EZ-Link sulfo-NHS-LC biotin cell-impermeative biotinylation reagent (Pierce Chemical), which reacts with primary amines on cell surface proteins. Cells were washed with TBS, fixed with electron microscopy grade 3% paraformaldehyde (Electron Microscopy Sciences, Fort Washington, PA) for 15 min, washed twice with TBS, and permeabilized with 1.0% Triton X-100. Monolayers were subsequently blocked for 1 h with 7% normal goat serum and 7% BSA in TBS (TBS blocking buffer) before incubation for 5 h with either anti-V5 (Invitrogen, Carlsbad, CA) for recognition of gK, anti-FLAG for recognition of UL20p, anti-gD, or anti-gB (Rumbaugh-Goodwin Institute, Plantation, FL) diluted 1:500 in TBS blocking buffer. Cells were then washed extensively and incubated for 1 h with Alexa Fluor-conjugated anti-immunoglobulin G diluted 1:500 in TBS blocking buffer. After incubation, excess antibody was removed by washing five times with TBS. For cell surface labeling, biotinylated cells were reacted with 1:1,000-diluted Alexa Fluor 647-conjugated streptavidin for 20 min. For Golgi apparatus and ER organelle labeling, cell monolayers were incubated with 1:750 dilutions of Alexa Fluor 488-conjugated lectins GSII and concanavalin A, respectively (7, 16, 27). The TGN was identified with a donkey anti-TGN46 primary antibody and an Alexa Fluor 488-conjugated sheep anti-donkey secondary antibody (30). Specific immunofluorescence was examined using a Leica TCS SP2 laser-scanning confocal microscope (Leica Microsystems, Exton, PA) fitted with a CS APO 63× Leica objective (1.4 numerical aperture). Individual optical sections in the z axis, averaged six times, were collected in series in the different channels at a 512- by 512-pixel resolution. Images were compiled and rendered in Adobe Photoshop.
Electron microscopy.
Cell monolayers were infected with the indicated virus at an MOI of 5. All cells were prepared for transmission electron microscopy examination at 16 hpi. Infected cells were fixed in a mixture of 2% paraformaldehyde and 1.5% glutaraldehyde in 0.1 M NaCaC buffer, pH 7.3. Following treatment with 1% OsO4 and dehydration in an ethanol series, the samples were embedded in Epon-Araldyte resin and polymerized at 70°C. Thin sections were made on an MTXL Ultratone (RMC Products), stained with 5% uranyl acetate and CNA lead, and observed with a Zeiss 10 transmission electron microscope as described previously (14).
Complementation assay for virus-induced cell-to-cell fusion.
Subconfluent Vero cell monolayers in six-well plates were transfected with Lipofectamine 2000 and 5 μg of either UL20p or UL20 ER retention plasmids as described by the manufacturer (Invitrogen). At 18 h posttransfection, the monolayers were infected at an MOI of 0.1 with either Δ20/gKsyn1 or Δ20/gBsyn3 virus. At 24 hpi, virus-induced cell fusion was visualized by light and fluorescence microscopy.
RESULTS
Construction and characterization of recombinant viruses carrying in-frame epitope and ER retention tags within UL20p and gK.
UL20p and gK are highly hydrophobic, multiple-membrane-spanning proteins that cannot be readily detected by immunological means due to the lack of exposed antigenic epitopes. To circumvent this problem, we have successfully utilized in-frame insertions of either V5, protC, or FLAG epitope tags inserted within different UL20p or gK domains without adversely affecting the structure and function of these proteins (12, 15, 17). To facilitate experiments examining physical interactions between gK and UL20p, a set of new recombinant viruses carrying different combinations of epitope tags within either gK or UL20p was constructed (Fig. 1). First, a new recombinant virus, Δ20/gKD3prtC, was constructed by replacing the UL20 gene of the recombinant virus gKD3prtC (17) with a gene cassette expressing the EGFP gene under control of the cytomegalovirus immediate-early promoter, as we described previously for the construction of the Δ20/gKD1V5 virus, which has a V5 epitope inserted within gK domain 1 (12) (Fig. 1B and C). The Δ20/gKD3prtC virus has the protein C epitope tag inserted within gK cytoplasmic domain III (Fig. 1E), which has been previously shown to not affect gK functions (17). The UL20 deletion of the Δ20/gKD3prtC and Δ20/gKD1V5 viruses was rescued by homologous recombination using UL20 genes expressing a 3× FLAG-epitope tagged UL20p (Fig. 1D) inserted at the very amino terminus of UL20p (Fig. 1E). Similarly, two recombinant viruses specifying the 3× FLAG epitope-tagged UL20p were constructed, but in addition they contained at their carboxyl termini either the ER retention amino acid motif KKSL or the control ER retention motif KKSLAL, which includes two additional carboxyl-terminal amino acids. The addition of these two terminal amino acids has been shown to abrogate the ability of the KKSL ER motif to retain proteins within the ER (5). As we have reported previously, the insertion of epitope tags within either gK or UL20p did not adversely affect virus replication (12, 15, 17). Similarly, the presence of epitope tags simultaneously within both gK and UL20p did not affect plaque morphology and virus spread (Fig. 2A and B) or infectious virus production (not shown).
FIG. 2.
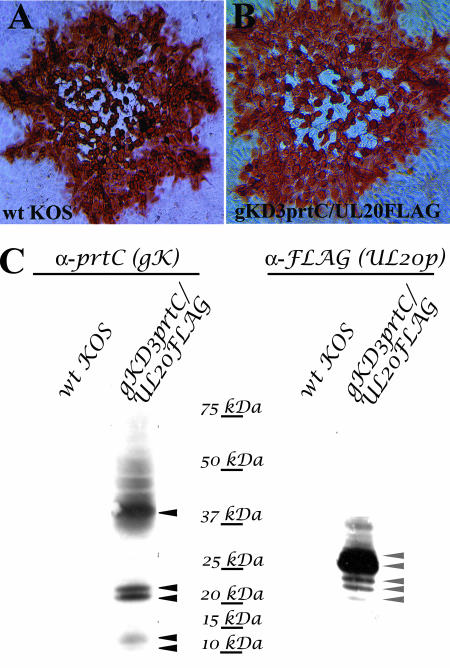
Insertion of antigenic tags in both gK and UL20p does not adversely affect virus replication, cell-to-cell spread, or plaque formation. (A and B) Confluent Vero cell monolayers were infected with either parental wild-type KOS virus (A) or the isolated dual-epitope-tagged recombinant D3gKprtC/UL20FLAG virus (B) at an MOI of 0.001, and viral plaques were visualized by immunohistochemistry at 48 hpi. (C) Detection and characterization of the protC-tagged gK or FLAG-tagged UL20p expressed in D3gKprtC/UL203×FLAG virus-infected cells. Specific detection of either gK (left) or UL20p (right) was achieved using antibodies against prtC (gK) or 3× FLAG (UL20p).
The tagged UL20p and gK were specifically detected using anti-FLAG and anti-protC antibodies, respectively, in Western immunoblots of virus-infected cell extracts (Fig. 2C). As we have reported previously (17), gK appeared as three distinct protein species: the fully glycosylated species, including proteins ranging from 36 to 50 kDa; a doublet of proteins with apparent molecular masses of 21 and 22 kDa; and a doublet of proteins with apparent molecular masses of 11 and 12 kDa appearing as faint bands in the lower portion of the Western immunoblot (Fig. 2C). Detection of the UL20p with the anti-FLAG antibody revealed two major protein species with apparent molecular masses of 27 and 28 kDa, as well as several protein species with either higher or lower apparent molecular masses (Fig. 2C). The apparent molecular mass of UL20p appears to be higher than the predicted mass of 22 kDa, suggesting potential posttranslational modifications. Overall, these Western immunoblot experiments showed that gK and UL20p could be detected with high specificity, as indicated by the absence of any nonspecific interactions with other viral or cellular proteins (Fig. 2C).
Determination of specific UL20p-gK interactions.
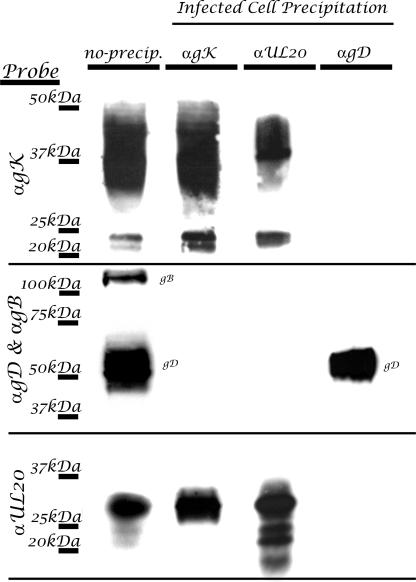
We have previously shown a strict interdependence of gK and UL20p for intracellular transport, cell surface expression, and TGN colocalization, suggesting that they physically interact (11, 12, 14, 18, 31). To demonstrate physical interactions between gK and UL20p, coimmunoprecipitation experiments were performed with cells infected with the doubly tagged recombinant virus gKD3prtC/UL20am3×FLAG. Specifically, immunoprecipitation of gK with anti-protC antibody and subsequent probing of Western immunoblots with either anti-protC (gK), or anti-FLAG (UL20p) revealed that gK and UL20p coimmunoprecipitated. Conversely, coimmunoprecipitation using anti-FLAG (UL20p) antibody and subsequent probing for either UL20p or gK revealed the presence of both proteins in the coimmunoprecipitates (Fig. 3). To ascertain the specificity of UL20 interaction with gK, the same immunoprecipitates were tested for the presence of either gD or gB, which are abundantly expressed in virus-infected cells. Neither gB or gD was detected in the coimmunoprecipitates. In addition, probing of anti-gD immunoprecipitates for the presence of gK, UL20p, or gB revealed the absence of any specific interactions with these proteins, while gD was readily detected (Fig. 3). Immunoprecipation of gK coimmunoprecipitated all UL20p-associated protein species. Similarly, immunoprecipitation of UL20p coimmunoprecipitated all major gK-associated protein species; however, it appeared to cause the enrichment of the gK derivative proteins with apparent molecular masses of 21 to 22 kDa.
FIG. 3.
HSV gK and UL20p interact in infected cells. Vero cells were infected with gKD3prtC/UL20FLAG virus, which specifies the differentially epitope-tagged gK (prtC) and UL20p (FLAG) proteins. Infected cell lysates were either directly loaded (no precipitation) or immunoprecipitated with antibodies to gK (αprtC), UL20p (αFLAG), or gD (αgD). Blots of SDS-PAGE-separated immunoprecipitates were probed with anti-prtC (αgK), anti-FLAG (αUL20), or αgB and αgD in combination in order to detect coimmunoprecipitated proteins. For control purposes, blots of SDS-PAGE-separated cellular extracts were probed for the presence of gK, UL20p, gD, and gB (no precipitation).
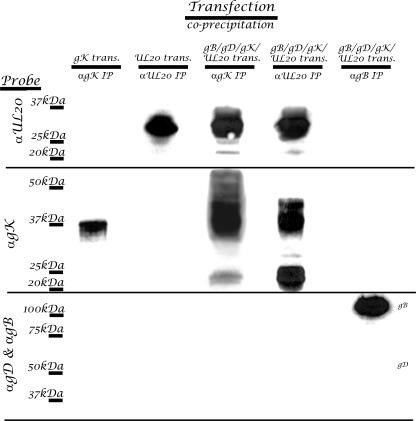
To ascertain whether the above-described UL20p interactions with gK could occur in the absence of other viral proteins or virus replication, transient-transfection experiments were performed in which a combination of plasmids expressing viral glycoproteins gB, gD, gK, and UL20p was used. gK-UL20p interactions were analyzed by immunoprecipitation followed by Western immunoblot experiments as described above. Coexpression of UL20p with gK caused the appearance of a wide range of gK protein species appearing as a smear on the Western immunoblot relative to the detection of gK alone (Fig. 4, lane 3 versus lane 1). This significant increase in the apparent molecular mass of gK, presumably caused by extensive glycosylation, confirms our previous confocal colocalization experiments (15), which showed that expression of UL20p was necessary and sufficient for intracellular transport of gK and TGN localization. Coimmunoprecipitation experiments using transfected cell extracts revealed that gK and UL20p coimmunoprecipitated with each other in a fashion similar to that seen in previous experiments with infected cell extracts (Fig. 4). Specifically, immunoprecipitation of UL20p enriched the gK-associated protein species of 21 to 22 kDa. Neither gB nor gD was detected in the gK or UL20p coimmunoprecipitates. As a negative control, immunoprecipitation of gB failed to immunoprecipitate gD, gK, or UL20p. These results showed that gK specifically interacted with UL20p in the presence or absence of virus infection.
FIG. 4.
HSV gK and UL20p interact in transfected cells. Vero cells were transfected with epitope-tagged gK (lane 1) or UL20p (lane 2) or cotransfected with gB, gD, UL20p, and gK (lanes 3 to 5). Transfected cell lysates were immunoprecipitated with anti-prtC (αgK) (lanes 1 and 3), anti-FLAG (αUL20) (lanes 2 and 4), or anti-gB (αgB) (lane 5). Blots of SDS-PAGE-separated precipitates were probed with either anti-prtC (αgK), anti-FLAG (αUL20), or αgB and αgD in combination in order to detect coimmunoprecipitated proteins.
The UL20p amino terminus directly interacts with gK.
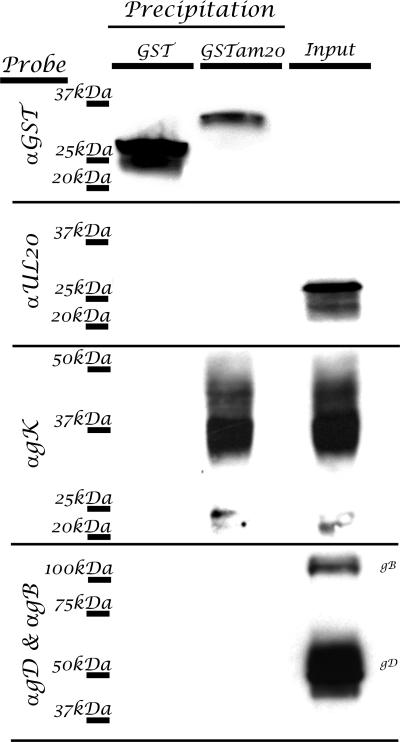
Previously, we described the topology of UL20p and mapped UL20p domains that functioned in infectious virus production, intracellular transport, and colocalization with gK in TGN compartments (31, 32). This work revealed that the amino terminus of UL20p contained domains that were necessary for gK transport to TGN compartments. To ascertain whether the amino terminus of UL20p, located in the cytoplasm, specifically interacted with gK, the amino terminus of UL20p was expressed as a GST fusion protein in E. coli (Fig. 5 ). Bacterial extracts revealed the presence of UL20 protein species as overrepresented protein species after staining of SDS-polyacrylamide gels (Fig. 5B). The GST and GST-UL20am proteins were purified on glutathione columns, and the purified proteins were detected by both SDS-PAGE and Western immunoblotting (Fig. 5C and D). To test whether the GST-UL20am purified protein physically interacted with cellular extracts obtained from virus-infected cells, purified protein was mixed with virus-infected cell extracts, and subsequently, the mixtures were precipitated using glutathione-linked agarose beads. The GST-UL20am precipitation specifically coprecipitated gK and not UL20p, gB, or gD (Fig. 6). The purified GST protein alone failed to precipitate any viral protein including gK, indicating that the amino terminus of UL20p specifically interacted with gK.
FIG. 5.
(A) Schematic of UL20p membrane topology. The intracellular amino terminus (domain I) of UL20p that was cloned in frame with GST for bacterial expression is highlighted within the boxed region. (B) SDS-PAGE and Coomassie blue staining of crude cell lysates from bacterially expressed GST and GST fused with the amino terminus of UL20p. (C) SDS-PAGE and Coomassie blue staining of glutathione column-purified bacterially expressed GST and GST fused with the amino terminus of UL20p. (D) Immunoblot specific detection of either GST or GST fused with the amino terminus of UL20p using anti-GST antibody.
FIG. 6.
The amino terminus of UL20p directly interacts with gK in transfected and infected Vero cells. Infected cell lysates were directly loaded (no precipitation) (lane 3) or incubated with GST (lane 1) or the purified UL20 amino terminus fused with GST (lane 2) and pulled down with glutathione-conjugated agarose. Blots of SDS-PAGE-separated precipitates were probed with anti-GST, anti-FLAG (αUL20), anti-prtC (αgK), or αgB and αgD in combination in order to detect GST pull-down proteins.
Retention of UL20p in ER compartments forces retention of gK in the ER and abrogates virus spread, infectious virus production, and virus-induced cell fusion.
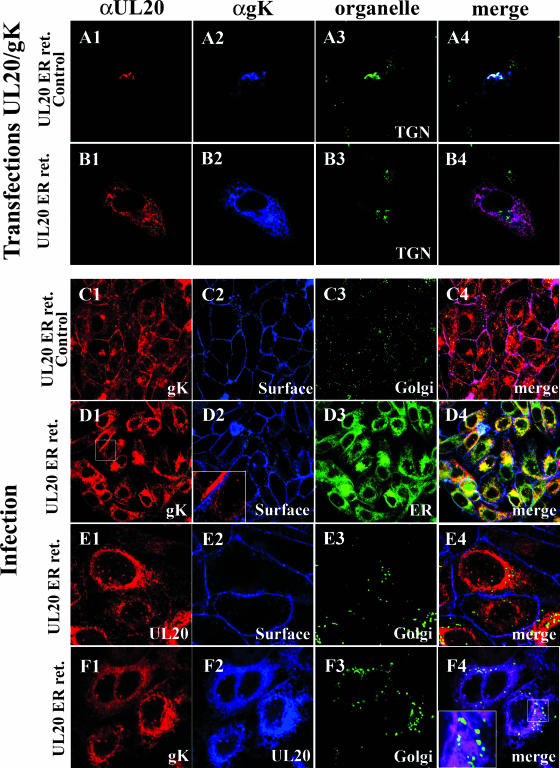
Previously, we showed that gK and UL20p were interdependent for intracellular transport, cell surface expression, and TGN localization (15). To further explore the physical association of UL20p and gK and its implications in infectious virus production and spread, recombinant viruses expressing UL20p having the ER retention peptide KKSL at its carboxyl terminus or the control peptide KKSLAL were produced. The latter construct has been shown to abrogate the ability of the ER motif to cause the retention of tagged proteins in the ER (5). Confocal microscopy was utilized to determine the fate of UL20p and gK after the addition of the ER and ER control peptides at the carboxyl terminus of UL20p. As we have shown previously, coexpression of gK and UL20p under transient-expression conditions resulted in intracellular transport and colocalization of gK and UL20p to TGN compartments (Fig. 7A1 to A4). However, coexpression of the UL20p-ER protein with gK caused retention of both gK and UL20p within ER compartments and their absence from TGN compartments (Fig. 7B1 to B4). Similar experiments were performed with virus-infected cells. Expression of UL20p tagged at its carboxyl terminus with the ER control peptide KKSLAL showed accumulation of gK on cell surfaces, as well as within Golgi compartments, as we have reported for wild-type KOS infections (15) (Fig. 7C1 to C4). A similar pattern of expression was observed for UL20p (not shown). In contrast, expression of the UL20p tagged with the ER retention motif KKSL produced an accumulation of gK within ER (Fig. 7D3 and D4), while gK was not detected on infected cell surfaces (Fig. 7D2 to D4). Similarly, UL20p was not detected on cell surfaces (Fig. 7E2 to E4) or within Golgi compartments (Fig. 7E3 and E4). UL20p and gK colocalized within ER-like compartments and were not transported to the Golgi compartments (Fig. 7F1 to F4).
FIG. 7.
UL20p transport from ER to TGN is required for the coordinate transport of gK and UL20p to post-ER cellular membranes. (A and B) Vero cells were cotransfected with plasmids expressing gK and either UL20 specifying the ER-retained UL20p(KKSL) (B) (UL20 ER ret.) or the UL20p(KKSLAL) control protein (A) (UL20 ER ret. control). (A) Coexpression of UL20 ER ret. control protein (red) and gK (blue), showing gK and UL20p cotransport to the TGN (green) membranes. (B) Coexpression of UL20 ER ret. protein (red) with gK (blue), showing lack of UL20p and gK transport to TGN (green) membranes. (C to F) Vero cells were infected with viruses that expressed either an ER-retained UL20 (D, E, and F) (UL20 ER ret.) or UL20 ER ret. control proteins (C). (C) Infection with the virus specifying the wild-type like UL20 ER ret. control protein, showing accumulation of gK (red) on cell surfaces (blue) and Golgi compartments (green). (D) Infection with the virus expressing UL20 ER ret. protein exhibited retention of gK (red) in the ER (green) and not on cell surfaces (blue). (E) Infection with the virus specifying UL20 ER ret. control protein, showing that UL20 (red) was transported to neither cell surfaces (blue) nor Golgi compartments (green). (F) Infection with a virus specifying UL20 ER ret. protein (blue), showing retention of both the UL20 ER ret. protein (blue) and gK (red) in the ER and lack of UL20 and gK transport to the Golgi compartments (green). Magnification, ×63 (zoom, ×4).
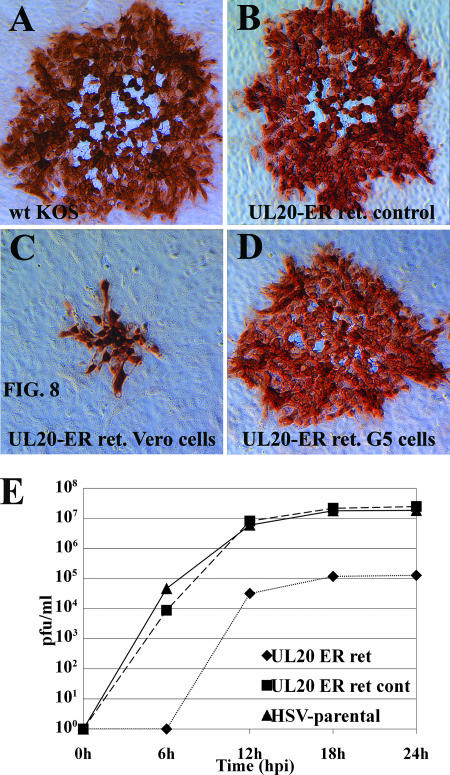
To investigate the significance of UL20p ER retention on infectious virus production, spread, and egress, the plaque morphologies of the above-mentioned recombinant viruses were examined. Infection of Vero cells with the recombinant virus carrying the UL20p ER motif KKSL caused the appearance of a UL20-null or gK-null plaque phenotype (Fig. 8C), while infection of Vero-G5 cells, which complement UL20-null viruses, produced wild-type-like viral plaques (Fig. 8D). In contrast, Vero cells infected with the recombinant virus expressing the ER control peptide KKSLAL produced viral plaques that appeared indistinguishable from those produced by the wild-type KOS virus (Fig. 8B and A, respectively). These results indicate that UL20p transport past the ER is absolutely required for infectious virus production and spread.
FIG. 8.
Retention of UL20p within the ER blocks efficient plaque formation and virus cell-to-cell spread. Confluent Vero (A to C) or UL20-complementing G5 (D) cell monolayers were infected with either parental wild-type KOS virus (A), UL20p ER retention control virus (B), or UL20p ER retention virus (C and D) at an MOI of 0.001, and viral plaques were visualized by immunohistochemistry at 48 hpi. (E) Comparison of replication kinetics of parental wild-type, UL20 ER retention control, and UL20 ER retention viruses.
The effect of UL20p retention in the ER on virus replication was further explored by obtaining one-step replication kinetics for the recombinant viruses expressing either the UL20p ER motif KKSL or the UL20p ER control peptide KKSLAL. These results showed that the virus expressing UL20p(KKSL) replicated approximately 2 log units less efficiently than either the UL20p(KKSLAL) or the parental HSV-1 virus. The parental HSV-1 and the UL20p(KKSLAL) viruses replicated with similar replication kinetics, revealing that the addition of the terminal two amino acids AL to the KKSL ER retention motif completely abrogated the negative effect of UL20p retention to the ER on viral replication (Fig. 8E).
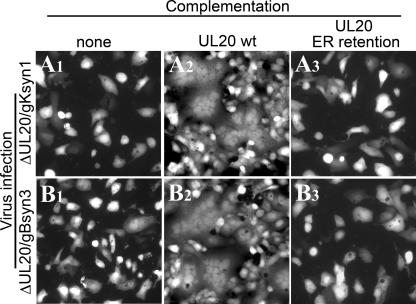
Complementation experiments were performed to assess the effect of UL20p retention in the ER on virus-induced cell fusion caused by syncytial mutations in either gB or gK. In these experiments, cells transfected with either the UL20p or UL20p expressing the ER retention motif KKSL were infected with either the ΔUL20/gBsyn3 or ΔUL20/gKsyn1 virus. In the absence of UL20p, neither gBsyn3 nor gKsyn1 was able to cause virus-induced cell fusion (Fig. 9A1 and B1), while wild-type UL20p efficiently complemented for virus-induced cell fusion (Fig. 9A2 and B2). The UL20p tagged with the ER retention motif KKSL failed to complement for virus-induced cell fusion (Fig. 9A3 and B3), indicating that transport of UL20p and/or gK to cell surfaces was necessary for both gBsyn3 and gKsyn1 virus-induced cell fusion.
FIG. 9.
UL20 ER retention abrogates virus-induced cell-to-cell fusion. Vero cells were transfected with negative control plasmids (A1 and B1) or plasmids encoding wild-type (A2 and B2) or UL20p ER retention (A3 and B3) genes and then infected with either the Δ20gKsyn1 or the Δ20gBsyn3 virus. At 24 hpi, the ability of these plasmids to complement virus-induced cell fusion was determined by visualization of syncytia formation by fluorescence microscopy.
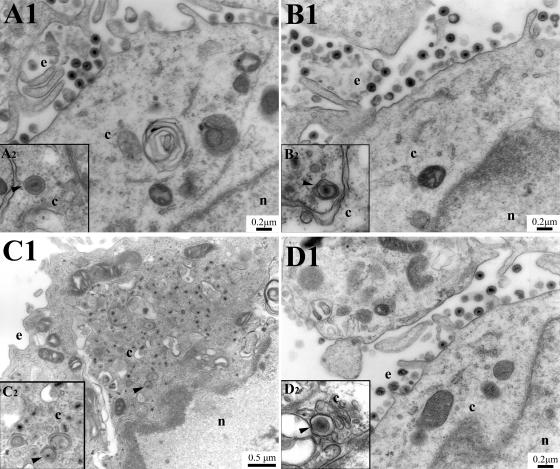
As expected from the plaque morphology, electron micrographs of Vero cells infected with the recombinant viruses expressing the UL20p tagged with the ER retention motif KKSL revealed the absence of enveloped virions in extracellular spaces and an accumulation of capsids in the cytoplasm (Fig. 10C1) with some capsids juxtaposed near curvilinear membranes (Fig. 10C2), as we have reported previously for the UL20-null and gK-null viruses (14, 26). This defect was fully reversed in the UL20-complementing cell line G5 (Fig. 10D1 and D2). For comparison purposes, the recombinant virus expressing UL20p tagged with the control peptide KKSLAL produced an ultrastructural phenotype indistinguishable from that of the wild-type KOS virus.
FIG. 10.
Retention of UL20p within ER membranes causes a UL20-null defect in cytoplasmic virion envelopment. Vero (A, B, and C) or UL20-null complementing G5 (D) cells were infected with either parental wild-type KOS (A), control UL20p ER retention motif (B), or UL20p ER retention (C and D) virus. Cell monolayers were infected at an MOI of 5, incubated at 37°C for 20 h, and prepared for transmission electron microscopy. Nuclear (n), cytoplasmic (c), and extracellular (e) spaces are marked. Insets illustrate the inability of UL20p ER retention cytoplasmic capsids to attain an envelope and a transport vesicle.
DISCUSSION
We have shown previously that HSV-1 gK and UL20p are interdependent for intracellular transport, cell surface expression, and TGN localization in virus-infected cells, as well as in transient-coexpression experiments where gK was coexpressed with UL20p (15). These results strongly suggested that physical interactions between gK and UL20p in the rough ER (RER) were necessary for their intracellular trafficking. In this paper, we show for the first time that UL20p directly interacts with gK. This physical interaction is mediated by the amino terminus of UL20p interacting, most likely, with the carboxyl-terminal half of gK.
In previous publications, we reported the membrane topology of gK and UL20p, primarily through the use of epitope tags inserted in frame within each gK and UL20p domain (12, 14, 31). The membrane topologies of gK and UL20p are mirror images of each other, with gK having both amino- and carboxyl-terminal domains located extracellularly while UL20p has both amino- and carboxyl-terminal domains located intracellularly. Interestingly, both the gK and UL20p amino-terminal domains are quite large, composed of 134 and 63 amino acids, respectively. To facilitate protein-protein interaction experiments, we isolated and utilized the double-tagged virus gKD3prtC/UL20am3×FLAG containing the protC epitope tag within gK domain III and the 3× FLAG epitope within UL20 domain I. The presence of both of these epitope tags did not adversely affect infectious virus production and virus spread (see Results). Both gK and the UL20p were detected as multiple protein species, suggesting potential posttranslational processing and/or proteolytic fragmentation. In addition, the gK gene contains several internal ATGs, which have been hypothesized to code for truncated gK protein species through usage of internal codon initiations (35). The five internal, in-frame ATGs are located at amino acids 55, 57, 101, 106, and 145. Internal initiation from these ATGs is predicted to code for gK protein species of 283, 281, 237, 232, and 193 amino acids with apparent molecular masses of 28.3, 28.1, 23.7, 23.2, and 19.3 kDa. N-linked glycosylation is predicted to occur at amino acid 58, which could increase the apparent molecular mass of the gK derivative starting with amino acid 55. Also, other types of posttranslational modification may influence the apparent molecular mass of gK-derived peptides produced through usage of internal initiation codons. Alternatively, the experimentally observed 21-, 22-, and 11- to 12-kDa gK species could be produced by specific proteolytic cleavage of gK. Regardless of the mechanism by which these gK species have been produced, they all contain the protC epitope tag inserted in frame within gK domain III, indicating that they contain the carboxyl-terminal half of the gK molecule. The apparent molecular mass of UL20p is approximately 26 kDa, which is larger than the predicted molecular mass of 22 kDa based on the UL20p amino acid sequence. UL20p contains internal consensus motifs for phosphorylation, which may be responsible for the observed “ladder” pattern of UL20p-associated protein species. All UL20p species contained the amino terminus of UL20p, because they reacted with the anti-FLAG antibody inserted at the amino terminus of UL20p.
Immunoprecipitation experiments with virus-infected cell extracts revealed that UL20p and gK specifically coimmunoprecipitated. Immunoprecipitation with the anti-gK antibody precipitated UL20p enriched for the UL20p-associated protein species with the highest apparent molecular mass. In contrast, immunoprecipitation with the anti-UL20p antibody produced all UL20p-associated protein species and all gK-derived protein species. However, the smaller gK-associated proteins appeared to be overrepresented in comparison to immunoprecipitations with the anti-gK antibody alone. Neither gB nor gD was coimmunoprecipitated in these experiments, indicating that gK interacted specifically with UL20p. These experiments suggested that the largest UL20p species interacted with the gK carboxyl terminus. Similar results were obtained with transient coexpression of gK and UL20p, indicating that interactions of gK and UL20p occurred in the absence of viral replication and expression of viral proteins. Based on the membrane topology of UL20p and gK, it can be suggested that the amino terminus of UL20p specifically interacts with gK domain III, since this domain is the only gK domain which is located intracellularly.
The amino-terminal domain of UL20p was suspected as a potential interacting domain with gK due to its relatively large size, as well as the presence of the putative UL20p phosphorylation sites. The UL20p amino-terminal domain specifically interacted with gK, since it enabled coimmunoprecipitation of gK but not gB or gD. The GST-UL20am fusion protein did not contain the 3× FLAG epitope; therefore, any potential interaction of the 3× FLAG epitope with the ProtC epitope could not be responsible for the observed physical interaction between the epitope-tagged UL20p and gK. The cytoplasmic domain of UL20p is predicted to be composed of 63 amino acids. The purified GST-UL20am protein included 66 amino acids of the UL20p amino terminus. The inclusion of three more amino acids predicted to be part of the first transmembrane sequence of UL20p did not appear to influence the specificity of the UL20am interactions with gK, since no other viral glycoproteins were coimmunoprecipitated with the purified GST-UL20am protein. However, it cannot be ruled out that the additional three amino acids facilitated interactions with membrane-bound gK in these experiments. Previously, we showed that gK and UL20p were interdependent for intracellular transport (15). Apparently, UL20p and gK interactions occur within the RER, presumably forming as both proteins are translated. This conclusion is also supported by new evidence provided here that retention of either UL20p or gK in the RER by insertion of an ER retention signal at the carboxyl terminus of either protein forced the retention of both proteins in the ER, abrogating cytoplasmic virion envelopment and infectious virus production. We have shown previously that specific UL20p mutations in the amino terminus of UL20p including elimination of both putative phosphorylation sites located in the amino terminus of UL20p abrogated cytoplasmic virion envelopment, while they allowed UL20p and gK intracellular transport, cell surface expression, and TGN localization (31, 32). Therefore, potential phosphorylation of the amino terminus of UL20p is not required for UL20p-gK physical interactions.
We show here for the first time that forced retention of UL20p in the ER drastically inhibits cytoplasmic virion envelopment, producing an ultrastructural phenotype which is identical to that of the UL20-null or gK-null viruses. This evidence provides further support for a stochiometric and functional relationship between UL20p and gK revealed previously by quantitative confocal microscopy (15). Furthermore, these results support our previous observations that neither gK nor UL20p functions in capsid egress from the nucleus (33), since capsids readily accumulated in the cytoplasm when UL20p was forced to be retained in the ER. We reported previously that overexpression of gK in gK-transformed cells forced accumulation of gK in perinuclear spaces and inhibited viral glycoprotein transport. These phenomena were attributed to gK-induced collapse of the Golgi apparatus (16). Apparently, UL20p retention in the ER forced retention of gK without causing similar phenomena observed in gK-transformed cells. It is likely that the presence of sufficient amounts of UL20p interacting with gK prevented any gK-mediated deleterious effects, presumably produced by excessive amounts of misfolded gK within ER membranes. The importance of the observed UL20p interactions with gK in cytoplasmic virion envelopment and virus-induced cell fusion is under investigation.
Acknowledgments
We thank Olga Borkhsenious for her expert technical assistance with electron microscopy. We acknowledge the technical assistance of Ramesh Subramanian of Biommed.
This work was supported by grant AI43000 from the National Institute of Allergy and Infectious Diseases to K.G.K. We acknowledge financial support by the LSU School of Veterinary Medicine to Biommed.
Footnotes
Published ahead of print on 23 April 2008.
REFERENCES
- 1.Avitabile, E., G. Lombardi, and G. Campadelli-Fiume. 2003. Herpes simplex virus glycoprotein K, but not its syncytial allele, inhibits cell-cell fusion mediated by the four fusogenic glycoproteins, gD, gB, gH, and gL. J. Virol. 776836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., P. L. Ward, G. Campadelli-Fiume, and B. Roizman. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 656414-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balan, P., N. Davis-Poynter, S. Bell, H. Atkinson, H. Browne, and T. Minson. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J. Gen. Virol. 751245-1258. [DOI] [PubMed] [Google Scholar]
- 4.Bond, V. C., and S. Person. 1984. Fine structure physical map locations of alterations that affect cell fusion in herpes simplex virus type 1. Virology 132368-376. [DOI] [PubMed] [Google Scholar]
- 5.Browne, H., S. Bell, T. Minson, and D. W. Wilson. 1996. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J. Virol. 704311-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bzik, D. J., B. A. Fox, N. A. DeLuca, and S. Person. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137185-190. [DOI] [PubMed] [Google Scholar]
- 7.Cottin, V., A. A. Van Linden, and D. W. Riches. 2001. Phosphorylation of the tumor necrosis factor receptor CD120a (p55) recruits Bcl-2 and protects against apoptosis. J. Biol. Chem. 27617252-17260. [DOI] [PubMed] [Google Scholar]
- 8.Davis-Poynter, N., S. Bell, T. Minson, and H. Browne. 1994. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J. Virol. 687586-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debroy, C., N. Pederson, and S. Person. 1985. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology 14536-48. [DOI] [PubMed] [Google Scholar]
- 10.Desai, P., N. A. DeLuca, J. C. Glorioso, and S. Person. 1993. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J. Virol. 671357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietz, P., B. G. Klupp, W. Fuchs, B. Kollner, E. Weiland, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 745083-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster, T. P., X. Alvarez, and K. G. Kousoulas. 2003. Plasma membrane topology of syncytial domains of herpes simplex virus type 1 glycoprotein K (gK): the UL20 protein enables cell surface localization of gK but not gK-mediated cell-to-cell fusion. J. Virol. 77499-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster, T. P., and K. G. Kousoulas. 1999. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J. Virol. 738457-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster, T. P., J. M. Melancon, J. D. Baines, and K. G. Kousoulas. 2004. The herpes simplex virus type 1 UL20 protein modulates membrane fusion events during cytoplasmic virion morphogenesis and virus-induced cell fusion. J. Virol. 785347-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster, T. P., J. M. Melancon, T. L. Olivier, and K. G. Kousoulas. 2004. Herpes simplex virus type 1 glycoprotein K and the UL20 protein are interdependent for intracellular trafficking and trans-Golgi network localization. J. Virol. 7813262-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster, T. P., G. V. Rybachuk, X. Alvarez, O. Borkhsenious, and K. G. Kousoulas. 2003. Overexpression of gK in gK-transformed cells collapses the Golgi apparatus into the endoplasmic reticulum inhibiting virion egress, glycoprotein transport, and virus-induced cell fusion. Virology 317237-252. [DOI] [PubMed] [Google Scholar]
- 17.Foster, T. P., G. V. Rybachuk, and K. G. Kousoulas. 2001. Glycoprotein K specified by herpes simplex virus type 1 is expressed on virions as a Golgi complex-dependent glycosylated species and functions in virion entry. J. Virol. 7512431-12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 1997. The UL20 gene product of pseudorabies virus functions in virus egress. J. Virol. 715639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 753675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guggemoos, S., F. T. Just, and A. Neubauer. 2006. The equine herpesvirus 1 UL20 product interacts with glycoprotein K and promotes egress of mature particles. J. Virol. 8095-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haanes, E. J., C. M. Nelson, C. L. Soule, and J. L. Goodman. 1994. The UL45 gene product is required for herpes simplex virus type 1 glycoprotein B-induced fusion. J. Virol. 685825-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harley, C. A., A. Dasgupta, and D. W. Wilson. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 751236-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutchinson, L., K. Goldsmith, D. Snoddy, H. Ghosh, F. L. Graham, and D. C. Johnson. 1992. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J. Virol. 665603-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutchinson, L., and D. C. Johnson. 1995. Herpes simplex virus glycoprotein K promotes egress of virus particles. J. Virol. 695401-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson, J. G., S. H. Chen, W. J. Cook, M. F. Kramer, and D. M. Coen. 1998. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242161-169. [DOI] [PubMed] [Google Scholar]
- 26.Jayachandra, S., A. Baghian, and K. G. Kousoulas. 1997. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J. Virol. 715012-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, B. S., X. Alvarez, S. Ishido, A. A. Lackner, and J. U. Jung. 2000. Inhibition of intracellular transport of B cell antigen receptor complexes by Kaposi's sarcoma-associated herpesvirus K1. J. Exp. Med. 19211-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacLean, C. A., S. Efstathiou, M. L. Elliott, F. E. Jamieson, and D. J. McGeoch. 1991. Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J. Gen. Virol. 72897-906. [DOI] [PubMed] [Google Scholar]
- 29.McLean, G., F. Rixon, N. Langeland, L. Haarr, and H. Marsden. 1990. Identification and characterization of the virion protein products of herpes simplex virus type 1 gene UL47. J. Gen. Virol. 712953-2960. [DOI] [PubMed] [Google Scholar]
- 30.McMillan, T. N., and D. C. Johnson. 2001. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J. Virol. 751928-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melancon, J. M., T. P. Foster, and K. G. Kousoulas. 2004. Genetic analysis of the herpes simplex virus type 1 UL20 protein domains involved in cytoplasmic virion envelopment and virus-induced cell fusion. J. Virol. 787329-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melancon, J. M., P. A. Fulmer, and K. G. Kousoulas. 2007. The herpes simplex UL20 protein functions in glycoprotein K (gK) intracellular transport and virus-induced cell fusion are independent of UL20 functions in cytoplasmic virion envelopment. Virol. J. 4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melancon, J. M., R. E. Luna, T. P. Foster, and K. G. Kousoulas. 2005. Herpes simplex virus type 1 gK is required for gB-mediated virus-induced cell fusion, while neither gB and gK nor gB and UL20p function redundantly in virion de-envelopment. J. Virol. 79299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 761537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moyal, M., C. Berkowitz, A. Rosen-Wolff, G. Darai, and Y. Becker. 1992. Mutations in the UL53 gene of HSV-1 abolish virus neurovirulence to mice by the intracerebral route of infection. Virus Res. 2699-112. [DOI] [PubMed] [Google Scholar]
- 36.Muggeridge, M. I. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. 812017-2027. [DOI] [PubMed] [Google Scholar]
- 37.Pellett, P. E., K. G. Kousoulas, L. Pereira, and B. Roizman. 1985. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J. Virol. 53243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pogue-Geile, K. L., G. T. Lee, S. K. Shapira, and P. G. Spear. 1984. Fine mapping of mutations in the fusion-inducing MP strain of herpes simplex virus type 1. Virology 136100-109. [DOI] [PubMed] [Google Scholar]
- 39.Pogue-Geile, K. L., and P. G. Spear. 1987. The single base pair substitution responsible for the syn phenotype of herpes simplex virus type 1, strain MP. Virology 15767-74. [DOI] [PubMed] [Google Scholar]
- 40.Ramaswamy, R., and T. C. Holland. 1992. In vitro characterization of the HSV-1 UL53 gene product. Virology 186579-587. [DOI] [PubMed] [Google Scholar]
- 41.Ruyechan, W. T., L. S. Morse, D. M. Knipe, and B. Roizman. 1979. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J. Virol. 29677-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders, P. G., N. M. Wilkie, and A. J. Davison. 1982. Thymidine kinase deletion mutants of herpes simplex virus type 1. J. Gen. Virol. 63277-295. [DOI] [PubMed] [Google Scholar]
- 43.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment→deenvelopment→reenvelopment pathway. J. Virol. 755697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, Z., M. D. Gershon, Y. Hao, R. T. Ambron, C. A. Gabel, and A. A. Gershon. 1995. Envelopment of varicella-zoster virus: targeting of viral glycoproteins to the trans-Golgi network. J. Virol. 697951-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]